Abstract
Motility is required for Vibrio fischeri cells to interact with and specifically colonize the light-emitting organ of their host, the squid Euprymna scolopes. To investigate the influence of motility on the expression of the symbiotic phenotype, we isolated mutants of the squid symbiont V. fischeri ES114 that had altered migration abilities. Spontaneous hyperswimmer (HS) mutants, which migrated more rapidly in soft agar and were hyperflagellated relative to the wild type, were isolated and grouped into three phenotypic classes. All of the HS strains tested, regardless of class, were delayed in symbiosis initiation. This result suggested that the hypermotile phenotype alone contributes to an inability to colonize squid normally. Class III HS strains showed the greatest colonization defect: they colonized squid to a level that was only 0.1 to 10% that achieved by ES114. In addition, class III strains were defective in two capabilities, hemagglutination and luminescence, that have been previously described as colonization factors in V. fischeri. Class II and III mutants also share a mucoid colony morphology; however, class II mutants can colonize E. scolopes to a level that was 40% of that achieved by ES114. Thus, the mucoid phenotype alone does not contribute to the greater defect exhibited by class III strains. When squid were exposed to ES114 and any one of the HS mutant strains as a coinoculation, the parent strain dominated the resulting symbiotic light-organ population. To further investigate the colonization defects of the HS strains, we used confocal laser-scanning microscopy to visualize V. fischeri cells in their initial interaction with E. scolopes tissue. Compared to ES114, HS strains from all three classes were delayed in two behaviors involved in colonization: (i) aggregation on host-derived mucus structures and (ii) migration to the crypts. These results suggest that, while motility is required to initiate colonization, the presence of multiple flagella may actually interfere with normal aggregation and attachment behavior. Furthermore, the pleiotropic nature of class III HS strains provides evidence that motility is coregulated with other symbiotic determinants in V. fischeri.
The association of the luminous bacterium Vibrio fischeri and the sepiolid squid Euprymna scolopes is an established and easily manipulated model system for the study of bacterium-host interactions (reviewed in references 33 and 41). This benign association develops as a result of changes in gene expression in both partners. Juvenile squid hatch free of symbionts and must acquire cells of V. fischeri from a planktonic pool of bacteria in the environment. The colonization process is initiated when motile V. fischeri cells aggregate on host-derived, extracellular mucus-like structures that are induced by the presence of gram-negative bacteria in the ambient environment (30). An aggregate typically comprises tens to hundreds of V. fischeri cells that will migrate to the pores of the nascent light organ within several hours of aggregate formation. V. fischeri cells must then traverse ciliated ducts to enter the epithelium-lined crypt spaces where they can rapidly grow and proliferate. As cells of V. fischeri grow and reach a critical cell density within the crypts, their bioluminescence is induced. Concomitant with symbiont proliferation, newly hatched juvenile E. scolopes undergo a developmental program that is induced by the presence of V. fischeri cells and that results in the formation of the mature light-emitting organ (28).
Because transposon-induced motility mutants aggregate but do not migrate, it has been suggested that flagellum-mediated movement is at least one mechanism by which V. fischeri cells enter the light organ (30). However, while motile cells of other Vibrio species appear to be able to aggregate and migrate to the pores, only V. fischeri (and its closest relative, Vibrio logei) can traverse the ducts and produce a symbiotic infection (12). The list of V. fischeri genes whose expression is known to be required for normal symbiotic competence is growing and includes those encoding secreted proteins (1, 7, 18; B. Feliciano and E. G. Ruby, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. 99, p. 462, 1999), luminescence (40), siderophore production (19), catalase activity (42), and a sensor kinase (43). Thus, this association is not only highly specific but apparently involves host and bacterial signaling, as well as coordinated gene expression and development.
A possible mechanism by which V. fischeri cells recognize and respond to their symbiotic environment could be similar to those that coordinate the regulation of motility and colonization genes in some pathogenic associations (2). Because motility is both energetically expensive and often not required in a persistent, stable association, a number of bacterial pathogens and symbionts coordinately regulate the expression of motility-related and non-motility-related colonization genes in response to their environment. In gram-negative bacteria, such mechanisms typically involve two-component regulatory systems or flagellar master regulators, either of which can reciprocally control the expression of virulence genes and motility (31).
V. fischeri uses flagellum-mediated motility to initiate colonization of E. scolopes. Initial descriptions of the symbiosis proposed that motile cells enter the light organ, become nonmotile for the duration of the symbiosis, and re-elaborate flagella only after they have exited the organ as a result of the daily host-controlled expulsion event (34). However, recent analyses with green fluorescent protein (GFP)-labeled bacteria and imaging by confocal laser-scanning microscopy suggest a more involved scenario: different locations within the crypts house at least two distinct populations of V. fischeri cells that can be distinguished by their size and motility behavior (41). Thus, the role of motility in symbiosis is more complex than previously believed and encourages a better understanding of the mechanisms by which V. fischeri regulates the expression of motility genes within different regions of the crypt. Moreover, this observation suggests that a set of coordinately regulated genes may exist in V. fischeri that encodes not only motility but also other colonization factors.
To explore such a relationship, we isolated spontaneous mutants of V. fischeri that have alterations in their in vitro motility phenotype. We investigated the ability of hypermotile mutants to initiate colonization of juvenile E. scolopes. Furthermore, we found a class of these strains that are pleiotropic mutants, suggesting a link between the expression of normal motility behavior and symbiotic competence.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Wild-type V. fischeri ES114, isolated from an adult specimen of E. scolopes (6), was used as the parent strain for the isolation of hyperswimmer (HS) mutants. Mobilizable plasmid pKV111 carrying the gene for a red-shifted GFP derivative (30, 39) was used to label bacteria for confocal laser-scanning microscopy studies. V. fischeri cells were grown at 28°C in either seawater-tryptone (SWT) medium (6), which contains 0.5% tryptone, 0.3% yeast extract, and 0.3% glycerol in 70% natural seawater (SW), or LBS medium, which contains 1% tryptone, 0.5% yeast extract, and 2% NaCl in a 20 mM Tris-HCl buffer (pH 7.4). Chemotaxis assays were performed on plates made with tryptone medium, which contains 1% tryptone and 2% NaCl in a 20 mM Tris-HCl buffer (pH 7.4), while motility agar plates were prepared with SWT medium. Bacto-Agar (Difco, Detroit, Mich.) was added to 1.5% (wt/vol) for routine solid media or to between 0.3 and 0.7% for motility media. Chloramphenicol was added to growing cultures of V. fischeri cells in broth (5 μg ml−1) and to SW with colonized squid (1 μg ml−1) to maintain GFP-expressing plasmids. Where appropriate, supplements were added to media to the following final concentrations: V. fischeri acyl-homoserine lactone autoinducer (3-oxo-C6-HSL; Sigma, St. Louis, Mo.) at 250 ng ml−1 (in ethyl acetate) and decanal at 0.0025%. Ethyl acetate was evaporated from autoinducer solutions prior to addition of media and bacterial cultures. Decanal was diluted in filter-sterilized SW to a concentration of 0.05% immediately prior to use.
