Abstract
The dynamics and stability of four DNA duplexes are studied by means of molecular dynamics simulations. The four molecules studied are combinations of 4, 15 bases long, single-stranded oligomers, F1, F2, F3, and F4. The sequence of these single strand oligomers are chosen such that F1-F2 and F3-F4 form parallel (ps) DNA double helices, whereas F1-F4 and F2-F3 form antiparallel-stranded (aps) DNA double helices. Simulations were done at low (100 K) and room (300 K) temperatures. At low temperatures the dynamics are quasi-harmonic and the analysis of the trajectories gives good estimates of the low frequency vibrational modes and density of states. These are used to estimate the linear (harmonic) contribution of local fluctuations to the configurational entropy of the systems. Estimates of the differences in enthalpy between ps and aps duplexes show that aps double helices are more stable than the corresponding ps duplexes, in agreement with experiments. At higher temperatures, the distribution of the fluctuations around the average structures are multimodal and estimates of the configurational entropy cannot be obtained. The multi-basin, nonlinear character of the dynamics at 300 K is established using a novel method which extracts large amplitude nonlinear motions from the molecular dynamics trajectories. Our analysis shows that both ps DNA exhibit much larger fluctuations than the two aps DNA. The large fluctuations of ps DNA are explained in terms of correlated transitions in the beta, epsilon, and zeta backbone dihedral angles.
Full text
PDF


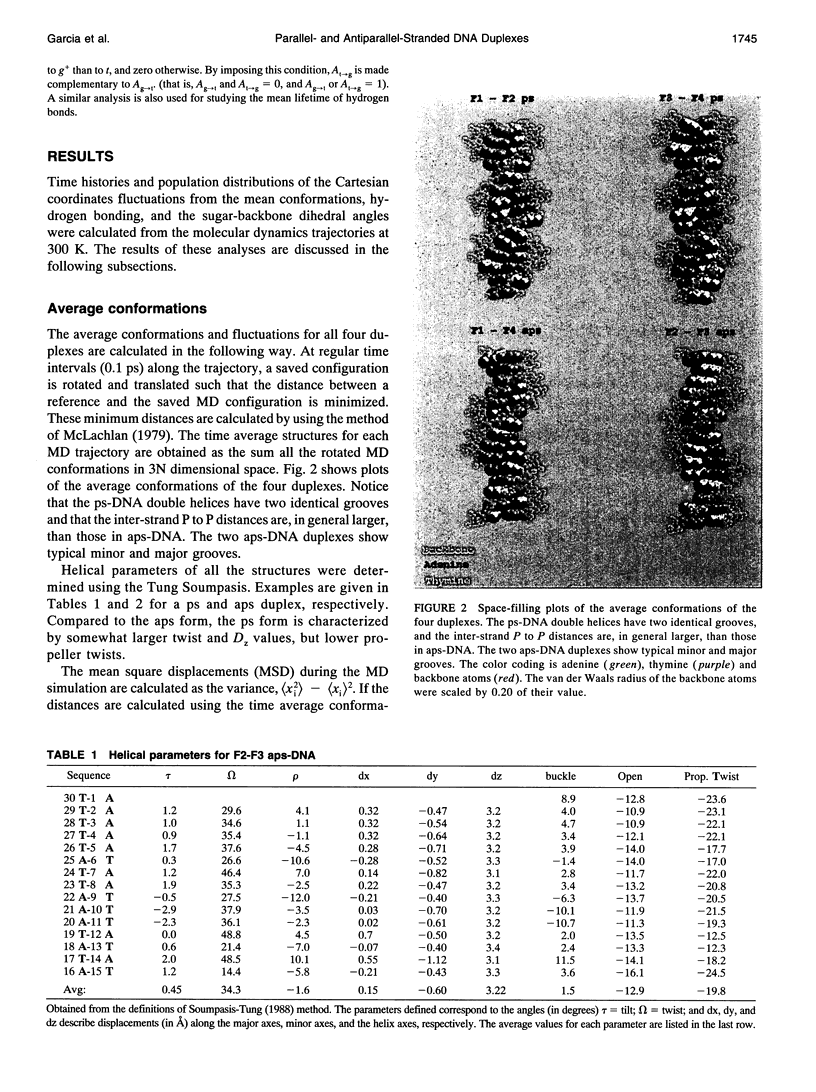
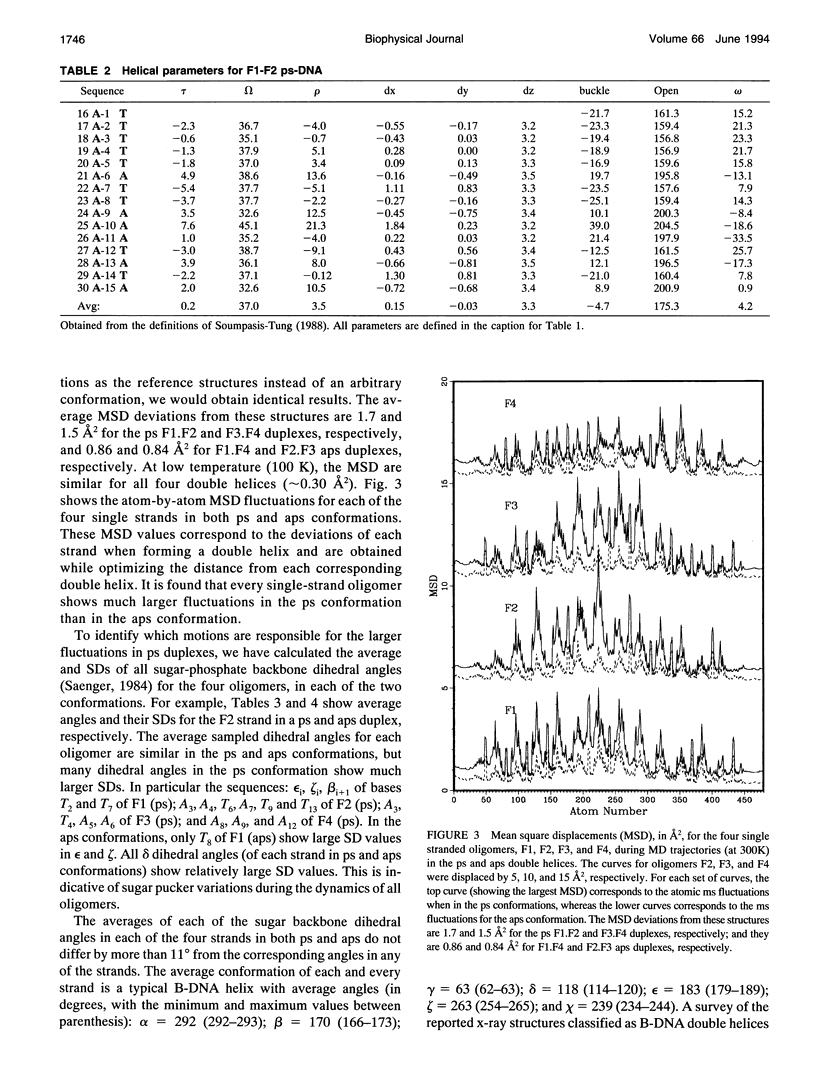
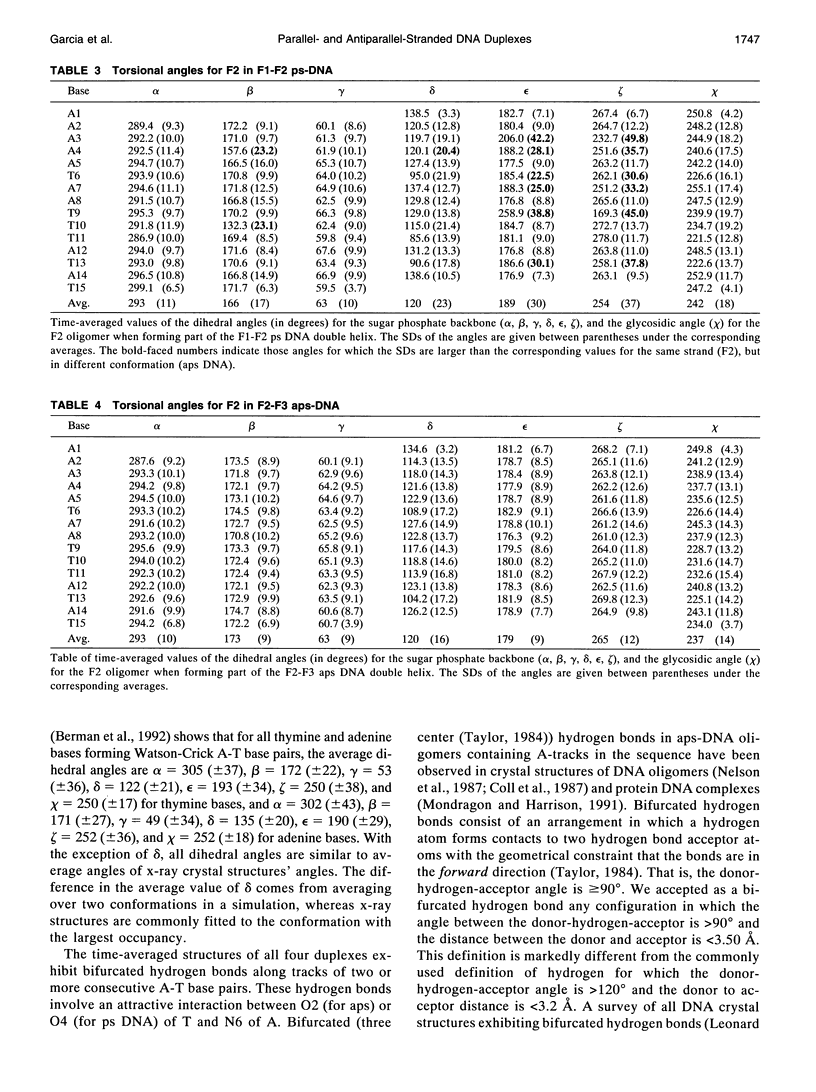




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catasti P., Gupta G., Garcia A. E., Ratliff R., Hong L., Yau P., Moyzis R. K., Bradbury E. M. Unusual structures of the tandem repetitive DNA sequences located at human centromeres. Biochemistry. 1994 Apr 5;33(13):3819–3830. doi: 10.1021/bi00179a005. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A. E., Gupta G., Soumpasis D. M., Tung C. S. Energetics of the hairpin to mismatched duplex transition of d(GCCGCAGC) on NaCl solution. J Biomol Struct Dyn. 1990 Aug;8(1):173–186. doi: 10.1080/07391102.1990.10507796. [DOI] [PubMed] [Google Scholar]
