Abstract
In the present study we have analyzed 44 Y-chromosome biallelic polymorphisms in population samples from northwestern (NW) Africa and the Iberian Peninsula, which allowed us to place each chromosome unequivocally in a phylogenetic tree based on >150 polymorphisms. The most striking results are that contemporary NW African and Iberian populations were found to have originated from distinctly different patrilineages and that the Strait of Gibraltar seems to have acted as a strong (although not complete) barrier to gene flow. In NW African populations, an Upper Paleolithic colonization that probably had its origin in eastern Africa contributed 75% of the current gene pool. In comparison, ∼78% of contemporary Iberian Y chromosomes originated in an Upper Paleolithic expansion from western Asia, along the northern rim of the Mediterranean basin. Smaller contributions to these gene pools (constituting 13% of Y chromosomes in NW Africa and 10% of Y chromosomes in Iberia) came from the Middle East during the Neolithic and, during subsequent gene flow, from Sub-Saharan to NW Africa. Finally, bidirectional gene flow across the Strait of Gibraltar has been detected: the genetic contribution of European Y chromosomes to the NW African gene pool is estimated at 4%, and NW African populations may have contributed 7% of Iberian Y chromosomes. The Islamic rule of Spain, which began in a.d. 711 and lasted almost 8 centuries, left only a minor contribution to the current Iberian Y-chromosome pool. The high-resolution analysis of the Y chromosome allows us to separate successive migratory components and to precisely quantify each historical layer.
Introduction
The systematic search for polymorphisms in the human Y chromosome, both by conventional techniques and by denaturing high-performance liquid chromatography (DHPLC), is producing a large number of new markers (Underhill et al. 1997, 2000; Shen et al. 2000), overcoming the initial dearth of available polymorphisms on that chromosome (Dorit et al. 1995). Among all the different types of Y-chromosome polymorphisms, base substitutions and insertion/deletion polymorphisms have proved to be especially useful in the reconstruction of the phylogeny of the 30-Mb Y-chromosome nonrecombining region. Given their nature, these mutations have probably arisen only once in evolutionary history and have created biallelic polymorphisms. In the absence of recurrence, the typing of such markers in nonhuman primates allows us to determine which is the ancestral allele. The knowledge of the ancestral and derived states of these markers, together with the fact that most of the Y chromosome does not recombine, allows the direct application of parsimony criteria to obtain its phylogeny. Underhill et al. (2000) developed a new set of markers and typed a large set of samples from different worldwide population, providing a well-established structure for Y-chromosome phylogeny and a wide context of very detailed information on Y-chromosome variation, against which any particular new population can be evaluated. A specific analysis of Europe (Semino et al. 2000) has shown the possibilities of the application of this marker set to a continental framework. Furthermore, because of this well-established phylogeny, we are able to characterize new populations by means of a fast hierarchical approach, in which markers are successively typed from the top to the bottom (from the root toward the branch tips) of the phylogenetic tree, as needed. Given the fine degree of paternal-lineage dissection achieved, the proper knowledge of the worldwide distribution and of patterns of variation of the haplotypes that constitute this phylogeny will pave the way for the elucidation of the patterns of male migration and admixture, among present and past human populations.
In addition to the geographical proximity of northwestern (NW) Africa and the Iberian Peninsula, which are separated only by the 15-km-wide Strait of Gibraltar, both regions are linked by historical events involving population movement. During the Upper Paleolithic (known as the “Late Stone Age” in the study of African prehistory), the Ibero-Maurusian industry, spanning the time period of 22,000–9,500 years ago (ya) (Newman 1995), is found throughout northern Africa. The prefix “Ibero-” refers to the presumption that this culture extended into Iberia, although an origin in the Nile River valley is now widely accepted (Camps 1974). The Ibero-Maurusian culture was followed, in the NW African Mesolithic, by the Capsian industry (10,000–4,700 ya; Desanges 1990). The Capsian culture persists well into the Neolithic (which began ∼5,500 ya), a fact that may indicate a persistence of the Mesolithic population and a cultural adoption of agriculture and husbandry with some Neolithic admixture, rather than a replacement by Neolithic populations originating in the Middle East. In general terms, prehistoric culture changes in NW Africa were quite independent of the change dynamics on the European shores of the Mediterranean. In Iberia, the first Upper Paleolithic settlements appear as early as 40,000 ya. Later local developments, until the spread of the Neolithic, followed traditions having European or northern Mediterranean distributions.
NW Africa enters history with the Phoenicians, who, originating in the Middle East, founded Carthage in 814 b.c. and established commercial relations with the local populations, who were the ancestors of the current Berbers. The Roman geographers documented the native kingdoms of the Mauri, Numidae, Gaetali, and Libii, all of whom were subsequently conquered, in ∼150 b.c.–a.d. 50, by the Romans, who established themselves, probably with a limited demographic impact, along a 100-km-wide strip along the Mediterranean coast. This has a parallel in the history of Iberia, where Phoenicians and Greeks established trading posts, and where the local populations (Iberians and Celts) were later conquered (starting ∼200 b.c.) by the Romans. Iberia (“Hispania”) then became a province of the Roman Empire.
During the 7th century a.d., the Arabs conquered northern Africa from east to west and spread their language and religion throughout the native Berber population. Although the cultural and political impact of this invasion changed the history of NW Africa profoundly, a precise estimation of the demographic contribution of the Arabs to NW Africa is not available. In a.d. 711, Berber troops under Arab leadership (Hitti 1990) crossed to the Iberian Peninsula, which they subsequently conquered. That date marks the start of an 8-century period during which the Iberian Peninsula was divided into the Christian kingdoms to the north and the Islamic kingdoms in the south. The border between the two moved southward until 1492, when the last Islamic kingdom was conquered. The demographic contribution of NW African populations to Iberia is not known precisely; it may have been on the order of tens of thousands of individuals in a total Iberian population of a few million (McEvedy and Jones 1978). After the first conquest of Iberia, two main Berber invasions swept through the peninsula: the Almoravids (a.d. 1056–1147) and the Almohades (a.d. 1121–1269). The empire founded by the former group extended into Africa as far south as present-day Senegal and Mali (Kasule 1998).
