Abstract
A cDNA encoding a eukaryotic translation initiation factor 5A (eIF-5A) homolog in heterotrophic dinoflagellate Crypthecodinium cohnii (CceIF-5A) was isolated through random sequencing of a cDNA library. The predicted amino acid sequence possesses the 12 strictly conserved amino acids around lysine 52 (equivalent to lysine 50 or 51 in other eukaryotes). A single 1.2-kb band was detected in Northern blot analysis. In synchronized C. cohnii cells, the transcript level peaked at early G1 and decreased dramatically on the entry to S phase. Although this has not been previously reported, studies of budding yeast (Saccharomyces cerevisiae) and certain mammalian cell types suggest a role for eIF-5A in the G1/S transition of the eukaryotic cell cycle. Phylogenetic trees constructed with 26 other published eIF-5A sequences suggest that CceIF-5A, while falling within the eukaryotic branches, forms a lineage separate from those of the plants, animals, and archaebacteria. The posttranslational modification of eIF-5A by a transfer of a 4-aminobutyl moiety from spermidine to conserved lysine 50 or 51, forming amino acid hypusine, is the only demonstrated specific function of polyamines in cell proliferation. It has been suggested that polyamines stimulate population growth of bloom-forming dinoflagellates in the sea. We demonstrate here putrescine-stimulated cell proliferation. Furthermore, ornithine decarboxylase inhibitor d-difluoromethylornithine and the specific hypusination inhibitor N-guanyl-1,7-diaminoheptane exhibited inhibitory effects in two species of dinoflagellates. The possible links of polyamines and saxitoxin synthesis to the arginine cycle are also discussed.
Dinoflagellates contribute significantly to primary production in the aquatic environment and are also a major causative agent of harmful algal blooms. Various environmental factors can contribute to the formation and decay of red tides, including water temperature, nutrient availability, turbulence, and water stratification. One of the ecophysiological differences between dinoflagellates and other phytoplankton species (e.g., diatoms) is their greater nutritional diversity, involving mixotrophic ability (43).
Eukaryotic translation initiation factor 5A (eIF-5A), formerly eIF-4D, is the only known protein to contain hypusine (8), which is synthesized posttranslationally by a two-step process in mammalian cells. A 4-aminobutyl moiety from spermidine is first transferred onto a specific lysine residue, followed by the hydroxylation of the aminobutyl group by the enzyme deoxyhypusine synthase (7, 32). Putrescine can be synthesized from ornithine by enzyme ornithine decarboxylase (ODC) and is itself a precursor of spermine and spermidine. Depleting polyamines in L1210 leukemia cells caused cytostasis with accumulation of unmodified eIF-5A (4), and it was suggested that eIF-5A content was a cellular mediator of polyamines in cell growth regulation (45). eIF-5A was first thought to be a translation initiation factor, as it can stimulate the formation of the first peptide bond in vitro (8) and was proposed to be responsible for the translation of a subset of proteins that are required for G1/S transition in eukaryotes (15). Recent studies reveal that eIF-5A is also involved in mRNA turnover (53).
The requirement of polyamines spermine, spermidine, and putrescine in eukaryotic cell proliferation is well documented (23). Polyamine depletion inhibits DNA synthesis (23) and causes cell cycle arrest in cultured mammalian cells (5, 17, 24). However, the detailed mechanisms of polyamine-stimulated growth are not well understood (16). The only proven and specific function of polyamines that may directly lead to growth stimulation is the modification of unusual amino acid hypusine (33). Recent data suggested that polyamines, especially putrescine from decaying fish, may stimulate the growth of flagellates and contribute to the formation of red tides (12). Polyamines can also be produced from the arginine pathway in the cell. Interestingly, the cellular concentration of arginine was shown to vary with the level of saxitoxins in the toxic dinoflagellate Alexandrium minutum (11).
It has been suggested that polyamines may play a role in the rapid growth of bloom-forming dinoflagellates (13). However, no data are available as to the polyamine requirement of dinoflagellates, especially in relation to possible growth stimulation. As eIF-5A was suggested to be a cellular monitor of polyamines for cell growth regulation (45), isolation of its gene would be important in studying the polyamine requirements of dinoflagellates. Also, dinoflagellates are well known to contain both eukaryotic and prokaryotic cytological features (reviewed in reference 35), including permanently condensed chromosomes and no nucleosomes. No full-length clones of eIF-5A from any protists have been reported. As polyamines are essential to growth in both prokaryotes and eukaryotes, it would be of interest to identify the phylogenetic relationship of the dinoflagellate eIF-5A homolog. Very little is known about the transcriptional and translational control of gene expression in dinoflagellates except for the genes regulated by the circadian rhythm. Circadian expression of the luciferin-binding protein and glyceraldehyde-3-phosphate dehydrogenase is regulated at the translational level, while their mRNAs remain constant in the light and the dark phases (10, 26).
In a general search for cell cycle and growth-regulatory genes in dinoflagellate Crypthecodinium cohnii, we obtained a cDNA clone of eIF-5A (CceIF-5A). We report here a characterization of the gene by Northern and Southern blot analysis. Interestingly, we observed a cell cycle-phased transcription of CceIF-5A, peaking at early G1. This may imply a role for CceIF-5A in the entry to S phase. We also describe the growth stimulation of dinoflagellates by putrescine and reduction of growth in the presence of inhibitors to hypusination and polyamine synthesis. The present study provides evidence for a role of polyamines in cell growth of the dinoflagellates. The application of molecular-biology techniques to the study of biological oceanography requires the development of molecular markers. Cell cycle probes have been used in conjunction with flow cytometry to estimate the population growth rate in dinoflagellates (6). With further work involving other dinoflagellate species, CceIF-5A can potentially be used as a molecular marker for G1 cells in oceanographic studies.