- García A. E., Soumpasis D. M. Harmonic vibrations and thermodynamic stability of a DNA oligomer in monovalent salt solution. Proc Natl Acad Sci U S A. 1989 May;86(9):3160–3164. doi: 10.1073/pnas.86.9.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992 Apr 27;68(17):2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., van de Sande J. H. Relative stability of parallel- and antiparallel-stranded duplex DNA. Biochemistry. 1988 Nov 1;27(22):8302–8306. doi: 10.1021/bi00422a002. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Vogel H. J., Pon R. T., van de Sande J. H. Characterization of a parallel-stranded DNA hairpin. Biochemistry. 1989 Jul 25;28(15):6220–6228. doi: 10.1021/bi00441a013. [DOI] [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il'ychova I. A., Lysov YuP, Chernyi A. A., Shchyolkina A. K., Gottikh B. P., Florentiev V. A. Parallel double helices of DNA. Conformational analysis of regular helices with the second order symmetry axis. J Biomol Struct Dyn. 1990 Feb;7(4):879–897. doi: 10.1080/07391102.1990.10508530. [DOI] [PubMed] [Google Scholar]
- Klement R., Soumpasis D. M., Kitzing E. V., Jovin T. M. Inclusion of ionic interactions in force field calculations of charged biomolecules--DNA structural transitions. Biopolymers. 1990 May-Jun;29(6-7):1089–1103. doi: 10.1002/bip.360290620. [DOI] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Parallel-stranded DNA under topological stress: rearrangement of (dA)15.(dT)15 to a d(A.A.T)n triplex. Nucleic Acids Res. 1991 Dec;19(25):7145–7154. doi: 10.1093/nar/19.25.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Reactivity of parallel-stranded DNA to chemical modification reagents. Biochemistry. 1990 Oct 23;29(42):9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- Lankhorst P. P., Haasnoot C. A., Erkelens C., Altona C. Carbon-13 NMR in conformational analysis of nucleic acid fragments. 2. A reparametrization of the Karplus equation for vicinal NMR coupling constants in CCOP and HCOP fragments. J Biomol Struct Dyn. 1984 Jun;1(6):1387–1405. doi: 10.1080/07391102.1984.10507527. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Evans D. H., Morgan A. R. Polypurine DNAs and RNAs form secondary structures which may be tetra-stranded. Nucleic Acids Res. 1980 Sep 25;8(18):4305–4320. doi: 10.1093/nar/8.18.