After 1492, first Jews and then Moslems were forced by the Spanish rulers to either convert to Catholicism or leave the country. Most of those who were expelled took refuge in NW Africa. However, the population substrate of the Moslem group is not well known; the extent to which this group was composed of converted Iberians rather than of the descendants of the Islamic invaders is difficult to ascertain.
In the present study, we have typed 44 biallelic polymorphisms and 8 microsatellites (also known as “short tandem repeats,” or “STRs”), to define the main Y-chromosome lineages in NW Africa and the Iberian Peninsula, in a well-established phylogeographical frame (Underhill et al. 2000), as well as to attempt to estimate the dates of both ancient and recent events in the history of those populations. Several hypotheses regarding population history are tested, such as those concerning the extent to which the Paleolithic genetic background may still be present in both regions, as well as the impact of the Neolithic wave of advance; we also quantify any gene flow between these two regions and from external populations into these regions. The present results are contrasted with those of previous analyses of classical polymorphisms (i.e., blood groups and protein polymorphisms), Alu-insertion polymorphisms, mtDNA control-region sequences, and other Y-chromosome studies. Previous analyses, of a smaller set of Y-chromosome polymorphisms in populations from NW Africa and the Iberian Peninsula, have been published by Bosch et al. (1999), who emphasized gene genealogy rather than population history, and by Rosser et al. (2000), who typed 11 biallelic polymorphisms in a broad survey of western-Eurasian populations and found that a principal-component analysis of haplotype frequencies separated NW African populations from European and Middle Eastern populations.
Subjects and Methods
Subjects
Different autochthonous samples from NW Africa and the Iberian Peninsula were typed. The NW African sample included blood from 29 Saharawis, 40 southern Moroccan Berbers, 44 Moroccan Arabs, and 63 north-central Moroccan Berbers. Samples from the Iberian Peninsula included blood from 37 Andalusians, 16 Catalans, and 44 Basques; the Basque individuals were also included in the study by Underhill et al. (2000). Appropriate informed consent was obtained from all participants in this study, and information about the geographical origin of their four grandparents and about their first language was recorded. DNA was extracted from fresh blood by standard phenol-chloroform protocols.
Polymorphism Typing
All samples in this study were characterized by means of a top-down approach, in which the markers indicated in figure 1 were successively typed, in hierarchical order, according to their position in the genealogy given by Underhill et al. (2000). The typing methods in our analysis would allow us to identify almost all haplotypes described by Underhill et al. (2000). Thus, the original haplotype notation of Underhill et al. (2000) has been kept.
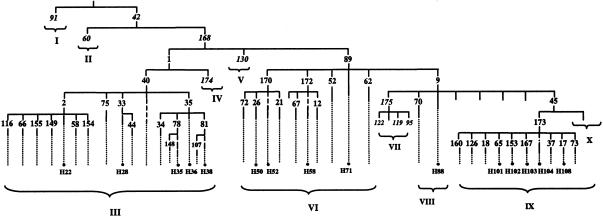
Figure 1.
Phylogenetic tree of Y-chromosome haplotypes and their absolute frequencies in NW Africa and the Iberian Peninsula. The identifiers of the genotyped markers, as well as the haplogroup and haplotype designations, are according to Underhill et al. (2000). Italicized markers were not typed, but their allelic states in the chromosomes analyzed can be deduced unequivocally from the phylogenetic tree.
DHPLC was used to type all biallelic markers, with the exception of YAP (also known as “M1”). Marker information such as primer sequences and PCR conditions for their amplification, whether alleles are ancestral or derived, as well as additional details for their typing conditions by DHPLC, have been provided by Underhill et al. (1997, 2000). YAP was assayed as described by Hammer and Horai (1995). It should be noted that a subset of the polymorphisms used in the present study has been typed in a number of European populations (Semino et al. 2000) and that a different notation has been given to those haplotypes: H22 is termed “Eu2”; H35, H36, and H38 are subsumed under “Eu4”; H52, H50, H58, and H71 are termed “Eu7,” “Eu8,” “Eu9,” and “Eu10,” respectively; H88 is termed “Eu15;” H101, H102, H103, and H104 are subsumed under “Eu18”; and, finally, H108 is termed “Eu19.”
Data for eight STRs (DYS388, DYS19, DYS390, DYS391, DYS392, DYS393, DYS389I, and DYS389II) were available for almost all chromosomes in the sample (Bosch et al. 1999, and additional typings reported here). Complete haplotypes (biallelic markers and STRs) are available from the authors.
Data Analysis
Haplotype-frequency differences among populations from NW Africa and the Iberian Peninsula were tested, by analysis of molecular variance (AMOVA), with the ARLEQUIN software package (Schneider et al. 2000). AMOVA was performed both separately, for NW African and Iberian populations, and as a joint analysis in which genetic variance was partitioned hierarchically as interregion (NW Africa vs. Iberia) variance, intraregion variance, and intrapopulation variance.