MATERIALS AND METHODS
Culture, cell cycle synchronization, and flow-cytometric analysis.
C. cohnii Biecheler cells were cultured in MLH medium (47) at 28°C in the dark. Photosynthetic Heterocapsa triquetra (CCMP449) dinoflagellates were cultured in f/2 medium at 17°C under daily cycles of 14 h of light and 10 h of darkness. C. cohnii cells were synchronized at early G1 by the cyst release filtration method as previously described (51). The C. cohnii culture was concentrated by centrifugation (1,200 × g, 15 min, 20°C) to around 1/200 of its original volume prior to being spread on MLH agar plates. The plates were incubated at 28°C in the dark for 48 h and filled with fresh MLH medium to allow the emergence of motile G1 cells. The medium was filtered with a 10-μm mesh, and the G1 cells were collected in the filtrate. Synchronous cells were harvested every 2.5 h (from 0 to 14 h) after synchronization for RNA extraction and flow-cytometric analysis. Synchronized harvested C. cohnii cells were fixed in 70% ethanol, rehydrated in phosphate-buffered saline, pH 7.4, and incubated at 37°C for 1 h with 200 μg of RNase H ml−1. The cells were stained with 25 μg of propidium iodide ml−1 (4°C, 3 h) before being analyzed with a Becton Dickinson Vantage flow cytometer. At least 10,000 events were measured for each flow cytogram.
eIF-5A cDNA cloning, sequencing, and phylogenetic analysis.
An eIF-5a cDNA clone was obtained through the random sequencing of a C. cohnii cDNA library. The fragment was further cloned into pGEM-T Easy vector (Promega Corporation, Madison, Wis.) and sequenced (AutoRead sequencing; Pharmacia Corporation, Peapack, N.J.) according to manufacturer's instructions. The deduced amino acid sequence was aligned and compared with amino acid sequences of eIF-5A/hypusine-containing proteins of 26 species from PubMed by using the ClustalX program (Center for Scientific Computing). Phylogenetic analysis was carried out using PHYLIP, version 3.5 (Joe Felsenstein, Department of Genetics, University of Washington) with elongation factor P of Escherichia coli as an outgroup. Five hundred bootstrap replicates were generated, and consensus trees based on protein parsimony and the unweighted pair group method with arithmetic averages (UPGMA) were constructed. For the species with more than one isoform of the eIF-5A gene cloned, such as yeast and chickens, only one of them was used in the alignment and the phylogenetic studies.
Northern blot and Southern blot analysis.
Genomic DNA was extracted from a mid-log-phase C. cohnii culture with cetyltrimethylammonium bromide buffer as previously described (51). Genomic Southern blotting was conducted using full-length C. cohnii eIF-5A cDNA as a probe. Probes were labeled by using the ECL direct nucleic acid labeling and detection system (Amersham). Total RNA was extracted from synchronous C. cohnii cells by LiCl precipitation and used for Northern blot analysis. 32P-labeled probes were prepared by random prime labeling using C. cohnii eIF-5A cDNA as a template. All standard molecular-biology techniques were based on reference 36.
Effects of d-DFMO, GC7, and putrescine on growth of dinoflagellates.
While there was suggestion that polyamine may stimulate population growth in dinoflagellates (13), there were no reports on experimental demonstration. Putrescine can be synthesized in many organisms from amino acid ornithine by ODC. Putrescine is itself the precursor for spermidine, which is further transformed to spermine. It is also a breakdown product of many marine organisms. In the present study, we tested the effects of exogenous putrescine on the cell proliferation of C. cohnii. ODC inhibitor difluoromethylornithine (DFMO), which effectively depletes polyamines in yeast and mammalian cells, was also used to evaluate the possible effects of depleting polyamines in dinoflagellates. The two enantiomers of DFMO differ in their abilities to inhibit ODC, with the l form being more potent than the d form (25). The d form is used here as a control for the ODC inhibitory function. N-Guanyl-1,7-diaminoheptane (GC7; a kind gift from Hans Johansson, Children's Hospital, Oakland Research Institute), the most effective specific inhibitor of deoxyhypusine synthase (33), was also tested for its effects on dinoflagellates in the present study. Exponentially growing dinoflagellate cells were cultured in the presence of 0.1, 1, and 10 μM putrescine; 1 mM d-DFMO and l-DFMO; and 1 and 10 mM GC7 (dissolved in dimethyl sulfoxide [DMSO]) in full-growth medium. These concentrations were observed to be effective in other eukaryotes. The control for GC7 is normal growth medium with the same percentage of DMSO added as for the treatment. Exponentially growing cells were inoculated at a 10-fold dilution with fresh medium before the addition of drugs at the beginning of each experiment. Each treatment was performed in triplicate. C. cohnii cells were collected every 12 h, while H. triquetra cells were collected every day for density determination with a Coulter Multisizer II.
RESULTS
Sequence analysis and comparisons of CceIF-5A (GenBank accession no. AF329432).
