Abstract
The identification of selective glucocorticoid receptor (GR) modifiers, which separate transactivation and transrepression properties, represents an important research goal for steroid pharmacology. Although the gene-activating properties of GR are mainly associated with undesirable side effects, its negative interference with the activity of transcription factors, such as NF-κB, greatly contributes to its antiinflammatory and immune-suppressive capacities. In the present study, we found that Compound A (CpdA), a plant-derived phenyl aziridine precursor, although not belonging to the steroidal class of GR-binding ligands, does mediate gene-inhibitory effects by activating GR. We demonstrate that CpdA exerts an antiinflammatory potential by down-modulating TNF-induced proinflammatory gene expression, such as IL-6 and E-selectin, but, interestingly, does not at all enhance glucocorticoid response element-driven genes or induce GR binding to glucocorticoid response element-dependent genes in vivo. We further show that the specific gene-repressive effect of CpdA depends on the presence of functional GR, displaying a differential phosphorylation status with CpdA as compared with dexamethasone treatment. The antiinflammatory mechanism involves both a reduction of the in vivo DNA-binding activity of p65 as well as an interference with the transactivation potential of NF-κB. Finally, we present evidence that CpdA is as effective as dexamethasone in counteracting acute inflammation in vivo and does not cause a hyperglycemic side effect. Taken together, this compound may be a lead compound of a class of antiinflammatory agents with fully dissociated properties and might thus hold great potential for therapeutic use.
Keywords: cytokine, glucocorticoid receptor, inflammation, NF-κB
Glucocorticoids (GC) are steroid hormones involved in the regulation of developmental and metabolic processes and stress responses. They act via binding to the glucocorticoid receptor (GR), a transcription factor belonging to the superfamily of thyroid/steroid nuclear hormone receptors. Steroidal ligands, such as the synthetic agonist dexamethasone (DEX), bind to GR and induce a conformational change in the cytoplasmic receptor, resulting in nuclear translocation of ligand-bound GR. GR can regulate its target genes in either a positive or negative way. Positive regulation is mainly mediated by direct binding of ligand-activated homodimerized GR onto inducible enhancer elements in the gene promoter, called glucocorticoid response elements (GREs). On the other hand, GR negatively interferes with the activity of transcription factors, such as NF-κB, which drive proinflammatory genes. It is widely accepted that the beneficial, antiinflammatory potential of the GR primarily resides in its ability to negatively modulate proinflammatory cytokines, whereas concomitant side effects are mainly the consequence of its transactivating capacities (1, 2).
NF-κB is a ubiquitous heterodimeric transcription factor, usually composed of a p65 and a p50 subunit, anchored to an inhibitor molecule I-κB. Upon stimulation of cells with TNF, LPS, irradiation, or viral infection, I-κB-α is degraded, allowing activated NF-κB to regulate its target genes in the nucleus (3). Genes activated by NF-κB are implicated in immune responses and code for cytokines, such as IL-6 and IL-8, enzymes, such as inducible nitric-oxide synthase and cyclooxygenase-2, and adhesion molecules, such as E-selectin and intracellular adhesion molecule (4, 5).
At present, GCs still remain the most effective drugs for the treatment of inflammatory disorders. However, their use is limited by the constellation of adverse effects associated with chronic steroid use. Side effects include osteoporosis, muscle wasting, hypertension, behavioral alterations, and disorders of glucose and lipid metabolism (6). Current research has focused on the identification of so-called dissociated GCs, which still elicit the antiinflammatory effects of GCs but exhibit reduced side effects (7, 8).
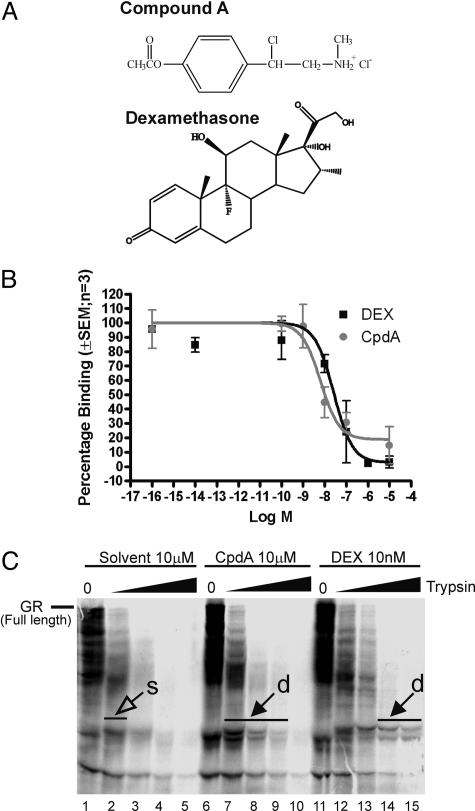
Compound A (CpdA), or 2-(4-acetoxyphenyl)-2-chloro-N-methyl-ethylammonium chloride, is a stable analogue of the hydroxy phenyl aziridine precursor found in the Namibian shrub Salsola tuberculatiformis Botschantzev (9, 10) (Fig. 1A).
Fig. 1.
