Abstract
Hybrid dysgenesis in Drosophila is a syndrome of gonadal atrophy, sterility, and male recombination, and it occurs in the progeny of crosses between males that harbor certain transposable elements (TEs) and females that lack them. Known examples of hybrid dysgenesis in Drosophila melanogaster result from mobilization of individual families of TEs, such as the P element, the I element, or hobo. An example of hybrid dysgenesis in Drosophila virilis is unique in that multiple, unrelated families of TEs become mobilized, but a TE designated Penelope appears to play a major role. In all known examples of hybrid dysgenesis, the paternal germ line transmits the TEs in an active state, whereas the female germ line maintains repression of the TEs. The mechanism of maternal maintenance of repression is not known. Recent evidence suggests that the molecular machinery of RNA interference may function as an important host defense against TEs. This protection is mediated by the action of endogenous small interfering RNAs (siRNAs) composed of dsRNA molecules of 21-25 nt that can target complementary transcripts for destruction. In this paper, we demonstrate that endogenous siRNA derived from the Penelope element is maternally loaded in embryos through the female germ line in D. virilis. We also present evidence that the maternal inheritance of these endogenous siRNAs may contribute to maternal repression of Penelope.
Keywords: hybrid dysgenesis, maternal effect, RNA interference, transposable element
In sexual organisms, genetic parasites, such as transposable elements (TEs), can proliferate in populations despite being deleterious to the host (1). Harmful effects include mutations in genes (2, 3), chromosomal abnormalities arising from ectopic recombination (4, 5), and costs associated with the production of transcripts that divert resources from the host (6). A prime example of the proliferative ability of TEs is the finding that half of the human genome is composed of these elements (7). Hybrid dysgenesis, a phenomenon in which mobilized TEs can induce sterility, represents an extreme example of the harm TEs can inflict as a result of their proliferation. In Drosophila melanogaster, when males carrying P, I,or hobo elements are mated with females lacking them, these elements become mobilized, resulting in sterility of the offspring (8-12). However, in the reciprocal cross between females carrying these elements and males lacking them, TE repression is maintained in the progeny. Thus, the salient pattern of TE-induced hybrid dysgenesis in Drosophila is that the maternal germ line maintains repression, whereas the male germ line does not. The mechanism by which maternal TE repression is epigenetically inherited is not well understood. However, it is clear that a genetic phenomenon known as the “homology effect” plays an important role. For P and I elements, TE repression can be maintained in a manner dependent on maternal copies of the particular TE, and maternal repression can be transmitted, even if the maternal copies are defective and unable to produce functional transcripts (13-15). Furthermore, TE repression can be epigenetically inherited maternally even if copies of the TE are not transmitted to the next generation (16).
Recent studies suggest a potential role for RNA interference (RNAi) in the maintenance of maternal repression of TEs. The RNAi machinery processes dsRNA into smaller 21- to 25-nt double-stranded fragments that mediate the destruction of homologous RNA molecules (17, 18). Mutations in genes necessary for RNAi often lead to increased TE activity, and it is now clear that the RNAi machinery functions as a defense mechanism to reduce the harmful effects of TEs and viruses in a wide variety of organisms, including nematodes (19, 20), fruit flies (21, 22), plants (23, 24), and Chlamydomonas (25). Endogenous silencing of TEs by the RNAi machinery likely depends on the action of 21- to 25-nt small interfering RNAs (siRNAs) derived from the same repeat sequences liable to silencing. These species have been designated repeat-associated siRNAs (26).
Given that repeat-associated siRNAs likely play a role in silencing endogenous TEs and that maternal repression can be mediated by TE copies unable to produce functional transcripts, maternal transmission of siRNA molecules may contribute to the maternal transmission of TE repression. Here we examine the hybrid dysgenesis syndrome in Drosophila virilis to determine patterns of TE-derived siRNA expression and test the hypothesis that they are maternally inherited.