The cDNA fragment we obtained contains 997 nucleotides, with two possible start codons, which may encode proteins differing by 32 amino acids in length (data not shown). Comparison with other species suggests that the first ATG is the probable start codon. Results from the Blast search suggest that the cDNA fragment is homologous to the known eIF-5A genes from other organisms, and we therefore name its product CceIF-5A. The 12 strictly conserved amino acids around the hypusination site known in other species are conserved in CceIF-5A. The deduced amino acid sequence has 53, 45, and 50% identity to, and 15.7, 21, and 19.3% conserved amino acid changes from, those of eIF-5As from thale cress, humans, and budding yeast, respectively. The cDNA obtained contains no 5′ untranslated region (UTR) upstream of the first possible start codon, while at the 3′ end, a putative UTR of 511 nucleotides, which is approximately the same length as the coding region, is present. A database search using only the 3′ UTR found no homology to any known genes. A poly(A) tail was found in the 3′ UTR, about 500 bases downstream of the predicted stop codon, but the usual eukaryotic poly(A) signal AATAAA and its major natural variant, ATTAAA, are absent. The absence of the AATAAA poly(A) signal from other dinoflagellate genes has been reported (1, 21).
Phylogenetic analysis.
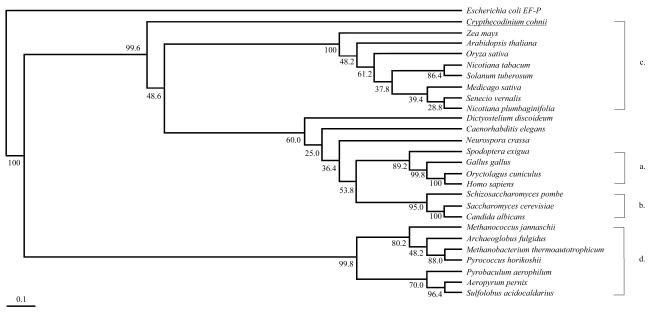
Only a few eIF-5A genes have been analyzed phylogenetically (9, 14). In the present study, we conducted phylogenetic analysis based on 26 eIF-5A amino acid sequences. Consensus trees from 500 bootstrap replicates generated from both the UPGMA (Fig. 1) and parsimony methods (data not shown) have an overall topology very similar to that inferred from rRNA genes. Among the group of eukaryotes, the trees show a clear distinction between eIF-5As from plants, fungi, and animals, except for Caenorhabditis elegans and slime mold. CceIF-5A is located in the eukaryotic group in both phylogenetic trees with almost 100% bootstrap support and forms a separate lineage at the root of plants, animals, and fungi. In some of the bootstrap replicates, CceIF-5A formed a lineage at the base of the plant group. However, this may be due to the lack of other protist sequences in the phylogenetic analysis.
FIG. 1.
Phylogenetic analysis (UPGMA) of EIF-5a genes. The phylogram presented was a consensus tree of 500 bootstrap replicates constructed with the PHYLIP program (version 3.5) with E. coli elongation factor P (EF-P; S34443) as an outgroup. Numbers at nodes are the percentages of trees with the same node among all the bootstraps. Both UPGMA and parsimony (tree not shown) methods essentially generated phylograms with similar topologies. Scale bar, 0.1 substitution per site. NCBI accession numbers of the gene sequences used for animals (a) are Q07460 (Gallus gallus), P10160 (Oryctolagus cuniculus), AAD14095 (Homo sapiens), AAF13315 (Spodoptera exigua), and Q20751 (Caenorhabditis elegans); those for fungi (b) are P38672 (Neurospora crassa), CAB16195 (Schizosaccharomyces pombe), AAD10697 (Candida albicans), BAA11826 (Saccharomyces cerevisiae), and FIDOA (slime mold; Dictyostelium discoideum); those for plants (c) are AAD39281 (Arabidopsis thaliana), CAA69225 (Zea mays), BAA20879 (Solanum tuberosum), P24921 (Nicotiana tabacum), AAC67555 (Oryza sativa), P26564 (Medicago sativa), P24922 (Nicotiana plumbaginifolia), and CAB65463 (Senecio vernalis); those for Archaebacteria (d) are Q58625 (Methanococcus jannaschii), O29612 (Archaeglobus fulgidus), O26955 (Methanococcus thermoautotrophicum), P28461 (Sulfolobus acidocaldarius), Q9YA53 (Aeropyrum pernix), O50089 (Pyrococcus horikoshii), and P56635 (Pyrobaculum aerophilum).
Genomic Southern blot and Northern blot analysis.
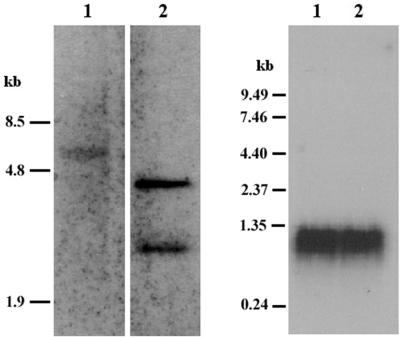
To confirm the presence of CceIF-5A in C. cohnii, genomic Southern blotting was conducted with the CceIF-5A cDNA as a probe. A single band of about 5 kb was detected in the HindIII-digested C. cohnii genomic DNA (Fig. 2, left, lane 1). Plasmid pGEM-eIF5A was used as a positive control (Fig. 2, left, lane 2). Another band of very high molecular size (>20 kb) was detected when the genomic DNA was digested for 5 h or less (not shown); this band may arise from the binding of the probe to incompletely digested DNA. Hybridization of the same cDNA fragment to total RNA extracted from a C. cohnii culture yielded a single 1.2-kDa band (Fig. 2, right), which was slightly larger than the cDNA but which is comparable to those of other species. The extra length detected probably corresponded to the 5′ UTR absent in the cloned fragment and the rest of the poly(A) tail.