Selection and assay of HS mutants.
HS mutants were obtained by stabbing a single colony of strain ES114 on a pointed round toothpick into tryptone medium containing 0.7% agar. The plates were incubated at 22°C for between 5 and 6 days. Cells were then removed from protrusions that occasionally were found to extend from the center of the inoculum and purified to single colonies by streaking on SWT agar. Single isolates were then screened for hypermotile activity on motility agar plates after incubation at temperatures between 22 and 24°C for 8 to 24 h. Relative motility rates were estimated by comparing the diameter of each swarm with the diameter ES114 produced on the same plate. Stable HS mutants consistently migrated at rates at least 1.3 times those of ES114 in 0.7% agar and often migrated faster than ES114 in 0.4% agar as well.
Hemagglutination assays.
Whole guinea pig red blood cells (Colorado Serum Company, Denver, Colo.) were washed by centrifugation and resuspended in phosphate-buffered saline containing 0.8% NaCl, 0.02% KCl, 0.12% Na2HPO4, and 0.024% KH2PO4 (pH 7.4). A 100-μl volume of 1% (vol/vol) packed blood cells in phosphate-buffered saline was mixed with 100 μl of a bacterial suspension containing ca. 108 cells in round-bottom wells of a 96-sample microtiter dish. Assays were performed with cells from either exponential or stationary-phase cultures in SWT medium. A mutant strain of Escherichia coli, ORN115, which constitutively expresses type I mannose-sensitive pili, was used as a positive control, and E. coli ORN103, a nonpiliated mutant, was used as a negative control (26). After 1 h of incubation at 25°C the plates were examined for evidence of an agglutination reaction.
Analysis of free-swimming patterns.
Free-swimming patterns of V. fischeri cells were analyzed by light microscopy. V. fischeri strains were grown in SWT at 28°C until cultures reached an optical density at 600 nm (OD600) of 0.5. Cells were viewed by using a light microscope with a ×40 objective and were recorded at 60 Hz on a model 6720 time-lapse recorder (Panasonic, Secaucus, N.J.). After this recording, the individual cells were tracked and their swimming speeds were estimated by measuring the distance of a single run per number of frames. A micrometer was used to calibrate measured distances with actual length. Swimming speeds were determined for at least 40 individual cells of each strain from at least two separate recordings and then calculated with the standard errors.
Electron microscopy.
V. fischeri cells were prepared for examination by transmission electron microscopy (TEM) as follows. Formvar-coated copper grids (Ted Pella Co., Tustin, Calif.) were floated on suspensions of cells grown to mid-exponential phase (OD600 = 0.4) in SWT medium and then transferred to a drop of fixative solution (2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 M sodium cacodylate buffer [pH 7.4]) for 10 min. The grids were washed twice with Nanopure (Millipore Corp., New Bedford, Mass.) water for 30 s and negatively stained for 1 min with freshly prepared and filtered 1% uranyl acetate. To examine cells grown on SWT agar, grids were gently placed directly onto an overnight colony and left for 10 min prior to washing, fixing, and staining as described above. All of the sample grids were examined by using a LEO 912 EF electron microscope at 100 kV of accelerating voltage. The number of flagella was determined from between 50 and 100 cells of each test strain grown either in liquid media or on the surface of an agar plate. Statistical analysis was performed with a two-sample Student's t test by using MINITAB software (MINITAB, Inc., State College, Pa.).
Preparation of flagellin proteins.
Flagella were isolated from cells grown to mid-exponential phase as described previously, with some modifications (27). Briefly, cells were pelleted by centrifugation at 8,000 × g for 10 min at 4°C, resuspended in 1 ml of chilled, 70% artificial seawater (ASW) (32), and transferred to 25-ml polycarbonate tubes containing 10 ml of chilled, 70% ASW. Flagella were sheared from cells by vortexing for 2 min. The resulting cell suspension was examined by phase-contrast microscopy to ensure that cells were neither motile nor lysed after this treatment. The cells were removed by two rounds of centrifugation at 6,000 × g for 10 min each at 4°C. Flagella were pelleted from the cell-free supernatant by centrifugation at 38,000 × g for 40 min at 4°C. The resulting pellet was resuspended in 80 μl of a standard loading buffer before the protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% polyacrylamide gel (35). Separated proteins were detected by Coomassie brilliant blue staining.
Measurement of luminescence.
V. fischeri strains were grown in SWT at 28°C either with or without the addition of 3-oxo-C6-HSL. At ca. 25-min intervals, 1-ml aliquots were removed from the culture flask and used to measure both luminescence and culture OD600. To determine the effects of added aldehyde substrate on the level of bioluminescence, 5 μl of a 0.05% suspension of decanal was added to 100 μl of culture immediately before assay. Luminescence levels were measured with a Turner 20/20 Luminometer (Turner Designs, Sunnyvale, Calif.) calibrated with a light standard.
Squid colonization.