Binding studies of DEX and CpdA to GR. (A) Chemical structures of CpdA and of DEX. (B) L929sA cells were incubated with 20 nM [3H]DEX in the absence or presence of varying concentrations of DEX (squares) or CpdA (circles). The results shown are typical of two independent experiments, where each condition was performed in triplicate (±SEM). (C) In vitro-translated 35S-radiolabeled WT hGRα was proteolysed with, respectively, 0, 2, 5, 10, and 25 μg/ml trypsin in the presence of the solvent vehicle ethanol, CpdA (10 μM), or DEX (10 nM). Protein fragments were separated by SDS/PAGE and detected by PhosphorImager technology. Protected bands, indicated by closed arrows and showing a doublet (d), correspond to segments of GR that are more resistant to increasing amounts of trypsin in the presence of ligand as opposed to the solvent signal [open arrow, single band (s)].
Our results demonstrate that CpdA, although lacking a steroidal structure, is capable of efficiently down-modulating NF-κB-driven genes via GR but, most interestingly, does not at all stimulate GRE-driven genes, suggesting that it is a completely dissociated agent. Both CpdA and DEX induce nuclear translocation of GR, a prerequisite for its functionality. Finally, we show that CpdA not only interferes with the DNA-binding capacity of NF-κB, but also directly inhibits the transactivation capacity of the NF-κB p65 subunit via activated GR. Most interestingly, apart from being an equally effective antiinflammatory agent as DEX in a mouse in vivo model, CpdA also displays a better side effect profile in vivo, because it does not stimulate hyperglycemia. Taken together, these results lead to the classification of this plant-derived compound as a nonsteroidal, NF-κB-inhibiting antiinflammatory agent and can thus be categorized as a so-called SGRM (selective GR modulator) (6).
Materials and Methods
Cytokines and Reagents. Recombinant murine TNF was produced in our laboratory (11). Dexamethasone was purchased from Sigma. CpdA was synthesized according to Louw and coworkers (9). A stock solution was prepared in ethanol, aliquoted, and stored at -70°C.
Anti-GR (H-300) and anti-p65 Abs were purchased from Santa Cruz Biotechnology and Biomol, respectively. Anti-GR phospho-Ser-211 and control anti-GR Abs were generous gifts from M. Garabedian (New York University School of Medicine, New York). Luciferase (luc) reagent was prepared as described in ref. 11. Human recombinant GR was purchased from Panvera (Madison, WI).
Plasmids. IL-6, IL-8, and E-selectin reporter gene plasmids are described in refs. 12 and 13. pSVhGRα and pMMTV-Luc were a generous gift from W. Rombauts (Katholieke Universiteit Leuven, Leuven, Belgium). Plasmids containing GR deletion variants were kind gifts from R. M. Evans (Howard Hughes Medical Institute, La Jolla, CA). pLT10hGRα cloning strategy can be obtained upon request. p(GRE)250hu.IL6P-luc+ was cloned by replacing the κB motifs in p(IL6κB)350hu.IL6P-luc+ with two consensus GRE sites. pGal4, pGal4-p65, and pGal4-VP16 plasmids were generously provided by M. L. Schmitz (University of Bern, Bern, Switzerland). p(Gal)2-50hu.IL6P-luc+ is described in ref. 11.
Cell Culture. Used cell lines were cultured in DMEM supplemented with 5% newborn calf serum and 5% FCS, 100 units/ml penicillin, and 0.1 mg/ml streptomycin.
Transfection Procedure. L929sA mouse fibroblast cells, HEK 293T human embryonic kidney cells or A549 and TC10 cells were either stably or transiently transfected with various reporter gene constructs (see figure legends) by the calcium phosphate precipitation procedure or by using Lipofectamine (Life Technologies), as described in refs. 11 and 14. Induction experiments were performed at least in triplicate in two independent experiments. Inductions with DEX or CpdA at the indicated concentration were performed at -2 h for a total of 8 h, and TNF (2,000 units/ml) was added at time point zero and left on the cells for 6 h.
After inductions, cells were lysed with lysis buffer (Tropix, Bedford, MA), and samples were assayed for their reporter gene activity according to the manufacturer's instructions (Promega).
RT-PCR. RNA was isolated from A549 cells by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The reverse transcriptase reaction was done by using MLV enzyme (Promega) followed by a PCR reaction with Taq polymerase (Promega) on the obtained cDNA. Primer sets for placental alkaline phosphatase (hPAP) and GAPDH multiplex PCR are available upon request.
ELISA. A murine IL-6 ELISA was performed, using a kit, according to the manufacturer's instructions (R & D Systems).
EMSA. NF-κB EMSA was performed as described in ref. 11.
Whole-Cell Binding Assays. Competitive whole-cell binding assays were performed as described by Bamberger et al. (15). Binding data were analyzed by using prism software (GraphPad), using nonlinear regression and assuming competitive binding to one class of binding sites, to obtain IC50 values ± SEM.
Partial Protease Digestion Assay. The protease digestion was performed as described by Miner and coworkers (16).
Immunofluorescence Analysis. The immunostaining technique was performed essentially as described in ref. 17 by using a 1:200 dilution of the anti-GR antibody (Santa Cruz Biotechnology). Nuclei were visualized by using propidium iodide staining including RNase. Samples were analyzed by using a Zeiss confocal LSM410 microscope, and assessment of intracellular localization of protein signal was performed in a double-blind fashion.
Chromatin Immunoprecipitation (ChIP) Assay. ChIP assays against p65 and GR were performed according to the ChIP kit instructions (Upstate Biotechnology, Lake Placid, NY). Cells were starved for 48 h in serum-free medium, then solvent-treated or treated as described in the figure legends. Quantitative PCR analysis of bound human GILZ and IL-8 promoter DNA was detected by specific primers described in refs. 18 and 19.