FIG. 2.
The presence of the CceIF-5A coding sequence in genomic DNA and total RNA of C. cohnii. (Left) Genomic Southern blot of C. cohnii eIF-5A DNA. Lane 1, HindIII-digested C. cohnii genomic DNA (10 μg); lane 2, positive control, plasmid pGEM-eIF5A (100 ng). The CceIF-5A cDNA (100 ng) was used by the ECL detection system (Amersham Corporation) to produce a probe. (Right) Northern blot of C. cohnii eIF-5A mRNA. Total RNA was extracted from an asynchronous culture of C. cohnii by the LiCl method. The CceIF-5A cDNA (200 ng) was used to produce the probe by random priming with [32P]dCTP.
eIF-5A transcript peaks at early G1 phase of the cell cycle.
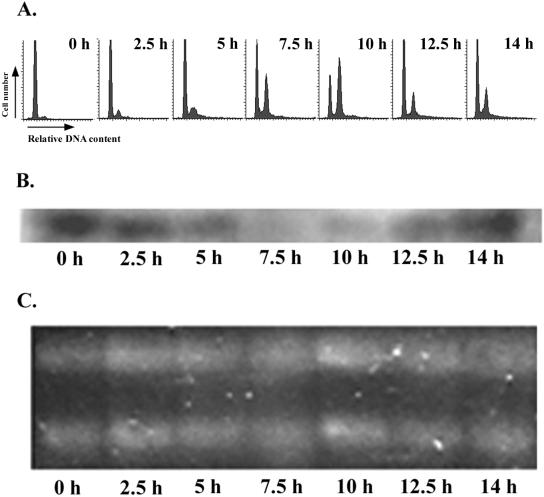
It has been suggested that eIF-5A plays a role in cell cycle control. However, there is little data as to its pattern of expression in the cell cycle. We used the cyst release filtration method to generate highly synchronized G1 C. cohnii cells (time zero; Fig. 3A) and monitored the progression of the cell cycle for one cell cycle (10 h). As previously described, the G2/M cells appeared after 5 h and the cell cycle was completed by 10 h, at which time cytokinesis occurred within the mother cell. Northern blotting with total-RNA samples from five different time points in the cell cycle was carried out with CceIF-5A cDNA as the probe. Interestingly, the level of CceIF-5A mRNA peaks at early G1 (0 and 2.5 h), and the signal detected dropped dramatically on the entry to S phase (5 to 7 h) and stayed low toward the end of the cell cycle (10 h; Fig. 3B), while the level of rRNA stayed relatively constant throughout the experiment (Fig. 3C). The level of CceIF-5A mRNA then increased again as the cells entered G1 of the next cell cycle. While the occurrence of multiple fission (MF) would affect the synchrony of cells after 14 h, the low percentage of MF in this strain (21) did not affect the synchrony of the first cell cycle.
FIG. 3.
Variation of CceIF-5A mRNA level during the cell cycle. Synchronized G1 C. cohnii cells were monitored through the cell cycle for 14 h. (A) Flow cytograms of samples collected at the five time points in the cell cycle; (B) Northern blot of the total RNA collected at the corresponding time points, with CceIF-5A cDNA as the probe; (C) ethidium bromide-stained gel of total RNA loading of each sample.
Effects of d-DFMO, GC7, and putrescine on growth of dinoflagellates.