The ability of different V. fischeri strains to colonize juvenile E. scolopes squid was determined as previously described (34) with several modifications. Specifically, each strain to be tested was added to 4 ml of SW at a final concentration of between 200 and 10,000 CFU ml−1. Animals were exposed to inoculating bacteria in SW for 12 h, at which time the animals were placed in V. fischeri-free SW for the remainder of the experiment. Bacterial bioluminescence in squid was measured either continuously, by using an automated, modified scintillation counter, or by determining the luminescence periodically over the course of the experiments with a luminometer. Initiation of the symbiosis was examined by incubating squid in the presence of an inoculum of V. fischeri cells and then continuously monitoring colonization for up to 48 h postinoculation. To determine colonization levels at specific times, individual animals were rinsed with filter-sterilized SW and homogenized. Dilutions of the homogenate were spread on SWT agar medium to determine the number of CFU per light organ. The ability of mutant strains to compete with ES114 for squid colonization was examined by combining equal cell numbers of mutant and wild-type strains in SW and by using this suspension as the inoculum for animal experiments. The actual ratio of the wild type and mutant was determined by plating a dilution of the inoculum and testing individual colonies (at least 100 for each animal) for their ability to migrate through soft agar. Individual colonies were stabbed into a 0.7% soft agar motility plate and scored at 18 h. HS strains were easily identified by their ability to migrate through 0.7% agar within 18 h at a substantially greater rate than the wild-type strain. The behavior of both HS and wild-type cells in 0.7% agar was consistent and appeared stable; reversion of the mutant phenotype was never observed. Only inocula in which the ratio of mutant to wild type was between 1.1 and 1.3 were used. At 15, 24, or 48 h after inoculation, animals were sacrificed and the number of colonizing bacteria was determined. To estimate the numbers of mutant and wild-type cells in competition experiments, individual colonies were assayed for motility as described above.
Confocal laser-scanning microscopy.
Newly hatched squid were exposed to between 1 × 105 and 5 × 105 cells ml−1 of either wild-type or HS strains of V. fischeri carrying the GFP-encoding plasmid, pKV111 (30, 39). Before examination, some infected animals were stained for 30 min in SW containing 0.005% CellTracker Orange (Molecular Probes, Eugene, Oreg.). At times ranging from 3 to 24 h postinoculation, animals were anesthetized in 2% ethanol in filter-sterilized SW and dissected prior to viewing by either differential interference contrast imaging for unstained animals or, for stained animals, by fluorescence on an LSM510 laser-scanning confocal fluorescence microscope (Zeiss, Jena, Germany). Bacterial GFP fluorescence was detected by using a 488-nm excitation wavelength, and the CellTracker Orange stain was detected by using a 543-nm excitation wavelength. Digital images were processed by using Zeiss LSM510 software.
RESULTS
Isolation and classification of V. fischeri mutants with altered motility phenotypes.
We began our investigation of the role of motility, and the potential for its coregulation with other symbiotic factors, by isolating mutants of V. fischeri ES114 with altered swimming abilities. Hypermotile (or HS) mutants arose spontaneously and were identified by a greater rate of penetration of soft motility agar than the wild-type strain. The independently derived mutants were grouped based on several phenotypes, including their rate of movement in both 0.7 and 0.4% soft agar, their ability to hemagglutinate red blood cells, and the production of a colony morphology that was both mucoid in appearance and more translucent relative to wild type. Classification based on these phenotypes resulted in three classes of HS strains, of which class I was the most common type isolated (Table 1). Three of the 125 HS strains belonged to class II, which was defined by a mucoid colony morphology and a diminished growth rate (data not shown). The class III HS strains were distinguishable from the other two classes because, while they produced the mucoid colony morphology of class II strains, they were also unable to hemagglutinate red blood cells. All of the HS strains isolated were stable and did not revert to wild type. Two representative strains of each class were chosen for further study (Table 1).
TABLE 1.
HS mutant classes and their phenotypes
| Strain or class | % Motility ina:
|
Colony morphology | Doubling timeb (%) | Hemagglutination | No. of HS mutants isolated (frequency of occurrence [%]) | Representative strains | ||
|---|---|---|---|---|---|---|---|---|
| 0.4% agar | 0.7% agar | Liquid | ||||||
| ES114 (parent) | [100] | [100] | [100] | Smooth | [100] | + | NAc | NA |
| Class I | 117 | 177 | 144 | Smooth | 100 | + | 119 (95.2) | DM66, DM69 |
| Class II | 115 | 140 | 149 | Mucoid | 80 | + | 3 (2.4) | DM73, DM74 |
| Class III | 110 | 164 | 151 | Mucoid | 100 | − | 3 (2.4) | DM61, DM70 |
The “100%” value (in brackets) is equivalent to 4.5 mm h−1 (in 0.4% agar) and 0.18 mm h−1 (in 0.7% agar).
The “100%” value (in brackets) is equivalent to 42 min.
NA, not applicable.
HS strains are hypermotile but do not appear to have additional swimming pattern defects.
Both wild-type and HS strains were found to produce the typical three rings of growth on chemotaxis agar (data not shown), including the one resulting from serine chemotaxis (A. J. Wolfe, E. J. Simel, and K. L. Visick, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. 99, p. 388, 1999). Free-swimming patterns were viewed microscopically and were analyzed in two ways: (i) qualitatively, by observing the length of runs and frequency of reversals of individual cells, and (ii) quantitatively, by measuring swimming speeds of individual cells. Swimming patterns were similar among the wild type and two representative strains of each of the three HS classes. However, when the average swimming speeds of individual cells were measured, the HS strains were capable of significantly greater swimming speeds. The average swimming speed of wild-type cells was 67 ± 3 μm s−1, while those of HS mutant strains representing classes I, II, and III showed greater rates of 98 ± 4, 100 ± 3, and 101 ± 4 μm s−1, respectively. These increased swimming speeds correspond to between 144 and 151% of the rate of wild-type cells (Table 1).
HS strains are hyperflagellated.