Coalescence analysis (Griffiths and Tavaré 1994) was used to test whether NW Africa and Iberia could be regarded as a panmictic unit, to estimate the amount of gene flow among the two regions, and to estimate the ages of M35, M78, and M81, under assumptions of both constant and exponential growth, by means of the Genetree program (available from the Genetree Web site). All biallelic polymorphisms constituting the haplotypes were given the same weight regardless of whether they were nucleotide substitutions or indels, given that they were all compatible with the infinite-sites model implemented in Genetree. First, the values of θ=Nμ (where N is effective population size and μ is mutation rate) that maximized the likelihood of the gene genealogy were obtained separately for the combined haplotype frequencies and for consideration of the two regions separately (in this case, maximum-likelihood estimates of the Nm migration parameter were also obtained, where m is migration rate per generation, were also obtained); next, the likelihood values obtained in the two scenarios were compared by a likelihood-ratio test, after application of the appropriate combinatorial factor (Bahlo and Griffiths 2000). Mutation-age estimates were obtained by use of the growth, θ, and migration parameters that maximized gene-genealogy likelihood and under the assumptions of an effective population size of 5,000 and a 20-year generation time. Genetree provides mutation-age estimates as multiples of θ; thus, either N or μ should be fixed a priori to transform ages in θ units to ages in generations. We fixed N at 5,000, which is close to the global value obtained by Goldstein et al. (1996). With our estimated θ values and with N set at 5,000, we obtained mutation rates of ∼6×10-9 per nucleotide, a result that is consistent with the nuclear-genome average (Li et al. 1985). All Genetree program executions were run for 1,000,000 iterations.
Phylogenetic relations for STR haplotypes within the haplotypes defined by biallelic polymorphisms were depicted by means of reduced median networks (Bandelt et al. 1995), as implemented in the Network 2.0c program (available from www.fluxus-engineering.com).
Separation times, within Y-chromosome lineages, between NW African and Iberian chromosomes, were estimated from STR haplotypes, by means of the average square distance (ASD) (Thomas et al. 1998), by use of a mutation rate of 2.1×10-3 (Heyer et al. 1997; Jobling et al. 1999) and a generation time of 20 years.
Results and Discussion
Male-Lineage Structure of NW African and Iberian Populations
Haplotype frequencies in Moroccan Arabs, north-central Moroccan Berbers, southern Moroccan Berbers, and Saharawis are given in table 1. Haplotype-frequency differences among those populations were tested via AMOVA. Only 0.8% of the genetic variance was found to be due to haplotype-frequency differences among the populations (statistically not significantly different from 0; P=.169). H38, which, according to Underhill et al. (2000), belongs to haplotype group III, is the most common haplotype in NW Africa (64%), with its highest frequencies found within the Saharawis (76%). H71, which belongs to group VI, is the second-most-frequent haplotype (11%) in this area. Other haplotypes, found at lower frequencies, are H22 and H35 (6% each) and H36 (5%), all belonging to group III. The remaining haplotypes, which jointly represent 8% of the NW African Y chromosomes, are found at frequencies of <3%. The genetic homogeneity of NW African Y chromosomes points to a common origin, for all populations analyzed, independent of ethnicity or language (Arab or Berber). These data support the interpretation of the Arabization and Islamization of NW Africa, starting during the 7th century a.d., as cultural phenomena without extensive genetic replacement.
Table 1.
Haplotype Frequencies in NW African and Iberian Populations[Note]
|
No. (%) of Occurrences |
|||||||||||
| Group III |
Group VI |
||||||||||
| Populationa | H22 | H28 | H35 | H36 | H38 | Total | H50 | H52 | H58 | H71 | Total |
| NW Africa: | |||||||||||
| Saharawis (N=29) | 1 | 1 | … | … | 22 | 24 | … | … | … | 5 | 5 |
| SM Berbers (N=40) | 1 | … | 5 | 3 | 26 | 35 | … | … | 1 | 3 | 4 |
| M Arabs (N=44) | 3 | … | 5 | 1 | 23 | 32 | 1 | … | 1 | 6 | 8 |
| NCM Berbers (N=63) | 6 | 2 | 1 | 5 | 41 | 55 | … | … | 2 | 5 | 7 |
| Total (%) | 11 (6.3) | 3 (1.7) | 11 (6.3) | 9 (5.1) | 112 (63.6) | 146 (83.0) | 1 (.6) | 0 | 4 (2.3) | 19 (10.7) | 24 (13.6) |
| Iberian Peninsula: | |||||||||||
| Andalusians (N=37) | … | … | 1 | 1 | 2 | 4 | 2 | … | 2 | 4 | 8 |
| Catalans (N=16) | … | … | … | … | … | 0 | … | 1 | … | 3 | 4 |
| Basquesb (N=44) | … | … | … | … | 1 | 1 | 2 | 1 | … | 1 | 4 |
| Total (%) | 0 |
0 |
1 (1.0) |
1 (1.0) |
3 (3.1) |
5 (5.1) |
4 (4.1) |
2 (2.1) |
2 (2.1) |
8 (8.2) |
16 (16.5) |
|
No. (%) of Occurrences |
|||||||
| Group IX |
|||||||
| Group VIII:H88 | H101 | H102 | H103 | H104 | H108 | Total | |
| NW Africa: | |||||||
| Saharawis (N=29) | … | … | … | … | … | … | 0 |
| SM Berbers (N=40) | … | … | … | … | 1 | … | 1 |
| M Arabs (N=44) | 1 | … | … | … | 3 | … | 3 |
| NCM Berbers (N=63) | … | … | … | … | 1 | … | 1 |
| Total (%) | 1 (.6) | 0 | 0 | 0 | 5 (2.8) | 0 | 5 (2.8) |
| Iberian Peninsula: | |||||||
| Andalusians (N=37) | … | … | 1 | 1 | 22 | 1 | 25 |
| Catalans (N=16) | … | … | … | 5 | 7 | … | 12 |
| Basquesb (N=44) | … | 2 | 7 | 5 | 25 | … | 39 |
| Total (%) | 0 | 2 (2.1) | 8 (8.2) | 11 (11.4) | 54 (55.7) | 1 (1.0) | 76 (78.4) |
Haplotype frequencies for Basques, Catalans, and Andalusians are also given in table 1. AMOVA showed that 2% of the genetic variance was attributable to haplotype-frequency differences among them (statistically not significantly different from 0; P=.08). Pairwise population comparisons via AMOVA did not yield any values significantly different from 0. The most frequent haplotype in these populations is H104 (56%), which belongs to group IX. Haplotypes H102 and H103, which also belong to group IX, are found at frequencies of ∼10%. The frequency of H71 (8%) is similar to that haplotype's frequency in NW Africa. The proportion of haplotypes belonging to group VI (which includes H71) is slightly higher in Iberia (16%) than in NW Africa (14%). H35, H36, and H38, the only haplotypes found to belong to group III, constitute 5% of the Iberian Y chromosomes.