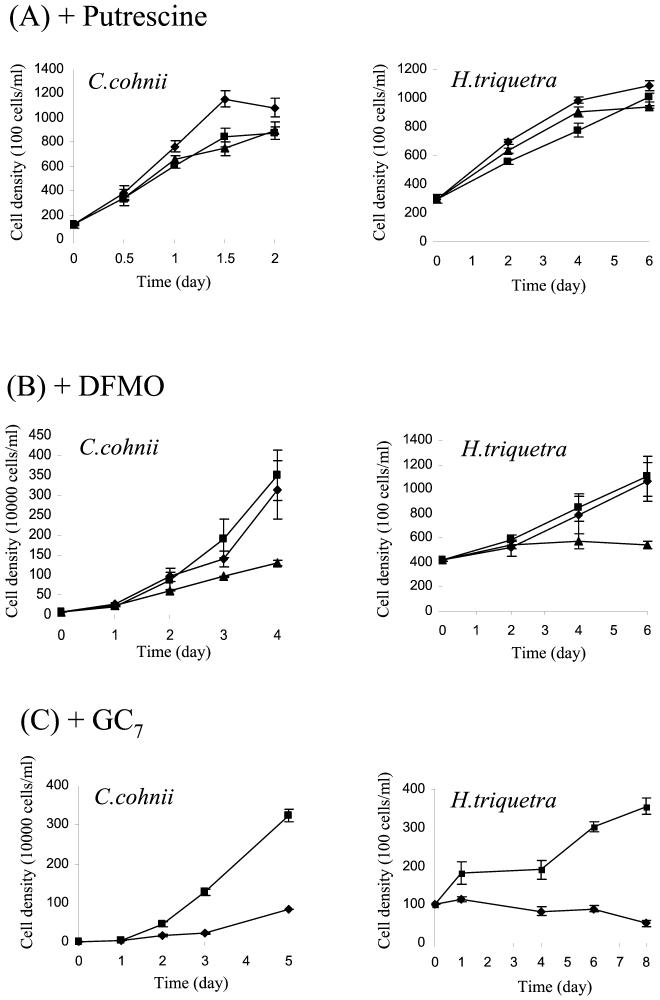
Despite the suggestion of the possible stimulation of dinoflagellate blooms, there are no published accounts of the requirements for polyamines by the group. To demonstrate a possible role of polyamines in dinoflagellate proliferation, we tested the effects of putrescine, the precursor of spermidine and the direct product of ODC, on the cell proliferation of C. cohnii. Addition of 10 μM putrescine induced a significant (t = 0.05) difference in cell number when compared to the control in both species of dinoflagellate (C. cohnii and H. triquetra) tested. At other concentrations of putrescine, there were no significant differences in cell number compared to the control culture (Fig. 4A). Cells treated with the less-potent enantiomer of DFMO, d-DFMO, were not significantly different in their numbers when compared to the control. A dramatic reduction of cell proliferation in both H. triquetra and C. cohnii cultures was observed after treatment with ODC inhibitor l-DFMO (Fig. 4B). The effects of l-DFMO on the two dinoflagellate species were also different. No cell number increase was observed for the photosynthetic species H. triquetra, while a much reduced increase in cell number was observed for C. cohnii. GC7 at 1 mM had a significant effect on cell proliferation of both C. cohnii and H. triquetra (Fig. 4C). As for l-DFMO, the inhibitor was more potent for H. triquetra than for C. cohnii.
FIG. 4.
Effects of polyamine and a polyamine synthesis inhibitor on cell proliferation of dinoflagellates. (A) The effects of putrescine (1 and 10 μM) on the cell proliferation of C. cohnii and H. triquetra. Shown are cells treated with 10 (♦) and 1 μM (▴) putrescine and control cells (▪). For the purpose of clear presentation, the results for 0.1 μM are not shown. They were essentially similar to the results obtained for the corresponding control cells. (B) Effects of d- and l-DFMO on cell proliferation of C. cohnii and H. triquetra. Shown are control cells (▪) and d-DFMO- (♦; 1 mM) and l-DFMO (▴; 1 mM)-treated cells,. (C) Effects of specific deoxyhypusine synthase inhibitor GC7 on the cell proliferation of C. cohnii and H. triquetra. ▪, control cells; ♦, GC7 (1 mM)-treated cells.
DISCUSSION
The CceIF-5A gene.
In many eukaryotes, different isoforms of eIF-5A genes and nonfunctional pseudogenes are present (18, 19, 38). The single band detected in the genomic Southern blot suggests that the CceIF-5A gene is a single-copy gene in C. cohnii; it has been found in Dictyostelium discoideum and described (37), and its presence in insect Spodoptera frugiperda has been suggested (49). However, we cannot completely rule out the possibility that the 5-kb fragment contains a cluster of more than one eIF-5A gene. The size of the CceIF-5A transcript is similar to those reported from other species (18, 19). However, the CceIF-5A mRNA is slightly larger than the cDNA, and the extra length probably corresponds to the 5′ UTR, which is not found in the cloned fragment, and the rest of the poly(A) tail. The single RNA species detected suggests that eIF-5A mRNAs of various lengths derived from different genes (37, 47) are unlikely to exist in C. cohnii. One major cytological characteristic of dinoflagellates is the lack of histones. Recent data suggested that the loss of the histone gene cluster occurred in the evolution of the dinoflagellates, during which some prokaryotic genome was acquired (35). This was supported by the discovery of the nucleus-encoded dinoflagellate RuBisCo (ribulose-1,5-biphosphate carboxylase-oxygenase) in Gonyaulax polyedra; this gene is more homologous to those of the proteobacteria than to those of eukaryotes (27). The definite presence of a eukaryotic eIF-5a in C. cohnii implies that the dinoflagellates have eukaryotic protein synthesis machinery. More sequences are required to reveal the relative position of CceIF-5A among the protists.
Cell cycle variation in the CceIF-5A transcript.
There are two important regulatory proteins in dinoflagellate C. cohnii, cyclin and PCNA, with cell cycle expression patterns resembling those of their counterparts in higher eukaryotes (2, 21, 22). The presence of these cyclic proteins suggests that the dinoflagellate cell cycle is also partly controlled by the cell cycle expression of regulatory genes. In the present study, the level of CceIF-5A mRNA peaked at early G1 and dramatically decreased toward the entry into S phase (Fig. 4B). It was proposed that eIF-5A acts on the G1/S transition in yeast and mammalian cells (13), and predominant expression at early G1 phase in C. cohnii provides circumstantial evidence for this hypothesis. In the somatic cell cycle in maize, eIf-5A was observed to be strongly induced in late-G1 cells, but not in S or G2/M cells, by using a hydroxyurea arrest-release method of synchronization (9). However, it would be difficult to investigate early-G1 cells as hydroxyurea is essentially an S phase inhibitor and maize eIF-5A was not observed during the early part of the release experiment (9). Information on cell cycle variation of eIF-5A transcripts in other organisms is limited. In addition to transcription, there are at least two major events that control eIF-5A activity: translation of the eIF-5A precursor and posttranslational modification of hypusine. Further research is required to investigate the role of eIF-5A in cell cycle control.