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. The stability of polypurine tetraplexes in the presence of mono- and divalent cations. Nucleic Acids Res. 1990 Oct 25;18(20):6057–6060. doi: 10.1093/nar/18.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard G. A., Booth E. D., Hunter W. N., Brown T. The conformational variability of an adenosine.inosine base-pair in a synthetic DNA dodecamer. Nucleic Acids Res. 1992 Sep 25;20(18):4753–4759. doi: 10.1093/nar/20.18.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard G. A., Thomson J., Watson W. P., Brown T. High-resolution structure of a mutagenic lesion in DNA. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9573–9576. doi: 10.1073/pnas.87.24.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M., Sharon R. Accurate simulation of protein dynamics in solution. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Guo Q., Kallenbach N. R. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992 Mar 10;31(9):2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D. Gene duplications in the structural evolution of chymotrypsin. J Mol Biol. 1979 Feb 15;128(1):49–79. doi: 10.1016/0022-2836(79)90308-5. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Otto C., Thomas G. A., Rippe K., Jovin T. M., Peticolas W. L. The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson-Crick. Biochemistry. 1991 Mar 26;30(12):3062–3069. doi: 10.1021/bi00226a012. [DOI] [PubMed] [Google Scholar]
- Pattabiraman N. Can the double helix be parallel? Biopolymers. 1986 Sep;25(9):1603–1606. doi: 10.1002/bip.360250903. [DOI] [PubMed] [Google Scholar]
- Powers R., Jones C. R., Gorenstein D. G. Two-dimensional 1H and 31P NMR spectra and restrained molecular dynamics structure of an oligodeoxyribonucleotide duplex refined via a hybrid relaxation matrix procedure. J Biomol Struct Dyn. 1990 Oct;8(2):253–294. doi: 10.1080/07391102.1990.10507805. [DOI] [PubMed] [Google Scholar]
- Powers R., Olsen R. K., Gorenstein D. G. Two-dimensional 1H and 31P NMR spectra of a decamer oligodeoxyribonucleotide duplex and a quinoxaline ((MeCys3, MeCys7)(TANDEM) drug duplex complex. J Biomol Struct Dyn. 1989 Dec;7(3):515–556. doi: 10.1080/07391102.1989.10508507. [DOI] [PubMed] [Google Scholar]
- Ramsing N. B., Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988 Jul 25;16(14A):6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsing N. B., Rippe K., Jovin T. M. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989 Nov 28;28(24):9528–9535. doi: 10.1021/bi00450a042. [DOI] [PubMed] [Google Scholar]