These results clearly show that the contemporary populations from both regions originated from different patrilineages: group III haplotypes prevail in NW Africa, whereas Iberian haplotypes belong mostly to group IX. The proportion of genetic variance that can be attributed to the difference between the NW African and Iberian populations is 35.2% (P=.024), the minimum possible value, given the number of populations and the permutation procedure employed to estimate statistical significance (Excoffier et al. 1992). Moreover, a coalescence analysis of the gene genealogy (Bahlo and Griffiths 2000), including haplotype frequencies in both regions, allowed us to reject the hypothesis that they behave jointly as a panmictic unit (χ2=271.69, 1 df, and P≈0, for constant population sizes; and χ2=266.47, 1 df, and P≈0, for expanding populations). The migration parameters that maximized gene-genealogy likelihood were Nm=1.25 from Iberia to NW Africa and Nm=2 from NW Africa to Iberia, which indicates that gene flow from NW Africa to Iberia may have been greater than that in the opposite direction. Other studies, which analyzed either classical genetic markers (Bosch et al. 1997; Kandil et al. 1999; Simoni et al. 1999), a set of up to 21 autosomal STRs (Bosch et al. 2000), or 11 polymorphic Alu insertions (Comas et al. 2000), showed important genetic differences between NW African and Iberian populations. Moreover, Bosch et al. (1997) and Simoni et al. (1999), analyzing, respectively, 13 and 20 populations from all around the Mediterranean basin, found that the sharpest genetic differences were between populations situated on either side of the Strait of Gibraltar. However, beyond the identification of differences in allele frequencies, the use of a system such as high-resolution biallelic-polymorphism Y-chromosome haplotypes, with a well-established gene genealogy and clear geographical structure, allows us to recognize patterns of origin and diffusion of haplotypes, which can then be used to quantify gene flow, as discussed below.
Neither the overall AMOVA nor any pairwise comparison among populations within either NW Africa or Iberia were significantly different from 0, implying that Y-chromosome biallelic haplotypes are highly homogeneous within each geographical region. Classical genetic markers, together with linguistic, paleoanthropological, and archaeological data, point to a Mesolithic (or older) origin of the Basques (Calafell and Bertranpetit 1994). However, this degree of differentiation is not reached by Y-chromosome polymorphisms (Hurles et al. 1999). For further discussion on how different kinds of genetic markers reflect the Basque differentiation, see the report by Comas et al. (2000).
Geographical and Historical Origins of Y-Chromosome Haplotypes in NW Africa and the Iberian Peninsula
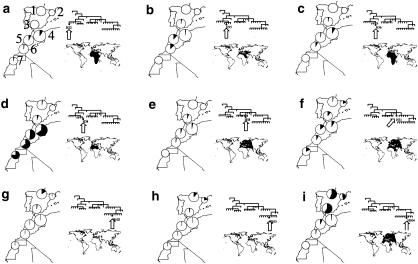
Analysis of the worldwide distribution of Y-chromosome haplotypes may help to establish the putative origins of the haplotypes that contributed to the present NW African and Iberian populations. Figure 2 shows the detailed frequencies of haplotypes H22, H35, H36, H38, H58, H71, H102, H103, and H104, for the populations studied, as well as their worldwide distributions. This type of descriptive analysis allows us to recognize the haplotypes either as being autochthonous or as having originated elsewhere (in regions such as sub-Saharan Africa, Europe, or the eastern Mediterranean).
Figure 2.
Frequency of Y-chromosome haplotypes H22 (a); H35 (b); H36 (c); H38 (d); H58 (e); H71 (f); H102 (g); H103 (h); and H104 (i), in NW Africa and the Iberian Peninsula. The phylogenetic position of each haplotype is found in the upper right of each panel. The worldwide distribution, according to Underhill et al. (2000), is found in the lower right of each panel. Areas of circles are proportional to sample size. In panel a, 1 = Basques; 2 = Catalans; 3 = Andalusians; 4 = north-central Moroccan Berbers; 5 = Moroccan Arabs; 6 = southern Moroccan Berbers; and 7 = Saharawi.
Specific founder effect for some NW African haplotypes: an Upper Paleolithic differentiation?
Although group III haplotypes H35, H36, and H38 are found in eastern and southern Africa, southern Europe, and the Middle East, their overall frequencies in NW Africa are, by far, the highest reported to date (Semino et al. 2000; Underhill et al. 2000). This is particularly true for H38, which clearly constitutes the male population core of NW Africa. By contrast, haplotype H35 is found mainly in Ethiopia (22.7%) and Sudan (17.5%), and H36 is most frequent among Khoisans (10.3%) and Ethiopians (6.5%) (Underhill et al. 2000). Given that H36 is directly ancestral to H35 and H38 and is found at moderate frequencies in Ethiopia and in southern Africa, this branch of the haplotype phylogeny may have been introduced into NW Africa from eastern Africa. On the other hand, the dramatic discontinuity in frequencies of group III haplotypes (especially H38) that is seen in northern Africa suggests that such differences originated under strong genetic drift in small, isolated populations. Such demographic conditions were probably found only before the population surge brought by the Neolithic, which may have prevented further significant differentiation by drift (Cavalli-Sforza et al. 1994), as shown by computer simulations (Rendine et al. 1986; Calafell and Bertranpetit 1993).