Polyamines, dinoflagellate cell proliferation, and toxin production.
Cells treated with 10 μm putrescine were observed to have a higher rate of proliferation than the control cells in both C. cohnii and H. triquetra. However, we cannot determine whether this is due to the requirements of polyamines or to the use of polyamines as a nitrogen source (30). Reduction of cell proliferation due to l-DFMO, but not to d-DMFO, in both dinoflagellates suggests the presence of ODC and a polyamine metabolic pathway in the dinoflagellates similar to that in higher eukaryotes. However, no complete growth arrest was observed in C. cohnii after 2 days of d-DFMO treatment. This reduced inhibitory activity of l-DFMO may possibly be attributable to unexpectedly low stability of l-DFMO in MLH medium, up-regulation of ODC activity, or replenishment of the depleted polyamines from other sources, as C. cohnii is a heterotrophic species. Similarly, a high level of GC7 was required to inhibit cell proliferation in dinoflagellates. However, the specificity of GC7 suggests that hypusination is essential for cell proliferation of dinoflagellates. Both dinoflagellates are thecated species, and the reduced effect observed in C. cohnii was unlikely to be due to the reduced permeability of the dinoflagellate amphiesma in this species. In the toxic-bloom-forming prymnesiophyte Chrysochromulina leadbeateri, putrescine was found to stimulate both cell proliferation and toxin production (12). Putrescine was demonstrated to be present in the dinoflagellate Alexandrium tamarensis (29), and it was also suggested that putrescine stimulates the growth of red tide-forming dinoflagellate Gymnodinium nagasakiense (13). The present study demonstrated the stimulatory effects of putrescine in the proliferation of dinoflagellates. In mammalian cells, excess putrescine accumulation inhibits the formation of hypusine in eIF-5A and induces apoptosis (3, 45, 46). The cloning of CcEIF5a and the demonstration of its G1 expression is significant also for its application as a potential probe in biological oceanography for identifying active dinoflagellate cells in G1 versus inactive ones. Development of antibodies to CcEIF5ap and further work on immunocytochemistry in conjunction with flow cytometry for other dinoflagellate species will be required.
The synthesis of both polyamines and saxitoxin is linked to the intracellular arginine cycle. Interestingly, homospermidine synthase, an enzyme responsible for the synthesis of some plant alkaloids, was shown to have evolved from deoxyhypusine synthase (29). While the pathways for the biosynthesis of the alkaloid-based saxitoxins have yet to be dissected, there is circumstantial evidence for their links to polyamine biosynthesis (11). The production of saxitoxins in Alexandrium was also shown to exhibit a cell cycle-phased pattern, occurring mainly in G1 and before the entry to S phase (42, 44). As eIF-5A was suggested to have regulatory roles in the entry to S phase and polyamine-mediated cell growth (45), the cloning of a dinoflagellate eIF-5A will provide the necessary reagents for further research into cellular growth and cell proliferation, as well as their relationship to toxin production in bloom-forming dinoflagellates.
Acknowledgments
The present project was partly supported by grants CERG:HKUST6175/97 M and DAG99/00.SC13 to J.T.Y.W. from the Research Grant Council of the Government of the Hong Kong Special Administrative Region.
GC7 was kindly provided by Hans Johansson, Children's Hospital, Oakland Research Institute. We thank Francis Chan for technical assistance in flow cytometry and Patrick K. K. Yeung for his help in the preparation of the figures. We also thank Donald Anderson for discussion in relation to possible links between polyamines and saxitoxin production.