Zymosan-Induced Inflamed Paw Model. Eight-week-old C57BL/6J mice were purchased from Iffa Credo. The experimental setup contained four groups with seven animals per group. Group 1 received an injection i.p. of 500 μl of low endotoxin PBS, followed 30 min later by a s.c. injection of 20 μl of zymosan solution (15 mg/ml in PBS, sterilized) in the right footpad and 20 μl of PBS in the left footpad. Group 2 was treated i.p. with 500 μl of 20% ethanol solution (solvent for CpdA and DEX) followed 30 min later by zymosan–PBS injections. Group 3 received 50 μg of DEX i.p. (500 μl of 0.1 mg/ml in 20% EtOH) 30 min before zymosan–PBS treatment, and group 4 was treated with 250 μg of CpdA (500 μl of 0.5 mg/ml in 20% EtOH) followed 30 min later by a zymosan–PBS treatment.
Twenty-four, 48, and 72 h after the zymosan–PBS treatment, the thickness of both footpads was measured by using a caliper, and the difference between zymosan- and PBS-injected footpads was compared for all four experimental groups. The study was performed in a double-blind fashion.
Blood Glucose Determination. Food was removed overnight, and blood samples were taken by sinus retroorbital punction under isoflurane anesthesia from C57Bl6 mice after a 16-h fasting period that was followed by treatment with solvent control PBS, CpdA (12.5 mg/kg), or DEX (2.5 mg/kg) i.p. injected (six mice per group). Blood glucose levels were determined according to the instructions of the Glucotrend 2 kit (Roche Diagnostic).
Results
CpdA Interacts with GR. To investigate whether CpdA directly binds to GR, two independent competitive whole-cell binding assays were performed in L929sA cells (Fig. 1B). Both DEX and CpdA were able to compete with [3H]DEX for binding to the endogenous GR, with a percentage of displacement up to 81%. Homologous/heterologous curves were analyzed, and similar ratios of EC50 values between DEX and CpdA were obtained for both experiments. In the presented experiment, IC50 values of 25.9 nM (95% confidence levels are 7.9–84 nM) for DEX and 6.4 nM (95% confidence levels are 1.9–20.5 nM) for CpdA were obtained. CpdA thus displays a slightly higher binding affinity, with an IC50 ≈4-fold lower than that of DEX.
To additionally detect whether this interaction leads to a change in receptor conformation, we performed a trypsin protease digestion experiment (Fig. 1C). The solvent control displays little (one single band at low trypsin) or no specific band fragmentation after longer proteolysis. In contrast, both CpdA- and DEX-treated GR not only resisted longer to high trypsin concentration but was also cleaved in a different pattern (doublets), as compared with solvent. These results suggest that, in a manner similar to the positive control DEX, CpdA does interact with GR.
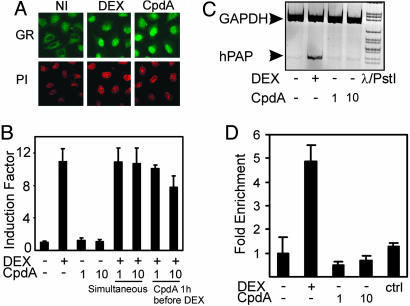
CpdA Induces Nuclear Translocation of GR but Exhibits No Transactivation Potential on GRE-Driven Gene Expression. Because our data in Fig. 1 B and C suggest that CpdA can interact with GR (9), we asked whether this interaction would result in nuclear translocation of GR. By indirect immunofluorescence analysis, we demonstrated that incubation with DEX or CpdA leads to a clear shift of endogenous GR from a predominant cytoplasmic phenotype to a predominant nuclear phenotype in A549 cells (Fig. 2A; and see Figs. 7 and 8, which are published as supporting information on the PNAS web site). Moreover, it was tempting to investigate whether CpdA could also stimulate GRE-driven gene expression.
Fig. 2.
Effect of DEX and of CpdA on GRE-dependent gene expression and translocation of GR. (A) A549 cells were serum-starved in phenol red-free medium for 8 h before induction with solvent control (NI), 1 μM DEX, or 10 μM CpdA for 45 min. After fixation, cells were subjected to immunostaining with anti-GR, followed by anti-rabbit-Alexa 488 as a secondary Ab (GR). Propidium iodide staining (PI) was used to visualize nuclei. (B) L929sA cells with stably integrated p(GRE)2-50-luc+ were solvent-treated or induced with DEX (1 μM), CpdA (1 or 10 μM), or various combinations thereof for a total period of 8 h. Cell lysates were assayed for luciferase activities and normalized for protein content. Promoter activities are expressed as relative induction factor, i.e., the ratio of expression levels recorded either at induced and noninduced conditions, with the latter taken to be 1. Assays were performed in triplicate, and results are representative of at least two independent induction experiments. (C) A549 cells were solvent-treated or induced with DEX (1 μM) or CpdA (1 or 10 μM) for a total period of 8 h. RNA was isolated and reverse transcribed. The resulting cDNA was subjected to PCR analysis with primers to detect the household gene GAPDH (loading control) or the gene coding for human placental alkaline phosphatase in the same sample. (D) ChIP on the GILZ promoter was performed with the GR Ab on A549 cells treated for 2 h with either solvent, DEX (1 μM) or CpdA (1 and 10 μM). GR enrichment on the GRE was determined by correcting the SYBR green quantitative PCR signal for the bound fraction to that of the input fraction. The reaction was performed in triplicate. ctrl, IgG Ab control.