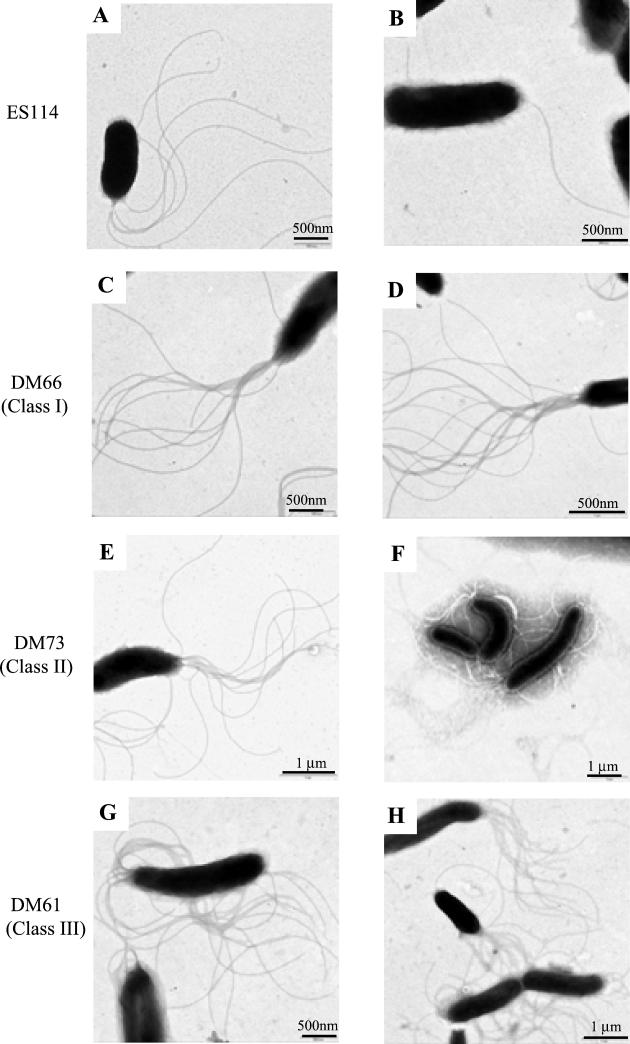
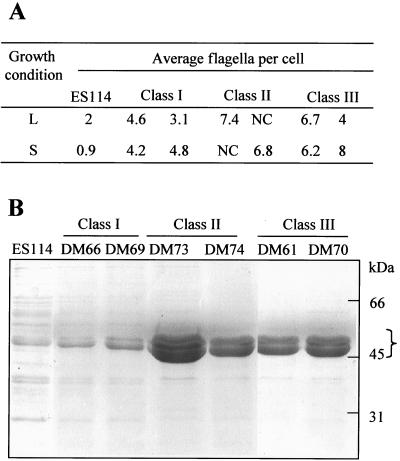
To investigate the physical basis for the hypermotility behavior, we compared the extent of flagellation of wild-type and mutant cells by TEM visualization and by comparing their relative levels of flagellin protein production. V. fischeri wild-type cells typically contain a single tuft of between one and three polar, sheathed flagella, but as many as six flagella have been observed. When grown in liquid medium, all three classes of HS strains showed a dramatic increase in the average number of flagella present (Fig. 1), bearing polar tufts of up to 16 flagella per cell. In contrast to the wild-type strain, which apparently downregulates flagellin expression when grown on a surface (Fig. 1B), HS strains produced either the same number of flagella, or more flagella, per cell when grown on a surface (Fig. 1D, F, and H) than when grown in liquid (Fig. 1C, E, and G). The average number of flagella per cell on at least 50 cells of each strain was determined (Fig. 2A), and the results were subjected to a statistical assessment by using a two-sample Student's t test. The null hypothesis, that the flagellar densities of the wild-type strain ES114 and HS cells are equivalent, was rejected in all comparisons at a high level of confidence (P < 0.005%). These results indicate that HS strains produce a significantly higher level of flagellation than the wild-type strain.
FIG. 1.
Electron micrographs of V. fischeri wild-type strain ES114 and a representative strain from each of the three HS mutant classes: DM66 (class I), DM73 (class II) and DM61 (class III). Cells were grown either in liquid media (A, C, E, and G) or on agar surfaces (B, D, F, and H).
FIG. 2.
Quantitative analysis of flagellin production by V. fischeri wild-type strain ES114 and HS mutant strains. (A) Average number of flagella per cell determined from cultures of ES114 and two representative strains from each of the three classes of HS mutants grown either in liquid media (L) or on a surface (S). At least 50 cells for each strain and condition were assayed by using TEM. Cells of class II strains displayed a flagellar density that often could not be counted but was >10 flagella per cell. (B) SDS-PAGE analysis of flagellin proteins produced by V. fischeri strain ES114 and HS strains. Purified flagellar preparations from 4 × 108 cells were denatured and subjected to PAGE, followed by Coomassie brilliant blue staining. The bracket indicates the location of flagellin subunits.
Similar results were obtained by estimating the relative abundance of flagellin proteins isolated from flagella preparations of HS and wild-type strains. When grown in SWT liquid broth all of the HS strains produced more flagellin proteins than the wild-type strain (Fig. 2B). In SDS-PAGE analyses the dominant protein bands present at ca. 45 kDa appear to be multiple flagellin subunits based on analysis of the predicted protein sequence of the V. fischeri flagellin-encoding genes and on the absence of these bands in flagellum synthesis mutant strains (unpublished data). Each class of HS mutants appears to differ in the relative amount of flagellin proteins produced (Fig. 2B), and this relationship (class II > class III > class I > wild type) correlates with the number of flagella per cell observed in each of the three classes (Fig. 2A).
Class III HS strains are defective in luminescence.
In comparing levels of luminescence emission per cell by the class III HS strain DM61 in culture to levels emitted by strain ES114, we found that the mutant could also be distinguished from other mutant classes by its inability to produce light. The addition of exogenous autoinducer and/or aldehyde, both limited in V. fischeri ES114 cultures, induced luminescence of DM61, although not to wild-type levels (Table 2), suggesting that such additions alone are not sufficient to restore normal levels of luminescence to this strain. Similar results were obtained with DM70, another class III strain (data not shown). We were also unable to detect V. fischeri autoinducer, 3-oxo-C6-HSL, from culture supernatants of class III HS strains (data not shown). Under the same assay conditions, wild-type culture supernatants contained at least four times more 3-oxo-C6-HSL.
TABLE 2.