REFERENCES
- 1.Bae, Y. M., and J. W. Hastings. 1994. Cloning, sequencing and expression of dinoflagellate luciferase DNA from a marine algae, Gonyaulax polyedra. Biochim. Biophys. Acta 1219:449-456. [DOI] [PubMed] [Google Scholar]
- 2.Barbier, M., M. Albert, M.-L. Géraud, Y. Bhaud, A. Picard, and M.-O. Soyer-Gobillard. 1995. Cell cycle regulation of the primitive dinoflagellate Crypthecodinium cohnii Biecheler: evidence for the presence of an homolog of cyclin B. Biol. Cell 84:35-42. [Google Scholar]
- 3.Bergeron, R. J., W. R. Weimar, R. Muller, C. O. Zimmerman, B. H. McCosar, H. Yao, and R. E. Smith. 1998. Effect of polyamine analogues on hypusine content in JURKAT T-cells. J. Med. Chem. 41:3901-3908. [DOI] [PubMed] [Google Scholar]
- 4.Byers, T. L., J. R. Lakanen, J. K. Coward, and A. E. Pegg. 1994. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine. Biochem. J. 303:363-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casero, R. A., Jr., R. J. Bergeron, and C. W. Porter. 1984. Treatment with alpha-difluoromethylornithine plus a spermidine analog leads to spermine depletion and growth inhibition in cultured L1210 leukemia cells. J. Cell. Physiol. 121:476-482. [DOI] [PubMed] [Google Scholar]
- 6.Chang, J., and E. J. Carpenter. 1991. Species-specific phytoplankton growth rates via diel DNA synthesis cycles. V. Application to natural populations in Long Island Sound. Mar. Ecol. Prog. Ser. 78:115-122. [Google Scholar]
- 7.Chen, K. Y., and Q. P. Dou. 1988. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 229:325-328. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, H. L., M. H. Park, J. E. Folk, B. Safer, and R. Braverman. 1983. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc. Natl. Acad. Sci. USA 80:1854-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dresselhaus, T., S. Cordts, and H. Lörz. 1999. A transcript encoding translation initiation factor 5A eIF-5A is stored in unfertilized egg cells of maize. Plant Mol. Biol. 39:1063-1071. [DOI] [PubMed] [Google Scholar]
- 10.Fagan, T., D. Morse, and J. W. Hastings. 1999. Circadian synthesis of a nuclear-encoded chloroplast glyceraldehyde-3-phosphate dehydrogenase in the dinoflagellate Gonyaulax polyedra is translationally controlled. Biochemistry 38:7689-7695. [DOI] [PubMed] [Google Scholar]
- 11.Flynn, K., J. Franco, P. Fernandez, B. Reguera, M. Zapata, K. J. Flynn, P. Lassus, and G. Arzul. 1995. Nitrogen and phosphorus limitation in cultured Alexandrium minutum Halim does not promote toxin production, p. 439-444. In E. E. Le Denn, P. Gentien, and C. M. Le Baut (ed.), Harmful marine algal blooms. Lavoisier, Paris, France.
- 12.Geir, J., D. Runar, E. Wenche, L. Catherine, A. Jan., and S. H. Rune. 1999. Eco-physiology, bio-optics and toxicity of the ichthyotoxic Chrysochromulina leadbeateri (Prymnesiophyceae). J. Phycol. 35:1465-1476. [Google Scholar]
- 13.Gentien, P. 1998. Bloom dynamics and ecophysiology of the Gymnodinium mikimotoi species complex, p. 155-173. In D. M. Anderson, A. D. Cembella, and G. M. Hallegraeff (ed.), Physiological ecology of harmful algal blooms. Springer-Verlag KG, Berlin, Germany.
- 14.Gerloff, D. L., M. Joachimiak, F. E. Cohen, G. M. Cannarozzi, S. G. Chamberlin, and S. A. Benner. 1998. Structure prediction in a post-genomic environment: a secondary and tertiary structural model for the initiation factor 5A family. Biochem. Biophys. Res. Commun. 251:173-181. [DOI] [PubMed] [Google Scholar]
- 15.Hanauske-Abel, H. M., B. Slowinska, S. Zagulska, R. C. Wilson, L. Staiano-Coico, A.-R. Hanauske, T. McCaffrey, and P. Szabo. 1995. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF-5A in the onset of DNA replication. FEBS Lett. 366:92-98. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271:559-564. [DOI] [PubMed] [Google Scholar]
- 17.Kapyaho, K., R. Sinervirta, and J. Janne. 1985. Effects of inhibitors of polyamine biosynthesis on the growth and melanogenesis of murine melanoma cells. Cancer Res. 45:1444-1448. [PubMed] [Google Scholar]
- 18.Koettnitz, K., B. Kappel, T. Baumruker, J. Hauber, and D. Bevec. 1994. The genomic structure encoding human initiation factor eIF-5A. Gene 144:249-252. [DOI] [PubMed] [Google Scholar]
- 19.Koettnitz, K., T. Wöhl, B. Kappel, T. F. Lottspeich, J. Hauber, and D. Bevec. 1995. Identification of a new member of the human eIF-5A gene family. Gene 159:283-284. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. H., M. Mittag, S. Sczekan, D. Morse, and J. W. Hastings. 1993. Molecular cloning and genomic organization of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J. Biol. Chem. 268:8842-8850. [PubMed] [Google Scholar]
- 21.Leveson, A., F. Wong, and J. T. Y. Wong. 1997. Cyclins in a dinoflagellate cell cycle, Mol. Mar. Biol. Biotechnol. 6:172-179. [PubMed] [Google Scholar]
- 22.Leveson, A., and J. T. Y. Wong. 1999. PCNA-like proteins in dinoflagellates. J. Phycol. 35:798-805. [Google Scholar]
- 23.Luk, G. D., and R. A. Casero, Jr. 1987. Polyamines in normal and cancer cells. Adv. Enzyme Regul. 26:91-105. [DOI] [PubMed] [Google Scholar]
- 24.Mamont, P. S., P. Bohlen, P. P. McCann, P. Bey, F. Schuber, and C. Tardif. 1976. Alpha-methyl ornithine, a potent competitive inhibitor of ornithine decarboxylase, blocks proliferation of rat hepatoma cells in culture. Proc. Natl. Acad. Sci. USA 73:1626-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McWilliams, M. L., G.-D. Chen, and L. D. Fechter. 2000. Characterization of the ototoxicity of difluoromethylornithine and its enantiomers. Toxicol. Sci. 56:124-132. [DOI] [PubMed] [Google Scholar]