Fig. 2B shows that CpdA lacks activity at either concentration on a p(GRE)2-50-luc+ construct or a pMMTV-Luc construct (data not shown), stably integrated in L929sA cells. In contrast and as expected, DEX, a synthetic steroid ligand for GR, is able to strongly transactivate this construct. However, when added before DEX, CpdA is able to functionally compete for GR, leading to lower transactivation levels of the former (last lane of Fig. 2B). When looking at the expression of the endogenous GC-inducible enzyme human placental alkaline phosphatase (hPAP) in A549 cells (Fig. 2C) or the gluconeogenic enzyme glucose-6-phosphatase in BWTG3 liver cells (data not shown) by RT-PCR analysis, we only found stimulation by DEX. Finally, we performed a ChIP assay using a specific primer set for the GILZ promoter, recently described to contain two functional GRE elements (18). We did not detect any GR occupancy in CpdA-stimulated cells, whereas we did see a significant GR occupancy of DEX-treated cells (Fig. 2D).
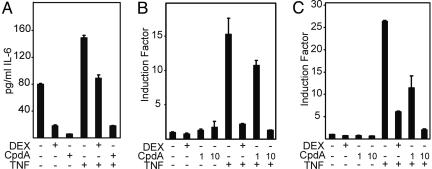
CpdA Inhibits TNF-Induced IL-6 Protein Production and Transrepresses TNF-Induced Cytokine Promoter Activities by Means of Negative Interference with NF-κB. The activated GR exerts its antiinflammatory actions by inhibiting the production of proinflammatory cytokines. Because CpdA induces nuclear translocation of GR, we tested whether this effect could result in repression of proinflammatory gene expression.
Fig. 3A shows that CpdA is able to efficiently lower the basal IL-6 production as well as the TNF-induced IL-6 protein levels in TC10 cells. To determine whether the inhibitory effect of CpdA is directed at the transcriptional level of NF-κB-driven genes, we additionally tested the NF-κB-dependent physiological promoter construct, pE-selectin-Luc, stably integrated into L929sA cells, and observed CpdA-mediated inhibition of TNF-induced gene expression in a dose-responsive manner (Fig. 3B). The same results were obtained for a human IL-6 promoter construct (p1168hu.IL6P-luc+), as well as a for human IL-8 promoter construct (p1481.IL8P-luc+), stably integrated in L929sA cells (data not shown). Previous studies in our group have designated NF-κB as the most important transcription factor involved in TNF-mediated IL-6 gene induction (13). Accordingly, we found that CpdA efficiently inhibits the TNF-activated recombinant reporter gene construct p(IL6κB)350hu.IL6Pluc+ in a dose-dependent manner (Fig. 3C).
Fig. 3.
Effect of DEX or CpdA on constitutive and TNF-induced IL-6 protein levels and on activated NF-κB-dependent reporter gene expression. (A) A murine IL-6 ELISA was performed with the supernatant culture medium of subconfluent TC10 endothelial cells. Cells were treated with 1 μM DEX or 10 μM CpdA in the absence or presence of 2,000 units/ml TNF. DEX or CpdA was added 2 h before TNF for a total period of 8 h. This figure is representative of two independent experiments. L929sA cells with the stably integrated reporter gene construct pE-selectin-Luc (B) or p(IL6κB)350hu.IL6P-luc+ (C) were either treated with DEX (1 μM) or CpdA (1 or 10 μM), as indicated, in the absence or presence of 2,000 units/ml TNF. DEX or CpdA was added 2 h before TNF for a total induction period of 8 h. Plotting of the results is as in Fig. 2 A.
CpdA Repression of NF-κB-Driven Genes Depends on the Presence of GR. Regardless of the translocation experiment, the fact that CpdA does not stimulate GRE-dependent gene expression and selectively represses NF-κB-driven genes made us wonder whether this mechanism could occur independently of GR.
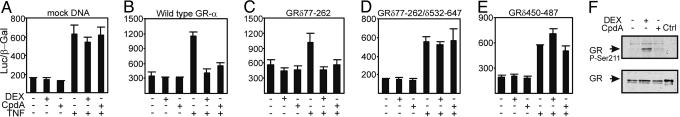
HEK 293T cells have negligible amounts of endogenous GR (data not shown). Therefore, we used these cells to test whether the transrepressive effect of CpdA on NF-κB requires the presence of functional GR. We looked at the effect of DEX and CpdA on NF-κB-driven gene expression with or without introduction of GR into these cells. Fig. 4 A and B demonstrates that TNF-activated p(IL6-κB)350hu.IL6P-luc+ expression can be repressed by either DEX or CpdA but that in both cases the presence of GR is an absolute prerequisite. Fig. 4 C–E demonstrates that, although the N-terminal part of GR appears to be dispensable, both the intact DNA- and ligand-binding domains are important both for CpdA and DEX to mediate their transrepressive effects.
Fig. 4.