Effects of autoinducer and aldehyde additions on the bioluminescence of V. fischeri wild-type strain ES114 and the class III HS strain DM61
| Strain | Specific luminescence (LU/OD)a | Fold increase of luminescence by the addition of:
|
||
|---|---|---|---|---|
| AIb | Aldc | AI + Ald | ||
| ES114 | 16 | 10,700 | 430 | 10,700 |
| DM61d | <0.01 | 210 | 130 | 1,500 |
One light unit (LU) = 1.3 × 104 quanta s−1.
Cells were grown with shaking in the presence of AI (3-oxo-C6-HSL).
Decyl aldehyde (Ald) was added to an aliquot of culture prior to reading the luminescence.
Similar results were obtained for DM70 (another class III strain).
HS strains are defective in normal colonization.
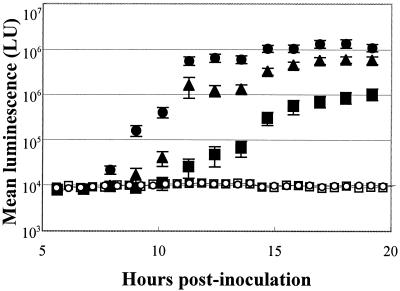
Because motility is required for V. fischeri cells to colonize the juvenile squid light organ (18), we hypothesized that the ability to swim faster might provide a competitive advantage, at least during the initial stages of colonization. To test this, we first examined how well animals became colonized when exposed to individual HS strains. Juvenile squid were placed in SW containing cells of either the wild-type strain ES114 or one of the HS mutant strains and then incubated for 12 h prior to transferring the squid to V. fischeri-free SW. Upon entering the light organ of newly hatched squid, V. fischeri cells grow rapidly, reaching a high cell density that induces light emission and allows the extent of colonization to be estimated by measuring the luminescence of the whole animal. Animals inoculated with either strain DM66 (class I) or DM73 (class II) were delayed in luminescence induction, and reduced in their level of light emission relative to ES114-infected animals (Fig. 3). While the differences in delay and final levels were small for animals colonized by the class I strain DM66, the appearance of light emission of squid inoculated with the class II strain DM73 was delayed 3 to 5 h, and by 19 h postinoculation had reached a level that was only 10% that of animals colonized by ES114. Because the luminescence capability of class III strains was severely limited in culture (Table 2), we predicted that the strains would also produce less light per cell than ES114 in the light organ. Juvenile squids colonized by the class III strain DM61 produced less than 0.1% of the light of animals infected by ES114 (Fig. 3).
FIG. 3.
Luminescence of squids infected with either V. fischeri wild-type strain ES114 or HS mutant strains. Luminescence of individual animals was determined approximately every hour over the first 20 h postinoculation. Shown are the mean values for groups of 20 animals for each strain and the respective standard errors. Animals were infected with wild-type strain ES114 (•) or one representative strain from each of the three HS classes, DM66 (class I; ▴), DM73 (class II; ▪), or DM61 (class III; ○). Control animals (□) were maintained in V. fischeri-free SW. Similar results were obtained in three separate experiments. One light unit (LU) = 11 quanta s−1.
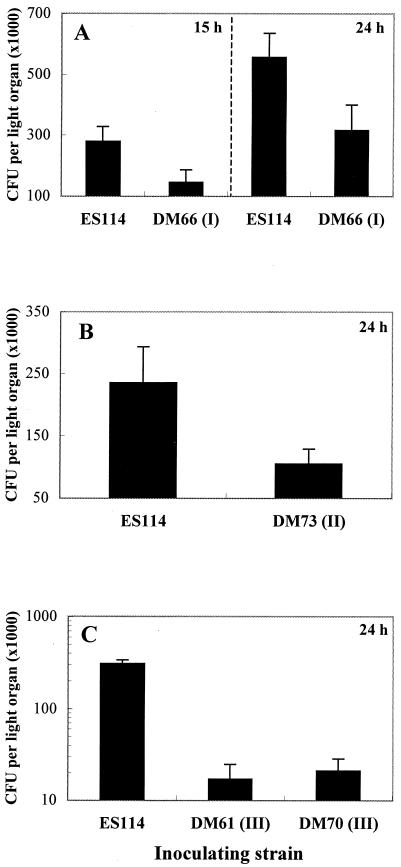
Because animal luminescence is only an indirect measure of the level of bacterial colonization (and provided little information in the case of class III strains), we determined the number of CFU present in light organs at different times postinoculation by spreading light-organ homogenates on SWT. All of the HS strains tested colonized to an extent that was significantly below wild-type levels (Fig. 4). The class I mutants, DM66 and DM69, colonized at a level between 50 and 60% that of the parent strain at both 15 and 24 h postinoculation. However, both luminescence and colonization levels of these HS strains were indistinguishable from wild type at 48 h (data not shown), suggesting that members of this class have a defect in initiating the infection, but one from which they eventually recover. Colonization levels achieved by class III HS strains were more variable relative to wild type and were between 0.1 and 10% that reached by ES114 at both 24 (Fig. 4C) and 48 h (data not shown). Class III mutants continued to show this dramatic defect in colonization beyond 48 h postinoculation (data not shown).
FIG. 4.
Symbiotic colonization levels of V. fischeri wild-type strain ES114 and HS mutant strains. (A) Colonization levels achieved by HS class I mutant strain DM66 at 15 and 24 h postinoculation. Similar results were obtained with a second class I mutant, DM69 (data not shown). (B) Colonization levels achieved by HS class II mutant strain DM73 at 24 h postinoculation. (C) Colonization levels achieved by HS class III mutant strains DM61 and DM70 at 24 h (similar results were obtained at 48 h). Each bar represents the mean values obtained with groups of at least 15 animals, and the error bars indicate the standard error of the mean. Similar results were obtained in three separate experiments.