Use of classical genetic markers has suggested (Bosch et al. 1997) that the NW African populations may have a sizeable Upper Paleolithic component. This hypothesized Upper Paleolithic expansion may be represented today by the descendants of the haplotypes that share mutation M35 and that are further characterized by M78 (H35) and M81 (H38). It remains to be resolved whether the latter two haplotypes arose independently from H36 or share a common ancestor, yet to be discovered, that distinguishes them from the remaining haplotypes derived from H36.
Assuming a constant population size, an infinite-sites model, and population subdivision between NW Africa and Iberia, we used Genetree (Griffiths and Tavaré 1994) to estimate the age of M35 (giving H36) to be 53,000±21,000 years ago (ya), that of M78 (giving H35) to be 16,000±10,000 ya, and that of M81 (giving H38) to be 32,000±11,000 ya. Under the more likely condition of population growth (Thomson et al. 2000), the respective estimated ages were 30,000±6,000 ya, 7,600±6,000 ya, and 19,000±4,000 ya. Hence, the expansion that brought the ancestors of H35 and H38 (or even those haplotypes themselves) into NW Africa could have happened at any time after 30,000 ya, and, more specifically, it could have happened during the Upper Paleolithic. However, confidence intervals for those dates are large, even without the uncertainty in the effective population size or in generation time. Thus, any interpretation derived from these dates should be regarded with caution. The lower limit for the differentiation event that brought H35 and H38 to such high frequencies in NW Africa is set by the demographic conditions that are compatible with this magnitude, as discussed above, as well as by the genetic evidence, from classical genetic markers (Bosch et al. 1997), that suggests a strong Paleolithic background in NW Africa.
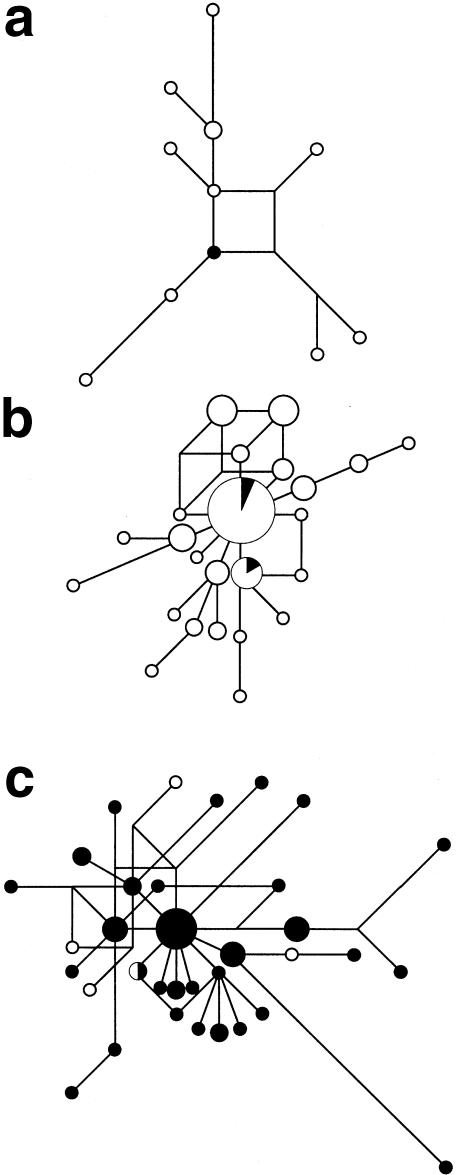
Haplotypes H35, H36, and H38 were found at a low overall frequency (5%) in the Iberian populations. Eight-locus STR haplotypes for the five Iberian group III chromosomes showed that four of them were identical to group III chromosomes in NW Africans and that the fifth was one STR mutation step away. This is clearly depicted in the reduced median networks in figure 3a and b. Given the fast mutation rate of STRs, the extreme similarity between the STR haplotypes in the two regions can be explained only if Iberian and NW African group III chromosomes have a common origin. The time necessary to accumulate this small number of differences was estimated at 700±600 years. Thus, recent gene flow, rather than common ancestry in the distant past, may have brought those chromosomes from NW Africa into Iberia.
Figure 3.
Median-joining networks relating eight-locus STR haplotypes H35 (a); H38 (b); and H104 (c), within biallelic lineages. Areas of circles are proportional to absolute frequencies. White circles denote NW African individuals, black circles denote Iberians. For the sake of clarity, parallel lines do not always represent parallel mutations.