- Rippe K., Fritsch V., Westhof E., Jovin T. M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992 Oct;11(10):3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Substrate properties of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9542–9549. doi: 10.1021/bi00450a044. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Klement R., Jovin T. M. A parallel stranded linear DNA duplex incorporating dG.dC base pairs. J Biomol Struct Dyn. 1990 Jun;7(6):1199–1209. doi: 10.1080/07391102.1990.10508559. [DOI] [PubMed] [Google Scholar]
- Robinson H., Wang A. H. 5'-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5224–5228. doi: 10.1073/pnas.90.11.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Unusual DNA conformation at low pH revealed by NMR: parallel-stranded DNA duplex with homo base pairs. Biochemistry. 1992 Nov 3;31(43):10510–10517. doi: 10.1021/bi00158a014. [DOI] [PubMed] [Google Scholar]
- Soumpasis D. M. Statistical mechanics of the B----Z transition of DNA: contribution of diffuse ionic interactions. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5116–5120. doi: 10.1073/pnas.81.16.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumpasis D. M., Tung C. S. A rigorous basepair oriented description of DNA structures. J Biomol Struct Dyn. 1988 Dec;6(3):397–420. doi: 10.1080/07391102.1988.10506497. [DOI] [PubMed] [Google Scholar]
- Soumpasis D. M., Tung C. S., Garcia A. E. Rigorous description of DNA structures. II. On the computation of best axes, planes, and helices from atomic coordinates. J Biomol Struct Dyn. 1991 Feb;8(4):867–888. doi: 10.1080/07391102.1991.10507850. [DOI] [PubMed] [Google Scholar]
- Van Gunsteren W. F., Berendsen H. J., Geurtsen R. G., Zwinderman H. R. A molecular dynamics computer simulation of an eight-base-pair DNA fragment in aqueous solution: comparison with experimental two-dimensional NMR data. Ann N Y Acad Sci. 1986;482:287–303. doi: 10.1111/j.1749-6632.1986.tb20962.x. [DOI] [PubMed] [Google Scholar]
- Zhou N., Germann M. W., van de Sande J. H., Pattabiraman N., Vogel H. J. Solution structure of the parallel-stranded hairpin d(T8<text text>C4A8) as determined by two-dimensional NMR. Biochemistry. 1993 Jan 19;32(2):646–656. doi: 10.1021/bi00053a033. [DOI] [PubMed] [Google Scholar]