Abstract
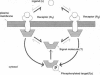
We have trained a computer model of a simple cell-signaling pathway to give specified responses to a pulse of an extracellular ligand. The pathway consists of two initially identical membrane receptors, each of which relays the concentration of the ligand to the level of phosphorylation of an intracellular molecule. Application of random "mutational" changes to the rate constants of the pathway, followed by selection in favor of certain outputs, generates a variety of wave forms and dose-response curves. The phenotypic effect of mutations and the frequency of selection both affect the efficiency with which the pathway achieves its target. When the pathway is trained to give a maximal response at a specific concentration of the stimulating ligand, it gives a consistent pattern of changes in which the two receptors diverge, producing a high-affinity form with excitatory output and a low-affinity form with inhibitory output. We suggest that some high- and low-affinity forms of receptors found in present-day cells might have originated by a similar process.
Full text
PDF
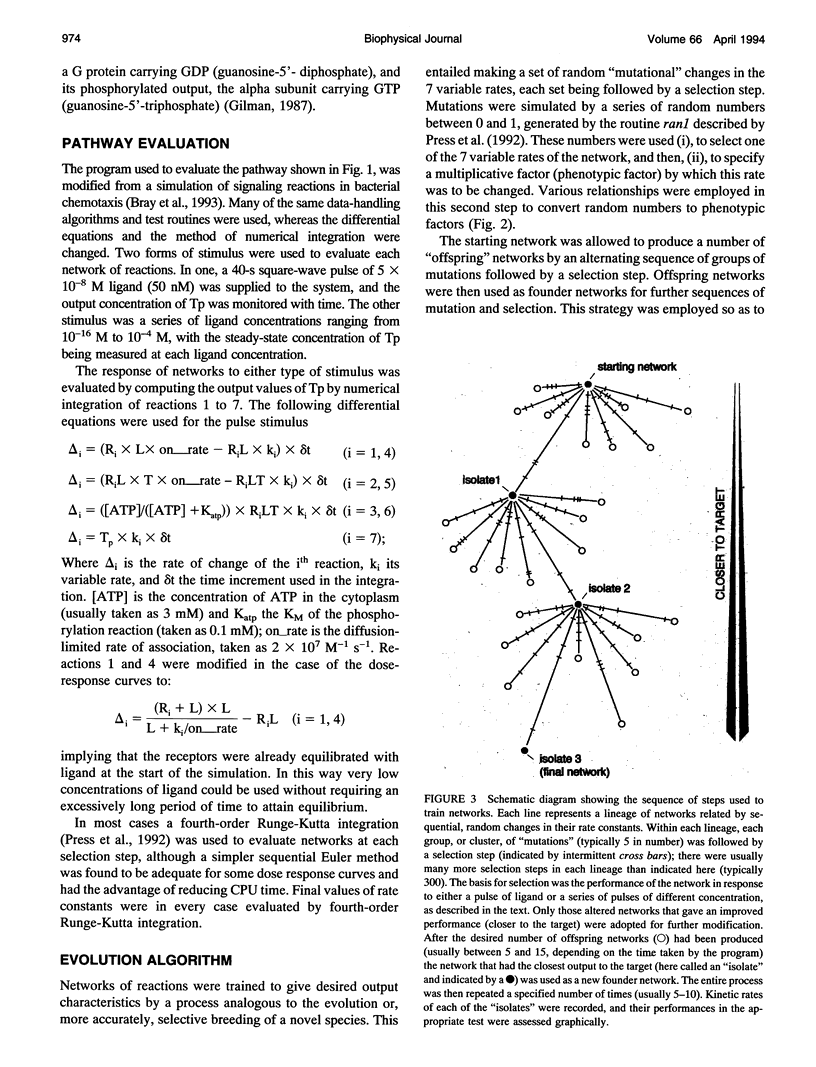
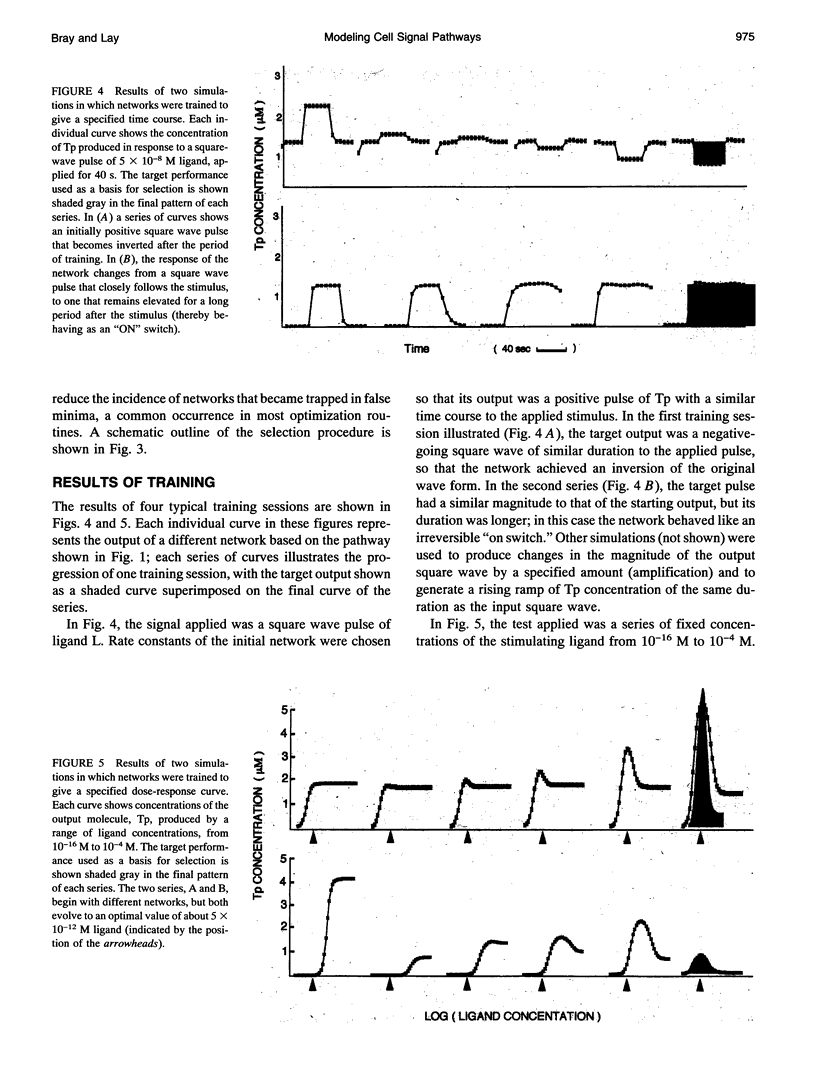


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkers J. A., van Bergen en Henegouwen P. M., Boonstra J. Three classes of epidermal growth factor receptors on HeLa cells. J Biol Chem. 1991 Jan 15;266(2):922–927. [PubMed] [Google Scholar]
- Bray D., Bourret R. B., Simon M. I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993 May;4(5):469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. Intracellular signalling as a parallel distributed process. J Theor Biol. 1990 Mar 22;143(2):215–231. doi: 10.1016/s0022-5193(05)80268-1. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Howell B. The when and how of Src regulation. Cell. 1993 Jun 18;73(6):1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- Fantl W. J., Johnson D. E., Williams L. T. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Heldin C. H., Westermark B. Platelet-derived growth factor: mechanism of action and possible in vivo function. Cell Regul. 1990 Jul;1(8):555–566. doi: 10.1091/mbc.1.8.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmfelt A., Schneider F. W., Ross J. Pattern recognition in coupled chemical kinetic systems. Science. 1993 Apr 16;260(5106):335–337. doi: 10.1126/science.260.5106.335. [DOI] [PubMed] [Google Scholar]
- Hjelmfelt A., Weinberger E. D., Ross J. Chemical implementation of neural networks and Turing machines. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):10983–10987. doi: 10.1073/pnas.88.24.10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Lindberg R. A., Middlemas D. S., Tracy S., van der Geer P. Receptor protein tyrosine kinases and phosphatases. Cold Spring Harb Symp Quant Biol. 1992;57:25–41. doi: 10.1101/sqb.1992.057.01.005. [DOI] [PubMed] [Google Scholar]
- Kacser H., Burns J. A. The control of flux. Symp Soc Exp Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- MONOD J., JACOB F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- Meakin S. O., Shooter E. M. The nerve growth factor family of receptors. Trends Neurosci. 1992 Sep;15(9):323–331. doi: 10.1016/0166-2236(92)90047-c. [DOI] [PubMed] [Google Scholar]
- Northrup S. H., Erickson H. P. Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Pawson T., Schlessingert J. SH2 and SH3 domains. Curr Biol. 1993 Jul 1;3(7):434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- Stoddard B. L., Biemann H. P., Koshland D. E., Jr Receptors and transmembrane signaling. Cold Spring Harb Symp Quant Biol. 1992;57:1–15. doi: 10.1101/sqb.1992.057.01.003. [DOI] [PubMed] [Google Scholar]