Neolithic Y-chromosome traces in NW Africa and Iberia
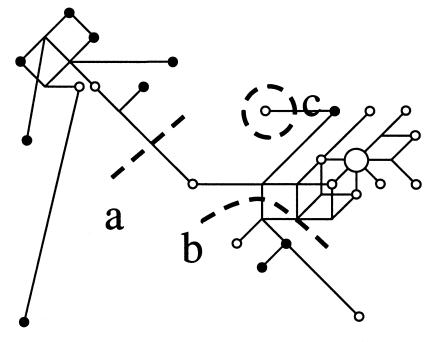
H58 and H71(fig. 2e and f) are part of group VI, which is defined by the presence of M89 and by the absence of M9 and subsequently derived mutations. These haplotypes constitute 10% of the Iberian and 13% of the NW African Y chromosomes and are likely to have spread, with the Neolithic expansions, from the Middle East (Semino et al. 2000). Both haplotypes include chromosomes with the derived 12f2-TaqI*8kb allele (Casanova et al. 1985), as confirmed by Bosch et al. (1999) in a subset of the samples in the present study. Y chromosomes bearing that allele have been found all around the Mediterranean basin, with higher frequencies in the Middle East, and have been interpreted to have spread with the Neolithic wave of advance (Semino et al. 1996; Rosser et al. 2000). A steep cline in the frequency of both H58 and H71, with maxima in the Middle East and frequencies declining with geographical distance from the Middle East, is evidence for a diffusion from the Middle East westward through Europe. The presence of H58 and H71 in both regions could be due to two, not necessarily mutually exclusive, historical processes: (1) the parallel, independent advance of the Neolithic expansion from the Middle East, along the northern and southern shores of the Mediterranean, and (2) an early arrival of the Neolithic in either NW Africa or Iberia and the subsequent crossing of the Strait of Gibraltar. Two independent regional analyses of large sets of classical polymorphisms (Bosch et al. 1997; Simoni et al. 1999) found parallel gradients of genetic differentiation, along the northern and southern shores of the Mediterranean, which make the first scenario more likely. In the present study, STR haplotypes for H58 and H71 chromosomes seemed to be associated with the history of their lineages rather than with population history. In a reduced median network (fig. 4), STR haplotypes clustered by lineage, and a main subdivision was linked to an additional biallelic polymorphism, 12f2 (Casanova et al. 1985; data from Bosch et al. 1999 and additional typings reported here), the 8-kb allele of which was found only in some H71 chromosomes and in all H58 chromosomes. Thus, 12f2*8kb seems to have appeared in the phylogeny after M89 but before M172 (fig. 1). Given that the 12f2*8kb allele was found more often in NW Africa than in Iberia, a comparison, between NW Africa and Iberia, of Y-chromosome STR haplotypes would be, in fact, a comparison of different lineages (as seen in fig. 4, where population origin is not random in the main sections of the network) and would confound attempts to differentiate the two scenarios. A confirmation by the Y chromosome would need to establish an independent correlation with distance to the Middle East. Unfortunately, samples from the appropriate populations are not yet available, particularly from countries along the southern shore, such as Libya and Tunisia.
Figure 4.
Median-joining network of STR haplotypes in H58 and H71. Chromosomes to the left of line a carry allele 12f2*10Kb; chromosomes to the right of line a carry allele 12f2*8Kb. Those under line b or within circle c belong to H58, whereas all others belong to H71. White circles denote NW African individuals; black circles denote Iberians. For the sake of clarity, parallel lines do not always represent parallel mutations.
The European Paleolithic background in Iberia
Group IX haplotypes (fig. 2g–i) are found in the Middle East and are most prevalent in Europe (Underhill et al. 2000). Group IX also contains three local Iberian haplotypes: H101, H102, and H103. The latter, which is defined by derived mutation M167 (also known as “SRY-2627”), is equivalent to Y-chromosome haplogroup 22 as described by Hurles et al. (1999). These authors examined haplogroup 22 worldwide and showed that it has a geographical distribution almost restricted to northern Iberia. Moreover, on the basis of the dating of microsatellite and minisatellite diversity within haplogroup 22, they suggested that it arose in Iberia a few thousand years ago.
Group IX is found at a low frequency (3%) in NW Africa. In Iberia, 56% of the Y chromosomes carry H104, which is found across Europe, with increasing frequencies toward the west; its defining mutation, M173, may have been introduced by the first Upper Paleolithic colonizations of Europe (Semino et al. 2000). It may not have been the only lineage introduced into Iberia during the Upper Paleolithic, but it seems to have been the only one that has persisted in the extant Iberian gene pool. Of five H104 NW African chromosomes, one had an STR haplotype identical to that in an H104 Iberian chromosome, one was one mutation step away from Iberian H104 chromosomes, and the remaining three were two mutation steps away. Moreover, the mean repeat-size difference within 53 H104 Iberian STR haplotypes was 2.8 (range 0–11). The phylogenetic relations among H104 STR haplotypes is shown by a reduced median network (fig. 3c), in which the NW African chromosomes appear to be clearly embedded within the Iberian diversity. The time necessary to accumulate the STR-allele differences between NW African and Iberian H104 chromosomes was estimated at 2,100±450 years. This close STR-haplotype similarity seems to indicate that H104 chromosomes found in NW Africa are a subset of the European gene pool and that they may have been introduced during historic times.
Sub-Saharan gene flow into NW Africa
H22 (defined by mutation M2, also referred to, by Seielstad et al. [1994], as “sY81”; see fig. 2a) and H28, which belong to group III, show a sub-Saharan distribution pattern (Seielstad et al. 1994; Hammer et al. 1997; Underhill et al. 2000). The highest frequency of H22 was found in Mali (30%), and the highest frequencies of H28 were found in southern (51%) and central Africa (57%). Both haplotypes together constitute 8% of the NW African Y chromosomes, and, given their geographical distribution, their presence in NW Africa can be interpreted as resulting from sub-Saharan gene flow. The NW African contact with the southern peoples was especially important during the Almoravid Berber expansion (a.d. 1056–1147), which reached as far south as present-day Senegal and Mali (Kasule 1998), and it has been maintained, until recently, by the trans-Saharan commercial routes.
mtDNA control-region sequence analysis (Rando et al. 1998) detected female-mediated gene flow from sub-Saharan Africa to NW Africa. In particular, 21.5% of the mtDNA sequences in a set of different NW African populations were found to belong to haplogroups L1, L2, and L3a, which constitute most of the sub-Saharan mtDNA sequences.