- 26.Mittag, M., D.-H. Lee, and J. W. Hastings. 1994. Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc. Natl. Acad. Sci. USA 91:5257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse, D., P. Salois, P. Markovic, and J. W. Hastings. 1995. A nuclear-encoded form II RuBisCo in dinoflagellates. Science 268:1622-1624. [DOI] [PubMed] [Google Scholar]
- 28.Naoyoshi, N., and N. Sachio. 1997. Occurrence of polyamines in the bloom forming toxic dinoflagellate Alexandrium tamarense. Fish. Sci. 63:319-320. [Google Scholar]
- 29.Ober, D., and T. Hartmann. 1999. Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc. Natl. Acad. Sci. USA 96:14777-14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palenik, B., and F. M. M. Morel. 1991. Amine oxidases of marine phytoplankton. Appl. Environ. Microbiol. 57:2440-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, M. H., H. L. Cooper, and J. E. Folk. 1981. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 78:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, M. H., H. L. Cooper, and J. E. Folk. 1982. The biosynthesis of protein-bound hypusine (N epsilon-(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon-(4-aminobutyl)lysine). J. Biol. Chem. 257:7217-7222. [PubMed] [Google Scholar]
- 33.Park, M. H., E. C. Wolff, and J. E. Folk. 1993. Is hypusine essential for eukaryotic cell proliferation? Trends Biochem. Sci. 18:475-479. [DOI] [PubMed] [Google Scholar]
- 34.Raikov, I. B. 1995. The dinoflagellate nucleus and chromosomes: the mesokaryote concept reconsidered. Acta Protozool. 34:239-347. [Google Scholar]
- 35.Rizzo, P. 1991. The enigma of the dinoflagellate chromosome. J. Protozool. 38:246-252. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual (2nd ed.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sandholzer, U., M. Centea-Intemann, A. A. Noegel, and F. Lottspeich. 1989. cDNA and derived amino acid sequence of the hypusine containing protein from Dictyostelium discoideum. FEBS Lett. 246:94-100. [DOI] [PubMed] [Google Scholar]
- 38.Schnier, J., H. G. Schwelberger, Z. Smit-McBride, H. A. Kang, and J. W. Hershey. 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3105-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheets, M. D., S. C. Ogg, and M. P. Wickens. 1990. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18:5799-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, X. P., K. C. Yin, J. Ahern, L. J. Davis, A. M. Stern, and L. Waxman. 1996. Effects of N1-guanyl-1,7-diaminoheptane, an inhibitor of deoxyhypusine synthase, on the growth of tumorigenic cell lines in culture. Biochim. Biophys. Acta 1310:119-126. [DOI] [PubMed] [Google Scholar]
- 41.Shivji, M. S., S. O. Rogers, and M. J. Stanhope. 1992. Rapid isolation of high molecular weight DNA from marine macroalgae. Mar. Ecol. Prog. Ser. 84:197-203. [Google Scholar]
- 42.Siu, G. K. Y., M. L. C. Young, and D. K. O. Chan. 1997. Environmental and nutritional factors which regulate population dynamics and toxin production in the dinoflagellate Alexandrium catenella. Hydrobiologia 352:117-140. [Google Scholar]
- 43.Smayda, T. J. 1997. Harmful algal blooms: their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol. Oceanogr. 42:1137-1153. [Google Scholar]
- 44.Taroncher-Oldenburg, G., D. Kulis, and D. M. Anderson. 1997. Toxin variability during the cell cycle of the dinoflagellate Alexandrium fundyense. Limnol. Oceanogr. 42:1178-1188. [Google Scholar]
- 45.Tome, M. E., and E. W. Gerner. 1997. Cellular eukaryotic initiation factor content as a mediator of polyamine effects on growth and apoptosis. Biol. Signals 6:150-156. [DOI] [PubMed] [Google Scholar]
- 46.Tome, M. E., S. M. Fiser, C. M. Payne, and E. W. Gerner. 1997. Excess putrescine accumulation inhibits the formation of modified eukaryotic initiation factor 5A (eIF-5A) and induces apoptosis. Biochem. J. 328:847-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuttle, R. C., and A. R. Loeblich. 1975. An optimal growth medium for the dinoflagellate Crypthecodinium cohnii. Phycologia 14:1-8. [Google Scholar]
- 48.van de Peer, Y., and R. de Wachter. 1997. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J. Mol. Evol. 45:619-630. [DOI] [PubMed] [Google Scholar]
- 49.van Oers, M. M., M. van Marwijk, M. S. Kwa, J. M. Vlak, and A. A. Thomas. 1999. Cloning and analysis of cDNAs encoding the hypusine-containing protein eIF5A of two lepidopteran insect species. Insect Mol. Biol. 8:531-538. [DOI] [PubMed] [Google Scholar]
- 50.Wöhl, T., H. Klier, H. Ammer, F. Lottspeich, and N. Magdolen. 1993. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutant. Mol. Gen. Genet. 241:305-311. [DOI] [PubMed] [Google Scholar]
- 51.Wong, J. T. Y., and A. Whiteley. 1996. An improved method for the cell cycle synchronization of the heterotrophic dinoflagellate Crypthecodinium cohnii. J. Exp. Mar. Biol. Ecol. 197:91-99. [Google Scholar]
- 52.Yeung, P. K. K., F. Wong, and J. T. Y. Wong. 1996. Sequence data of two large-subunit rRNA genes from an Asian strain of Alexandrium catenella. Appl. Environ. Microbiol. 62:4199-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuk, D., and A. Jacobson. 1998. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17:2914-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]