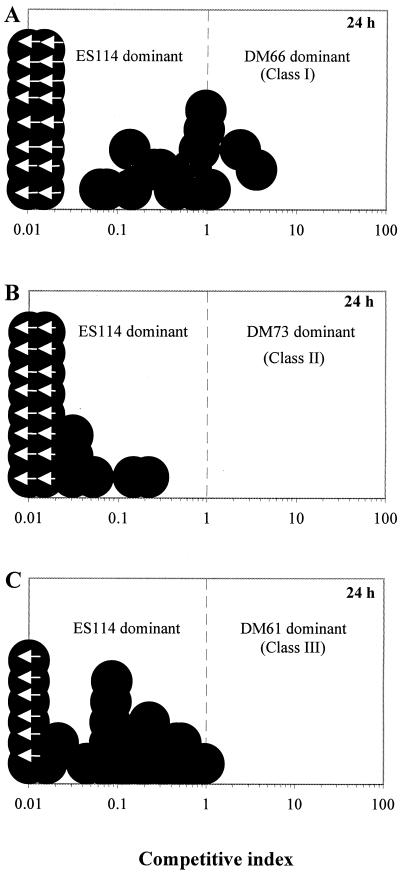
To determine the competitiveness of HS strains in the colonization of squid, animals were exposed to cell numbers of both the mutant and wild type at a ratio between 1.1 and 1.3, and at either 24 or 48 h, homogenates of their light organs were plated on SWT agar. The data are reported as a competitive index, which is the ratio of mutant to wild-type cells present within the light organ of each individual animal after exposure to an inoculum containing mutant and wild-type cells at a ratio of 1.1 to 1.3. Interestingly, we found that in almost all cases in which animals were coinoculated with an HS strain and the wild type, the predominant symbiont was the wild-type strain (Fig. 5). In fact, while the wild-type strain was always present in infected squid, in many of the animals (between 24 and 73%, depending on the class) the HS strain was undetectable. In the case of DM73, a class II strain that grows at only 80% the rate of wild type in culture (data not shown), it is possible that its competitive defect is due, at least in part, to a relatively slower growth rate in the light organ as well. Interestingly, the class III strains (e.g., DM61) express a much greater colonization defect than class I strains when they are colonizing animals by themselves (Fig. 4A and C); however, they appear not to be significantly less competitive than the class I strains (Fig. 5A and C).
FIG. 5.
Colonization of E. scolopes light organs by mixed inocula of V. fischeri wild-type strain ES114 and an HS mutant strain. Individual juvenile squid were coinoculated with a mixture (the ratio of mutant to wild-type cells was between 1.1 and 1.3) of either DM66 (A), DM73 (B), or DM61 (C) strains and their parent strain, ES114. At 24 h postinoculation, the animals were sacrificed and the numbers of the two strains present in the light organs of 33 (A), 24 (B), or 24 (C) animals were determined. The extent of dominance of one strain over the other was termed the competitive index (CI), which was calculated by dividing the number of mutant cells by the number of ES114 cells present in each animal. Each circle represents the CI of an individual animal after exposure to an inoculum containing mutant and wild-type cells at a ratio of 1.1 to 1.3. Animals with a CI of <1.0 are dominated by the wild-type strain ES114, and those with a CI of >1.0 are dominated by the mutant strain. Circles with arrows are values below the limit of detection (CI < 0.01). Results for each combination of strains are representative of three separate experiments.
HS strains are delayed in aggregate formation.
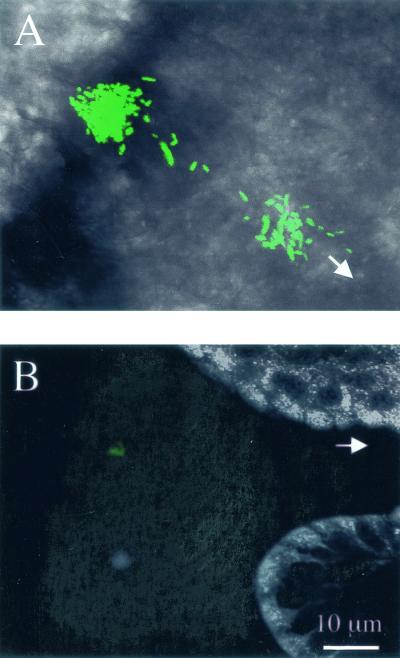
To further investigate the basis of the colonization defects observed in HS strains, we used confocal laser-scanning microscopy to visualize the behavior of GFP-expressing bacterial cells during their colonization of specific host tissues. Previous studies (30) have shown that V. fischeri cells aggregate outside of the pores of the nascent light organ on host-derived mucous structures and possibly respond to some as-yet-unknown cue to migrate into the light-organ crypts. Furthermore, these studies found that nonmotile and nonflagellated V. fischeri cells will form a typical aggregate outside of the nascent light organ but are unable to migrate to the light-organ pores and into the ducts and crypts. These observations suggest that flagellum-mediated motility is used by V. fischeri cells to migrate from the external environment into the light organ.
We investigated why hypermotile V. fischeri cells are delayed in their colonization of juvenile squid by asking three questions. (i) Do HS strains form normal aggregates? (ii) Are their timing of aggregate formation and pattern of migration normal? (iii) Do HS and wild-type strains behave similarly during colonization of the light organ? At between 4 and 6 h postinoculation, ES114 cells aggregated on the mucous strands and migrated along them toward the light-organ pores (Fig. 6A). During the same time period, HS strains did not accumulate on the strands but, instead, were detected on the epithelial surface of the adjacent light-organ tissue. These bacterial cells were highly motile and were spread in patches over the light-organ surface. This behavior is distinctly different from that of the ES114 cells within a mucous aggregate, which have not been observed to be motile during the early stages of aggregation.
FIG. 6.
Aggregation behavior of V. fischeri during colonization of squid. Confocal laser-scanning microscopy was used to compare the behavior of V. fischeri wild-type strain ES114 to HS mutant strains during the initial hours of symbiotic interaction. (A) At between 4 and 6 h postinoculation, animals exposed to wild-type cells were found to contain external aggregates of bacteria. Shown is a typical aggregate comprised of hundreds of GFP-labeled bacterial cells. (B) Animals exposed to the same concentration of HS strains contained smaller bacterial aggregates (tens of cells) that formed at a later time (between 8 and 10 h postinoculation). Shown is a typical aggregate from an animal exposed to GFP-labeled cells of the class III HS strain DM61; however, similar results were obtained for animals exposed to strains of any of the three HS mutant classes. White arrows indicate the location of the light-organ pores. Bacteria were visualized by the fluorescence of GFP (shown as green), and animal tissue (shown in grayscale) was visualized either by differential interference contrast imaging (A) or by fluorescence of CellTracker orange (B). Images are at the same magnification.