Assaying GR (domain) requirement for inhibition of NF-κB-driven gene expression and effect of DEX or CpdA on the specific phosphorylation of GR. HEK 293T cells were transiently transfected with 100 ng of p(IL6κB)350hu.IL6P-luc+, 100 ng of a β-galactosidase control plasmid, and 200 ng of Mock DNA (A), pSVhGRα (B), or plasmids harboring deletion variants of GR as indicated on the figure (C–E). After transfection, cells were pretreated for 2 h with 1 μM DEX or 10 μM CpdA, after which, TNF (2,000 units/ml) was added where indicated. All inductions were allowed to continue for another 6 h. (F) A549 cells were induced for 2 h with CpdA at 10 μM or DEX at 1 μM. Western blot analysis on total extracts was performed with an anti-GR phospho-Ser-211 Ab or an anti-GR nonphosphorylation site-specific Ab.
CpdA Does Not Phosphorylate Ser-211 of GR in Contrast to DEX. The group of Garabedian (20) recently showed that the transcriptional activity of GR upon agonist stimulation is correlated with a 10-fold increase in the phosphorylation status of the Ser-211 residue in the N terminus of GR. Because CpdA behaves as a completely dissociated, nontransactivating ligand for GR, we were interested to know how CpdA could affect phosphorylation of this residue.
Fig. 4F shows that DEX is able to enhance the phosphorylation status of this specific Ser residue at least 10-fold over background levels, as expected. Interestingly, induction with CpdA does not lead to a similar increase in the phosphorylation of Ser-211, nicely correlating with the lack of CpdA-induced GR-mediated transcriptional activation. Equal amounts of sample loading were verified by using a non-phosphorylation-specific Ab, which now also reveals the positive control of 500 ng of human recombinant GR (Fig. 4F Lower, last lane).
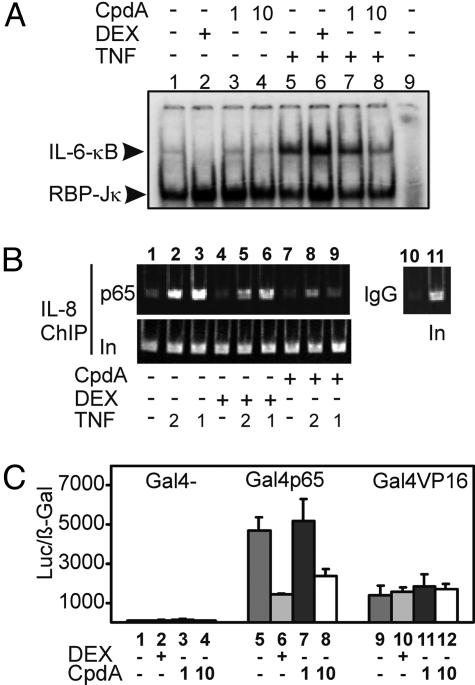
CpdA Interferes with the DNA-Binding Activity of NF-κB in Vitro and in Vivo and also Inhibits the Transactivation Potential of p65. We first investigated the effect of CpdA on NF-κB/DNA-binding activity by gel-shift analysis. From Fig. 5A it can be seen that CpdA at 10 μM affects NF-κB/DNA-binding activity (lane 8 vs. lane 5), which is in contrast to DEX (lane 6 vs. lane 5). Supershifts on the TNF-induced samples demonstrated that the complexes predominantly consist of p65 and p50 (data not shown). When performing a ChIP experiment to analyze the endogenous p65 recruitment on the IL-8 promoter in vivo, we observed a similar result, i.e., that CpdA pretreatment lowers p65 recruitment, as compared with TNF alone. Preincubation with DEX also seems to slightly affect the TNF-induced p65 recruitment at the NF-κB-binding site, although a substantial amount of p65 still remains attached onto the promoter (Fig. 5B). The activated GR can mediate transrepression of NF-κB by means of a nuclear interference mechanism (11). To investigate whether CpdA-activated and nuclear-translocated GR also has a direct inhibitory effect on the transactivation capacities of p65, we investigated the activity of a Gal4-p65 chimeric protein, stably transfected into L929sA cells. Fig. 5C demonstrates that CpdA (10 μM) is able to inhibit the activity of Gal4p65 almost to the same extent as DEX (1 μM). Gal4 serves as a negative control, whereas the specificity of Gal4-p65 repression is demonstrated by using Gal4 fused to the viral activator VP16, which is not subject to regulation by either DEX or CpdA.
Fig. 5.
Effect of DEX or CpdA on NF-κB/DNA binding and NF-κB transcriptional activity. (A) L929sA cells were either left untreated or treated with DEX (1 μM) or CpdA (1 or 10 μM), alone or together with TNF (2,000 units/ml). DEX and CpdA were added 2 h before TNF, for a total period of 4 h. The total protein extract was incubated with a 32P-labeled IL-6 NF-κB response element, and protein/DNA complexes were analyzed in an EMSA. Arrowheads indicate the activated κB complex and the constitutively expressed recombination binding protein (RBP)-Jκ. Lane 9 is a control without protein extract. (B) ChIP was performed with anti-p65 on A549 cells pretreated for 1 h with DEX (1 μM) or CpdA (10 μM) alone, or together with TNF for another 1 h (lanes 3, 6, and 9) or 2 h (lanes 2, 5, and 8). The inputs (In) show that the starting chromatin extracts had equal amounts of the probed regions; the bounds (p65) show enrichment of the IL-8 regions that contain NF-κB response elements. (B Right) ChIP with the IgG negative control and the corresponding input lane. (C) L929sA cell lines with various stably transfected Gal4 constructs, as indicated in the graph, were transiently transfected with the p(Gal4)2-50hu.IL6P-luc+ reporter gene and the β-galactosidase-expressing control plasmid. After transfection, cells were either left untreated or treated with DEX (1 μM) or with CpdA (1 or 10 μM) for 8 h.