So far, our analyses have allowed a clear dissection of almost all NW African and Iberian paternal lineages into several components with distinct historical origins. In this way, the historical origins of the NW African Y-chromosome pool may be summarized as follows: 75% NW African Upper Paleolithic (H35, H36, and H38), 13% Neolithic (H58 and H71), 4% historic European gene flow (group IX, H50, H52), and 8% recent sub-Saharan African (H22 and H28). In contrast, the origins of the Iberian Y-chromosome pool may be summarized as follows: 5% recent NW African, 78% Upper Paleolithic and later local derivatives (group IX), and 10% Neolithic (H58, H71). No haplotype assumed to have originated in subSaharan Africa was found in our Iberian sample. It should be noted that H58 and H71 are not the only haplotypes present in the Middle East and that the Neolithic wave of advance could have brought other lineages to Iberia and NW Africa. However, the homogeneity of STR haplotypes within the most ancient biallelic haplotypes in each region indicates a single origin during the past, with possible minor reintroductions, with the Neolithic expansion, from the Middle East. Thus, Neolithic contributions may be slightly underestimated.
Detection of Gene Flow across the Gibraltar Strait
The detection of gene flow between both geographical regions may provide a measure of the reciprocal contribution of Y chromosomes that has occurred during the past. In particular, we have shown that Iberian chromosomes carrying H35, H36, and H38 originated in NW Africa and were brought recently to the peninsula. Their frequency in Iberia will allow us to estimate the maximum NW African male contribution to the Iberian Y-chromosome pool. Since not all NW African Y chromosomes carry those haplotypes, gene flow from NW Africa must have brought other chromosomes. Thus, to estimate the NW African contribution, the proportion of H35, H36, and H38 chromosomes in NW Africa must be taken into account. Therefore, we estimated the overall NW African contribution to the Iberian Y-chromosome pool as being 5% (the frequency of H35, H36, and H38 in Iberia) divided by 75% (the frequency of those haplotypes in NW Africa)—that is, 7%, with the highest level of contribution (14%) being found in Andalusians from southern Iberia. Conversely, since group IX chromosomes in NW Africa may have an Iberian origin, the Iberian (or European) contribution to NW Africa can be estimated, as above, as being 4%.
A small NW African genetic contribution in Iberia is also detected with mtDNA, the female counterpart of the Y chromosome. Rando et al. (1998) suggested a NW African–specific origin for mtDNA haplogroup U6, which is found at frequencies of ∼10%–20% in NW Africans and is absent or nearly absent in Europeans and other Africans. The presence of this NW African mtDNA haplogroup in Iberia can be used as an indicator of NW African–female contribution. Such a contribution seems to be small, since haplogroup U6 is found at very low frequencies: it has been found in 3 of 54 Portuguese and in 2 of 96 Galicians and is absent in Andalusians and in 162 other Iberians (Bertranpetit et al. 1995; Côrte-Real et al. 1996; Pinto et al. 1996; Salas et al. 1998).
We have detected male-mediated gene flow from NW Africa to the Iberian Peninsula; gene flow in the opposite direction, as shown by the Nm and admixture estimates and by ages obtained from STR haplotypes, occurred at lower levels and is more ancient. However, date estimates integrate all the gene flow between the two regions and should be regarded as giving an average rather than as pinpointing a single event. In that respect, the more ancient age estimate for the north-to-south gene flow could have been caused by the fact that it occurred on a haplotype background, H104, that is slightly more diverse than its south-to-north counterpart, H38 (compare figs. 3c and 3b, respectively), thus carrying a more diverse set of Y chromosomes from Iberia into NW Africa.
The Islamic (Arab and Berber) occupation of the Iberian Peninsula, which began in a.d. 711 and, in the south, lasted until a.d. 1492, left a rich cultural heritage, from science and philosophy to agriculture and architecture. Islamic rule lasted longest, until 1492, in southern Iberia. Our results suggest that the demographic contribution linked to that occupation (and to movements in the opposite direction) must have been small but not at all negligible.
This study has demonstrated the unprecedented power of the use of Y-chromosome biallelic polymorphisms for the dissection of paternal lineages, which has allowed us to cut through the historic layers in the Iberian and NW African gene pools in much the same way as archaeologists excavate prehistoric layers at a site.
Acknowledgments
We express our appreciation to the original blood donors who made this study possible. We thank all persons involved in reaching the Saharawi donors, as well as Elisabeth Pintado (Sevilla), Josep Lluís Fernández-Roure, and Alba Bosch (Mataró), for their help in the contacting of Moroccan donors. We especially thank A. A. Lin and P. Shen, for technical support. Mark A. Jobling and two anonymous reviewers made fruitful comments on the manuscript. This research was possible thanks to E.B.’s stay at L. L. Cavalli-Sforza’s laboratory at Stanford University. This work was supported by Dirección General de Investigación Científica y Técnica in Spain grant PB98-1064, by Direcció General de Recerca, Generalitat de Catalunya grants 1998SGR00009 and 2000SGR00093, and by NIHGMS grant 28428 to L. L. Cavalli-Sforza. E.B. was supported by Comissionat per a Universitats i Recerca, Generalitat de Catalunya grant FI/96-1153.