At between 8 and 12 h postinoculation, a period during which ES114 cells have colonized the light-organ crypts and luminescence has been induced (Fig. 3), HS strains formed small aggregates on the mucous strands and were beginning to colonize the crypt spaces (Fig. 6B). Animals exposed to an inoculum of 5 × 105 cells ml−1 of ES114 typically form aggregates containing hundreds of cells. However, inoculation with a similar number of HS cells produced significantly smaller aggregates containing only 10 to 20 cells (Fig. 6B) that were more similar to aggregates formed with inoculations of 104 ES114 cells. The observed delay of initiation events, as well as evidence that smaller aggregates are formed with HS strains, could explain the observed lower numbers of CFU within the light organ at the early time points of 15 and 24 h (Fig. 4). Furthermore, the length of delay correlates with the 2- to 5-h delay in the onset of luminescence observed in animals infected with luminous HS strains (Fig. 3). Similar results were obtained with all HS strains tested, regardless of their class.
Possible explanations for both the observed delay in initiation and the presence of fewer aggregated cells could be (i) that mutant cells are less viable in SW than wild type or (ii) that mutant cells stick to the side of the incubation vials. Both of these possibilities would effectively reduce the concentration of free bacteria in the incubation chamber. However, cells of classes I and III showed a viability similar to that of the wild type over the incubation period used in this study and formed biofilms on glass coverslips that appeared indistinguishable from wild type (data not shown). Class II cells, on the other hand, were present after the incubation period at a level that was only 56% that of wild type cells in SW. This result suggested that either the reduced growth rate of class II strains or a decrease in their viability could contribute to lower inoculation levels.
DISCUSSION
We have identified three classes of spontaneous hypermotile V. fischeri mutants based on several associated phenotypes. All of the HS strains tested were distinguished from the parent strain both by an increase in the level of flagellation and by their diminished ability to colonize the light organ; i.e., (i) they were delayed in colonization; (ii) they attained lower cell numbers in the light organ; and (iii) they were outcompeted by the wild-type strain in mixed infections. While abnormal motility behavior has an effect on the initiation of symbiotic infection (18), in a wild-type infection most of the bacteria stop elaborating flagella within 12 h after entering the light organ (34); thus, it was unexpected that an altered motility defect would result in the inability of symbionts to proliferate normally beyond 12 h postinfection (Fig. 4). In addition to the defects of class I strains, class III mutants were further distinguished by several phenotypes, including luminescence, hemagglutination, and colony morphology. The pleiotropic nature of the class III mutants and the greater extent of their colonization defect suggest that nonmotility-related genes, whose expression may sometimes be linked with motility, are implicated in the proper expression and development of the colonization phenotype in V. fischeri.
In the V. fischeri-squid light-organ association, flagellum-mediated motility is required to initiate the colonization of host tissue (18). While a role for motility beyond this initial stage has not previously been investigated, it is likely that motility is downregulated within the light-organ environment, a location where motility apparently is not needed (34). In this study, we found that all HS strains tested displayed a delay in colonization that could reflect their slower aggregation and subsequent migration to the internal crypt spaces. One possibility is that these cells fail to downregulate flagellin production either on the external mucus strands or within the viscous environment of the light-organ crypts (20). In either case, such strains would continue to express the hyperflagellated phenotype, resulting in an inappropriate expression of motility that could prevent the cells from proper attachment or signaling to their host. While regulation of flagellar number in V. fischeri has not been investigated, inappropriate expression of motility in bacterial cells is known to affect virulence and colonization in pathogenic organisms such as Salmonella enterica serovar Typhimurium, Bordetella species, and V. cholerae (8, 9, 37). Another possible explanation for the delay in colonization exhibited by the HS strains (Fig. 3 and 6) could be a defect in their chemotactic ability. However, both wild-type and the HS mutant strains were found to produce the typical three rings of growth on chemotaxis agar (data not shown) and display swimming patterns similar to that of the wild type when viewed microscopically, suggesting that the HS strains do not have a general chemotactic defect.
The pleiotropic nature of class II and III HS strains isolated in this study suggests that they carry a genetic variation that results in a differential expression of multiple genes. While the role of phase variation in the ability of V. fischeri strains to colonize squid has not been described, a previous study reported the isolation of a more brightly luminous variant of the symbiotic strain ES114 in culture (11). Interestingly, this spontaneous variant was characterized by both an increase in luminescence in culture and a decrease in motility and flagellation compared to ES114; these traits are the inverse of the nonluminous, hyperflagellated HS mutant class found in the present study. The previous report, however, did not investigate the colonization ability of the variant. Similar to the class III strains isolated in this report, pleiotropic variation in the luminous bacterium V. harveyi can arise spontaneously, resulting in dark mutants that carry defects in other characteristics such as flagellation and colony morphology (10, 22). Recently, the luxO gene in V. harveyi was shown to coordinate expression of luminescence, siderophore production, and colony morphology (24), suggesting that a similar mechanism could control the conversion from the wild-type to the HS phenotype. V. fischeri LitR, a homolog of another V. harveyi transcriptional regulator, LuxR, has been identified recently and found to control luminescence, possibly by enhancing transcription of lux genes (P. Fidopiastis and E. G. Ruby, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. 101, p. 503, 2001). Because LitR represents another level of luminescence control in V. fischeri, we wondered whether LitR could complement the luminescence defect of the class III HS strains isolated in this study. However, carriage of a plasmid expressing litR did not alter the luminescence of strains DM61 and DM70 (unpublished data), suggesting that LitR alone is not sufficient to complement the genetic defect of these strains.