CpdA Displays Antiinflammatory Properties in Vivo Without Inducing Hyperglycemia. To study the possible antiinflammatory effects of CpdA in vivo, we opted for the zymosan-induced inflamed paw model. Mice are injected s.c. with zymosan in the footpad, and swelling is determined at several time points after the injection. Pretreatment with CpdA (i.p.) is performed to evaluate antiinflammatory effects, and DEX (i.p.) was chosen as positive control. In Table 1, we show the mean of the differences between zymosan- and PBS-treated footpad for all four groups (see Materials and Methods) and also indicate whether there are significant differences between the individual groups by statistic analysis with the Student t test.
Table 1. In vivo antiinflammatory effects of CpdA in a zymosan-induced inflamed paw mouse model.
| Group | Pretreatment | Difference zymosan–PBS | P value compared with PBS group | P value compared with ethanol group | P value DEX compared with CpdA group |
|---|---|---|---|---|---|
| 1 | PBS | 2.02 ± 0.30 | 0.381 (NS) | ||
| 2 | Ethanol | 1.89 ± 0.23 | 0.381 (NS) | ||
| 3 | DEX | 1.20 ± 0.39 | 0.0009 (***) | 0.0017 (**) | |
| 4 | CpdA | 1.07 ± 0.42 | 0.0004 (***) | 0.0007 (***) | 0.5596 (NS) |
Mice were injected s.c. with zymosan in the footpad, and swelling was determined at 24 h after the injection. Pretreatment with CpdA, DEX, or solvent was for 30 min. Means ± SD were compared with a Student t test. ** and ***, 0.001 ≤ P < 0.01 and P < 0.001, respectively. NS, not significant.
Both DEX and CpdA pretreatment show clear antiinflammatory activities because the swelling is significantly less as compared with the PBS and ethanol pretreatment groups. At all time points measured (measurements also performed at 48 and 72 h; data not shown), there were no significant differences between PBS and ethanol treatment and no significant differences between DEX and CpdA pretreatment.
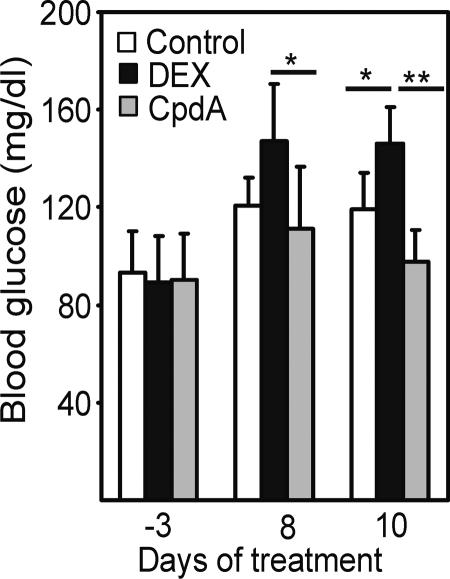
The transactivation-mediated increase in blood glucose concentration by GCs reflects the risk for induction of diabetes mellitus (8). After i.p. treatment in mice, we found that DEX, as expected, triggered a significant increase (P = 0.025) in blood glucose as compared with controls (Fig. 6). In contrast, CpdA did not at all lead to a significant increase in blood glucose level. The ANOVA is used for all analyses, followed by Scheffé post hoc tests for treated vs. control comparisons. The level of significance for all statistical analyses was set at P < 0.05.
Fig. 6.
Effect of DEX or CpdA on blood glucose levels. Mice were treated for up to 10 days with either solvent, 2.5 mg/kg/day DEX or 12.5 mg/kg/day CpdA. The blood glucose concentration was determined 3 days before the first treatment, as well at 8 and 10 days after treatment (mice were fasted 16 h before the blood samples were taken).
Discussion
Although the structure of CpdA is totally different from a normal steroidal ligand (Fig. 1A), it has been suggested that the biological effects of CpdA are most likely mediated by GR (9, 10, 21). Our binding results indicate that CpdA is indeed able to compete with the specific binding of [3H]DEX to GR for up to 81% (Fig. 1B). Protease digestions combined with transfection data also suggest that CpdA interacts with GR, and more specifically, with the ligand-binding domain of GR (Figs. 1C and 4 C–E). CpdA may, however, induce a subtly different conformational change of GR. This hypothesis would also fit with the fact that we found a differential phosphorylation status of GR (Fig. 4F), which may correspond to a different allosteric conformation, similarly as has been described for ER-α phosphorylation by the Rsk2 kinase (22). Because the affinity for GR is slightly higher with CpdA than with DEX, it seems rather unexpected that the effective concentration needed to mediate maximal physiological effects is higher for CpdA than for DEX. However, other examples of ligands exist that display a difference between receptor affinity and efficacy. For instance, the phytoestrogen genistein has a higher affinity for ERβ than for ERα, but its efficacy turns out to be greater with ERα than with ERβ (23). Even though CpdA can bind to GR and drive it into the nucleus (Fig. 2A), we found that CpdA does not stimulate endogenous GRE-dependent genes or different GRE-driven reporter genes (Figs. 2 B and C), nor does it induce GR occupancy on an endogenous GRE-dependent promoter in vivo. CpdA could partially repress DEX-mediated promoter activity on a GRE-driven reporter gene, when added beforehand, which implies some degree of competition for GR molecules in the cell. Why the order of addition may be important is not easily explained and needs further studying of binding parameters, such as association and dissociation kinetics, and/or even crystallographic studies. We rejected the hypothesis that CpdA would display a general irreversible reactivity (e.g., alkylation) toward GR and making the receptor dysfunctional by performing “wash-out” experiments (Fig. 8 A and B). Both GR translocation capability and functionality in reporter gene assays could be restored after washing out CpdA and replacing it with DEX.