Electronic-Database Information
URLs for software in this article are as follows:
- Genetree, http://www.maths.monash.edu.au/~mbahlo/mpg/gtree.html (software for coalescence analysis of gene genealogies)
- www.fluxus-engineering.com, http://www.fluxus-engineering.com (for Network 2.0c software for median-joining network construction)
References
- Bahlo M, Griffiths RC (2000) Inference from gene trees in a subdivided population. Theor Popul Biol 57:79–95 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertranpetit J, Sala J, Calafell F, Underhill P, Moral P, Comas D (1995) Human mitochondrial DNA variation and the origin of the Basques. Ann Hum Genet 59:63–81 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Pérez-Lezaun A, Clarimón J, Comas D, Mateu E, Martínez-Arias R, Morera B, Brakez Z, Akhayat O, Sefiani A, Harit G, Cambon-Thomsen A, Bertranpetit J (2000) Genetic structure of north-west Africa revealed by STR analysis. Eur J Hum Genet 8:360–366 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Pérez-Lezaun A, Comas D, Mateu E, Bertranpetit J (1997) A population history of northern Africa: evidence from classical genetic markers. Hum Biol 69:295–311 [PubMed] [Google Scholar]
- Bosch E, Calafell F, Santos FR, Pérez-Lezaun A, Comas D, Benchemsi N, Tyler-Smith C, Bertranpetit J (1999) STR variation is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 65:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafell F, Bertranpetit J (1993) A simulation of the genetic history of the Iberian Peninsula. Curr Anthropol 34:735–745. [Google Scholar]
- ——— (1994) Principal component analysis of gene frequencies and the origin of Basques. Am J Phys Anthropol 93:201–215 [DOI] [PubMed] [Google Scholar]
- Camps G (1974) Les civilisations préhistoriques de l’Afrique du Nord et du Sahara. Doin, Paris [Google Scholar]
- Casanova M, Leroy P, Boucekkine C, Weissenbach J, Bishop C, Fellous M, Purrello M, Fiori G, Siniscalco M (1985) A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230:1403–1406 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) History and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Comas D, Calafell F, Benchemsi N, Helal A, Lefranc G, Stoneking M, Batzer MA, Bertranpetit J, Sajantilla A (2000) Alu insertion polymorphisms in northwestern Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar Straits. Hum Genet 107:312–319 [DOI] [PubMed] [Google Scholar]
- Côrte-Real HBSM, Macaulay V, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes BC (1996) Genetic diversity in the Iberian Peninsula determined from mitochondrial sequence analysis. Ann Hum Genet 60:331–350 [DOI] [PubMed] [Google Scholar]
- Desanges J (1990) The proto-Berbers. In: Mokhtar G (ed) General history of Africa. Unesco, Paris, pp 236–245 [Google Scholar]
- Dorit R, Akashi H, Gilbert W (1995) Absence of polymorphism at the ZFY locus on the human Y chromosome. Science 268:1183–1185 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Zhivotovsky LA, Nayar K, Ruiz-Linares A, Cavalli-Sforza LL, Feldman MW (1996) Statistical properties of the variation at linked microsatellite loci: implications for the history of human Y chromosomes. Mol Biol Evol 13:1213–1218 [DOI] [PubMed] [Google Scholar]
- Griffiths RC, Tavaré S (1994) Ancestral inference in population genetics. Stat Sci 9:307–319 [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Spurdle AB, Karafet T, Bonner MR, Wood ET, Novelletto A, Malaspina P, Mitchell RJ, Horai S, Jenkins T, Zegura SL (1997) The geographic distribution of human Y chromosome variation. Genetics 145:787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P (1997) Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed] [Google Scholar]
- Hitti PK (1990) The Arabs: a short history. Gateway Editions, Washington, DC [Google Scholar]
- Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Pérez-Lezaun A, Bosch E, Shlumukova M, Cambon-Thomsen A, McElreavey K, Lopez De Munain A, Röhl A, Wilson IJ, Singh L, Pandya A, Santos FR, Tyler-Smith C, Jobling MA (1999) Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 65:1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Heyer E, Dieltjes P, de Knijff P (1999) Y-chromosome-specific microsatellite mutation rates re-examined using a minisatellite, MSY1. Hum Mol Genet 8:2117–2120 [DOI] [PubMed] [Google Scholar]
- Kandil M, Moral P, Esteban E, Autori L, Mameli GE, Zaoui D, Calò C, Luna F, Vacca L Vona G (1999) Rell cell enzyme polymorphisms in Moroccans and southern Spaniards: new data for the genetic history of the western Mediterranean. Hum Biol 71:791–802 [PubMed] [Google Scholar]
- Kasule S (1998) The history atlas of Africa. Macmillan, New York [Google Scholar]
- Li WH, Wu CI, Luo CC (1985) A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol 2:150–174 [DOI] [PubMed] [Google Scholar]
- McEvedy C, Jones R (1978) Atlas of world population history. Penguin, Hardmonsworth, UK [Google Scholar]
- Newman JL (1995) The peopling of Africa. Yale University Press, New Haven [Google Scholar]
- Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM (1996) Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann Hum Genet 60:321–330 [DOI] [PubMed] [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt HJ (1998) Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with Europeans, Near-Eastern, and sub-Saharan populations. Ann Hum Genet 62:531–550 [DOI] [PubMed] [Google Scholar]
- Rendine S, Piazza A, Cavalli-Sforza LL (1986) Simulation and separation by principal components of multiple demic expansions in Europe. Am Nat 128:681–706 [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN v 2.0: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Seielstad MT, Hebert JM, Lin AA, Underhill PA, Ibrahim M, Vollrath D, Cavalli-Sforza LL (1994) Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet 3:2159–2161 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti S (1996) A view of the neolithic diffusion in Europe through two Y chromosome–specific markers. Am J Hum Genet 59:964–968 [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Shen P, Wang F, Underhill PA, Franco C, Yang WH, Roxas A, Sung R, Lin AA, Hyman RW, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (2000) Population genetic implications from sequence variation in four Y chromosome genes. Proc Natl Acad Sci USA 97:7354–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni L, Gueresi P, Pettener D, Barbujani G (1999) Patterns of gene flow inferred from genetic distances in the Mediterranean region. Hum Biol 71:399–415 [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origin of Old Testament priests. Nature 394:138–140 [DOI] [PubMed] [Google Scholar]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW (2000) Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc Natl Acad Sci USA 97:7360–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatograpghy. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]