Interestingly, it is significant that the class III HS strains are defective in several traits that have been previously described as symbiotic determinants, including luminescence (40) and hemagglutination (Feliciano and Ruby, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999). However, class III HS strains show a greater colonization defect than has been reported for strains carrying either of the single mutations: both the luminescence and hemagglutination mutants colonize to a level that is 25% that achieved by wild type (40; Feliciano and Ruby, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999), while class III strains colonize to only 0.1 to 10% of wild-type levels (Fig. 4). These observations suggest that multiple mutations can result in an additive effect on colonization or that the class III strains carry additional unrecognized mutations that contribute to their inability to colonize squid to normal levels. In this regard, little is known about whether the mucoid colony morphology of these strains can be attributed to the overproduction of extracellular polysaccharide (data not shown); while the production of extracellular polysaccharide is important in symbiont recognition (16) and biofilm formation (45) and is linked to the expression of motility (4) in other bacteria, its role in the V. fischeri symbiosis has not been investigated. It would be interesting to determine the colonization effectiveness of a strain that displays the mucoid colony morphology of class II and III strains but displays normal motility behavior. However, to our knowledge this is the first report of mucoid colony morphology in V. fischeri. Because an increase in protease activity is a phenotype associated with HS strains in V. cholerae (14), we tested our HS strains for a similar phenotype but found that the level of protease activity in culture, as measured by a fluorescence-based assay, was indistinguishable among wild-type and HS strains (data not shown). Similarly, because the expression of motility genes has been correlated to phospholipase activity in other bacteria (36, 38), we investigated such a link in V. fischeri. However, in a qualitative plate assay of phospholipase activity, no differences were detected between the wild type and the HS mutant strains (data not shown).
While bacterial motility plays a variety of roles in host interactions, the greatest number of examples are those in which mutations in motility genes have been identified in screens for avirulence in pathogens (reviewed in reference 31). In recent years it has become evident that these mutants are avirulent not only as a result of a loss of tissue tropism but also because the expression of virulence genes requires a functioning flagellar regulon (5, 8, 14, 15, 25, 36, 46) or the presence of a global regulator that functions to coregulate virulence and motility (3, 13, 17, 21, 23, 29, 44). Thus, it appears that the influence of the flagellar regulatory cascade on virulence and colonization gene expression is a common phenomenon defining the role of motility in many bacterium-host interactions. For example, in V. cholerae, proper expression and activation of the motility regulator, FlrC, has been implicated not only in motility but also in colonization, suggesting that modulation of FlrC activity is required for pathogenesis (8). A motility regulator in V. fischeri, FlrA, has been found to regulate motility gene expression (D. S. Millikan and E. G. Ruby, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. 101, p. 428-429, 2001), and we are currently investigating the possibility that this regulator controls both motility and the expression of other colonization genes. Such studies could not only reveal a mechanism for hyperflagellation but also provide further evidence for a link between the regulation of motility and other symbiotic colonization genes in V. fischeri.
Acknowledgments
We thank B. Feliciano for assistance with the hemagglutination assays; C. Lupp for statistical analyses; T. Carvalho at the Pacific Biomedical Research Center's electron microscopy facility for assistance with TEM; and P. Fidopiastis, J. McCann, E. Stabb, and C. Whistler for comments on the manuscript.
This work was supported by NIH grant RR12294 to E.G.R. and M. McFall-Ngai and National Science Foundation grant IBN9904601 to M. McFall-Ngai and E.G.R. D.S.M. was supported by a National Science Foundation Postdoctoral Fellowship in Microbial Biology.
REFERENCES
- 1.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1996. Understanding signal transduction during bacterial infection. Trends Microbiol. 4:141-146. [DOI] [PubMed] [Google Scholar]
- 3.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badger, J. L., and V. L. Miller. 1998. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J. Bacteriol. 180:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182:2811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa, N., C. Lauriano, R. McGee, and K. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czyz, A., R. Zielke, G. Konopa, and G. Wegrzyn. 2001. A Vibrio harveyi insertional mutant in the cgtA (obg, yhbZ) gene, whose homologues are present in diverse organisms ranging from bacteria to humans and are essential genes in many bacterial species. Microbiology 147:183-191. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, P. V., K. Kita-Tsukamoto, J. B. Waterbury, and S. M. Callahan. 1995. Isolation and characterization of a visibly luminous variant of Vibrio fischeri strain ES114 from the sepiolid squid Euprymna scolopes. Arch. Microbiol. 164:194-202. [Google Scholar]
- 12.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frederick, R. D., J. Chiu, J. L. Bennetzen, and A. K. Handa. 1997. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol. Plant-Microbe Interact. 10:407-415. [DOI] [PubMed] [Google Scholar]
- 14.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, J. E., G. M. York, and G. C. Walker. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179:141-146. [DOI] [PubMed] [Google Scholar]
- 17.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 20.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 22.Keynan, A., and J. W. Hastings. 1961. The isolation and characterization of dark mutants of luminescent bacteria. Biol. Bull. 121:375. [Google Scholar]
- 23.Kitten, T., T. G. Kinscherf, J. L. McEvoy, and D. K. Willis. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28:917-929. [DOI] [PubMed] [Google Scholar]
- 24.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 25.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer, L., and P. E. Orndorff. 1987. Identification and characterization of genes determining receptor binding and pilus length of Escherichia coli type I pili. J. Bacteriol. 169:640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarter, L., M. Hilmen, and M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 28.McFall-Ngai, M. J. 1999. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu. Rev. Ecol. Syst. 30:235-256. [Google Scholar]
- 29.Nagasawa, S., S. Tokishita, H. Aiba, and T. Mizuno. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6:799-807. [DOI] [PubMed] [Google Scholar]
- 30.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 32.Reichelt, J. L., and P. Baumann. 1973. Taxonomy of the marine, luminous bacteria. Arch. Mikrobiol. 94:283-330. [DOI] [PubMed] [Google Scholar]
- 33.Ruby, E. G. 1999. The Euprymna scolopes-Vibrio fischeri symbiosis: a biomedical model for the study of bacterial colonization of animal tissue. J. Mol. Microbiol. Biotechnol. 1:13-21. [PubMed] [Google Scholar]
- 34.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, C. K., S. C. Darnell, V. L. Tesh, B. A. Stocker, and A. D. O'Brien. 1994. Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J. Bacteriol. 176:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Songer, J. G. 1997. Bacterial phospholipases and their role in virulence. Trends Microbiol. 5:156-161. [DOI] [PubMed] [Google Scholar]
- 39.Stabb, E., K. L. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-277. In N. Saxena (ed.), Recent Advances in Marine Science and Technology. Pacon International, Honolulu, Hawaii.
- 40.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, S. M., P. A. Carroll, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 1998. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 66:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]