Most interestingly, we discovered that CpdA was able to efficiently down-regulate the transcription of various TNF-induced and NF-κB-dependent proinflammatory genes, such as IL-6, E-selectin, intracellular adhesion molecule, and IL-8, as well as IL-6 protein production (Fig. 3 and data not shown), and thus displays a clear antiinflammatory action. As for the CpdA-mediated repression mechanism, we unambiguously established that the presence of functional GR, and more specifically its second zinc finger and its intact ligand-binding domain, are needed for this effect (Fig. 4 A–E), whereas the N-terminal part of the GR appears to be dispensable. The same domains have been reported to be necessary for GC-mediated repression (24). With regard to specificity, we further verified that none of the other nuclear receptors from class II (like PR, AR, MR, and ER) were translocated by CpdA or led to CpdA-mediated transrepression (see Fig. 9 A and B, which is published as supporting information on the PNAS web site).
Furthermore, CpdA substantially affects the in vivo NF-κB/DNA-binding activity on the IL-8 promoter in A549 cells (Fig. 5B), confirming the in vitro bandshift assay results of an IL-6 NF-κB probe, using L929sA cell extracts (Fig. 5A). DEX treatment slightly lowered the in vivo NF-κB-binding activity, which seems in contrast to earlier results by Nissen and Yamamoto (19). This differential effect may be due to the fact that we have pretreated the cells with DEX before adding TNF. Just as with GCs, we found that CpdA interferes with the transactivation potential of a nuclear Gal4-p65 fusion protein (Fig. 5C) and believe that this activity is the more important mechanism of CpdA-mediated transrepression.
Wang and coworkers (20) identified phosphorylation of Ser-211 in GR as a biomarker for agonist-activated GR, found predominantly in the nucleus. Our results now suggest that the phosphorylation of Ser-211 is not a prerequisite for nuclear transport of GR, because CpdA does not phosphorylate this residue to the same extent as DEX, whereas GR accumulated in the nucleus with both ligands. The lack of phosphorylation, however, nicely correlates with the lack of transactivation by CpdA on GRE-driven promoters. Thus, a difference in the phosphorylation status of Ser-211 may reflect differences in transrepression vs. transactivation potential of GR and consequently may be a valid test system to screen for dissociated compounds with a more beneficial action profile. Dissociating ligands for GR, with separate transactivation and transrepression capabilities, merit a lot of interest because of their improved pharmaceutical profiles (25). Some steroidal compounds with such properties have already been described and characterized in detail (26, 27). However, not all of these steroids have proven to be equally successful in vivo as in vitro, with regard to the side effect profile (28). Hence, the search continues for compounds that elicit marked antiinflammatory effects but have a minor impact on unwanted endocrine responses. The recently developed AL-438 (modified progestin scaffold) and ZK 216348 compounds demonstrate that the principle of an improved therapeutic index, i.e., by separation of therapeutic effect (transrepression) and side effects (transactivation) by GR, is possible (7, 8). Such compounds are called SGRMs (selective GR modulators) (6). In the present study, we demonstrate that CpdA clearly displays antiinflammatory characteristics not only in vitro but also in vivo, i.e., in an acute inflammatory mouse model (Table 1). Moreover, not only did we find a clear lack of stimulation of endogenous glucocorticoid-inducible genes by CpdA (Fig. 2C), we also confirmed that a truly improved side effect profile exists in vivo with regard to the risk of diabetes induction, by demonstrating a clear lack of hyperglycemia with CpdA as compared with DEX.
An extensive comparative study of CpdA with DEX on the other levels of GR-mediated side effects in vivo may offer insights or realistic possibilities to ascertain whether CpdA or other analogues can really contribute to human well-being as effective fully dissociated antiinflammatory drugs.
Supplementary Material
Acknowledgments
We thank I. Vanherpe and K. Van Wesemael for excellent technical assistance. I.M.E.B. is a predoctoral researcher and K.D.B., W.V.B., and W.V.M. are postdoctoral researchers at Fonds voor Wetenschappelijk Onderzoek–Vlaanderen. This work was supported by the Interuniversitaire Attractiepolen and a Bilateral Agreement between Flanders and South Africa (BIL99/39). This work was also supported (J.H. and A.L.) in part by grants from the Medical Research Council, the National Research Foundation, and the University of Stellenbosch.
Author contributions: K.D.B., B.S., A.L., C.L., and G.H. designed research; K.D.B., W.V.B., I.M.E.B., W.V.M., N.H., J.H., C.L., and A.L. performed research; K.D.B., W.V.B., A.L., and G.H. contributed new reagents/analytic tools; K.D.B., W.V.B., I.M.E.B., W.V.M., J.H., C.L., B.S., and A.L. analyzed data; and K.D.B. and G.H. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; CpdA, Compound A; DEX, dexamethasone; GC, glucocorticoid; GR, GC receptor; GRE, GC response element.
References
- 1.Reichardt, H. M., Tuckermann, J. P., Bauer, A. & Schütz, G. (2000) Z. Rheumatol. 59, 1-5. [DOI] [PubMed] [Google Scholar]
- 2.De Bosscher, K., Vanden Berghe, W. & Haegeman, G. (2003) Endocr. Rev. 24, 488-522. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh, S. & Karin, M. (2002) Cell 109, Suppl, S81-S96. [DOI] [PubMed] [Google Scholar]
- 4.Vanden Berghe, W., Vermeulen, L., De Wilde, G., De Bosscher, K., Boone, E. & Haegeman, G. (2000) Biochem. Pharmacol. 60, 1185-1195. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, P. J. & Adcock, I. M. (1998) Eur. Respir. J. 12, 221-234. [DOI] [PubMed] [Google Scholar]
- 6.Rosen, J. & Miner, J. N. (2005) Endocr. Rev. 26, 452-464. [DOI] [PubMed] [Google Scholar]
- 7.Coghlan, M. J., Jacobson, P. B., Lane, B., Nakane, M., Lin, C. W., Elmore, S. W., Kym, P. R., Luly, J. R., Carter, G. W., Turner, R., et al. (2003) Mol. Endocrinol. 17, 860-869. [DOI] [PubMed] [Google Scholar]
- 8.Schäcke, H., Schottelius, A., Docke, W. D., Strehlke, P., Jaroch, S., Schmees, N., Rehwinkel, H., Hennekes, H. & Asadullah, K. (2004) Proc. Natl. Acad. Sci. USA 101, 227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louw, A., Swart, P., de Kock, S. S. & van der Merwe, K. J. (1997) Biochem. Pharmacol. 53, 189-197. [DOI] [PubMed] [Google Scholar]
- 10.Louw, A. & Swart, P. (1999) Endocrinology 140, 2044-2053. [DOI] [PubMed] [Google Scholar]
- 11.De Bosscher, K., Schmitz, M. L., Vanden Berghe, W., Plaisance, S., Fiers, W. & Haegeman, G. (1997) Proc. Natl. Acad. Sci. USA 94, 13504-13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaisance, S., Vanden Berghe, W., Boone, E., Fiers, W. & Haegeman, G. (1997) Mol. Cell. Biol. 17, 3733-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanden Berghe, W., Plaisance, S., Boone, E., De Bosscher, K., Schmitz, M. L., Fiers, W. & Haegeman, G. (1998) J. Biol. Chem. 273, 3285-3290. [DOI] [PubMed] [Google Scholar]
- 14.De Bosscher, K., Vanden Berghe, W., Vermeulen, L., Plaisance, S., Boone, E. & Haegeman, G. (2000) Proc. Natl. Acad. Sci. USA 97, 3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamberger, C. M., Bamberger, A. M., de Castro, M. & Chrousos, G. P. (1995) J. Clin. Invest. 95, 2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miner, J. N., Tyree, C., Hu, J., Berger, E., Marschke, K., Nakane, M., Coghlan, M. J., Clemm, D., Lane, B. & Rosen, J. (2003) Mol. Endocrinol. 17, 117-127. [DOI] [PubMed] [Google Scholar]
- 17.Pierreux, C. E., Nicolas, F. J. & Hill, C. S. (2000) Mol. Cell. Biol. 20, 9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, J. C., Derynck, M. K., Nonaka, D. F., Khodabakhsh, D. B., Haqq, C. & Yamamoto, K. R. (2004) Proc. Natl. Acad. Sci. USA 101, 15603-15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissen, R. M. & Yamamoto, K. R. (2000) Genes Dev. 14, 2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, Z., Frederick, J. & Garabedian, M. J. (2002) J. Biol. Chem. 277, 26573-26580. [DOI] [PubMed] [Google Scholar]
- 21.Louw, A., Swart, P. & Allie, F. (2000) Biochem. Pharmacol. 59, 167-175. [DOI] [PubMed] [Google Scholar]
- 22.Clark, D. E., Poteet-Smith, C. E., Smith, J. A. & Lannigan, D. A. (2001) EMBO J. 20, 3484-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkhem, T., Carlsson, B., Nilsson, Y., Enmark, E., Gustafsson, J. & Nilsson, S. (1998) Mol. Pharmacol. 54, 105-112. [DOI] [PubMed] [Google Scholar]
- 24.Caldenhoven, E., Liden, J., Wissink, S., Van de Stolpe, A., Raaijmakers, J., Koenderman, L., Okret, S., Gustafsson, J. A. & Van der Saag, P. T. (1995) Mol. Endocrinol. 9, 401-412. [DOI] [PubMed] [Google Scholar]
- 25.Herrlich, P. (2001) Oncogene 20, 2465-2475. [DOI] [PubMed] [Google Scholar]
- 26.Vayssière, B. M., Dupont, S., Choquart, A., Petit, F., Garcia, T., Marchandeau, C., Gronemeyer, H. & Rèsche-Rigon, M. (1997) Mol. Endocrinol. 11, 1245-1255. [DOI] [PubMed] [Google Scholar]
- 27.Vanden Berghe, W., Francesconi, E., De Bosscher, K., Rèsche-Rigon, M. & Haegeman, G. (1999) Mol. Pharmacol. 56, 797-806. [PubMed] [Google Scholar]
- 28.Belvisi, M. G., Brown, T. J., Wicks, S. & Foster, M. L. (2001) Pulm. Pharmacol. Ther. 14, 221-227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.