Hybrid dysgenesis in D. virilis occurs in the progeny of a mating between two strains, known as strains 9 and 160. The hybrid dysgenesis is unique in that at least five unrelated families of TEs become mobilized (27, 28). When females of strain 9 are mated with males of strain 160, a large proportion of male and female progeny are sterile, but in the reciprocal cross the progeny are normal (29). The element Penelope, a member of a recently described class of retroelements that possess an intron (30), appears to play a primary role (31). It is abundant in strain 160, but aside from several ancient and highly degraded copies (ref. 32 and our unpublished data), is entirely absent from strain 9. Furthermore, when Penelope is injected into a strain lacking the element, mobilization of unrelated TEs has been reported (31). Aside from Penelope, at least four other unrelated elements are mobilized in the dysgenic cross: Paris, Helena, Telemac, and Ulysses. Paris is a DNA element in the mariner/Tc1 family, and Helena is a long interspersed nuclear element-like non-LTR retroelement. Each of these families is more abundant in strain 160 and may play some causal role in comobilization. Telemac, a BEL-like LTR-retrotransposon, and Ulysses, a gypsy-like LTR retroelement, are evenly distributed between the strains and, thus, are unlikely to play a role (27, 28). The mechanisms of maternal repression and comobilization are not known. Here we examine the pattern of TE-derived repeat-associated siRNA in the hybrid dysgenic cross. We show that Penelope siRNA is produced in both sexes but only maternally loaded in embryos. This finding is consistent with a role for maternal transmission of Penelope siRNA in the repression of transposition.
Methods
Fly Stocks. Strain 9 is a wild-type strain that was collected in Batumi (Republic of Georgia) in 1970. Strain 160 is a long-established laboratory culture that was generated by combining the recessive gl (glossy eye) marker in chromosome VI from strain 104 (collected by W. Stone in 1967) with strain 149 (obtained from Japan in 1968) that carried the following recessive markers: b (broken crossveins) (chromosome II), tb (tiny bristles), and gp-L2 (gap in longitudinal wing vein 2) (chromosome III), cd (cardinal eye color) (chromosome IV), and pe (peach eye color) (chromosome V). A C(1)RM, w stock that was kindly provided by the Thomas Cline laboratory (University of California, Berkeley) was used to generate three more strains. Strain XX̂/160 was generated by placing the C(1)w into the strain 160 background through 15 generations of backcrossing to strain 160. Strain XX̂/9 was generated by placing the C(1)w stock into the strain 9 background through 12 generations of back-crossing to strain 9. A third strain, X9, was generated by crossing males from strain 9 with XX̂/160 females; male offspring of this cross were backcrossed twice to the XX̂/160 strain, and male progeny that possessed all recessive markers from strain 160 chromosomes (indicating homozygosity of the nearly pure 160 autosomes) were collected. These males were then mated with XX̂/160 females to produce the true-breeding strain X9. Males of the X9 strain possess the X chromosome from strain 9 on a nearly pure 160 background.
Detection of siRNA. Detection of siRNA was performed by Northern hybridization as described by Hamilton and Baulcombe (23), with some modifications. Total RNA without enrichment of small RNAs was extracted by using TRIzol (Invitrogen) according to the manufacturer's directions, and 50 μg of total RNA was loaded onto a 15% polyacrylamide gel. Where noted, enrichment was performed by using the mirVana kit (Ambion, Austin, TX), and 5 μg was loaded. After electrophoresis, the gels were sliced. The region of each gel containing 20-50 nt RNA was blotted, and the upper portion was stained in ethidium bromide and photographed for detection of 5S RNA as a loading control. To ensure that RNA transcripts used as probes were either only sense or antisense, oligonucleotides corresponding to sense and antisense polarity were run as size standards. RNA from adults was collected from 5- to 10-day-old flies maintained at 25°C. RNA from embryos was collected by gathering embryos from grape plates and freezing them immediately in liquid nitrogen.
The hallmark signature of siRNAs is that they are 21-25 nt in length and that they include both sense and antisense sequences. The terminal repeats of Penelope have been found in inverted orientation within the genome, and full-length transcripts of these particular copies will contain regions of RNA sequence corresponding to both polarities. Therefore, an RNA probe designed to detect full-length sense transcripts will also detect antisense transcripts. For this reason, probes were generated by in vitro transcription of cloned products corresponding only to the unique sequence of the Penelope ORF. A portion of the ORF from Penelope was PCR-amplified from strain 160 and cloned by using the TOPO TA cloning kit (Invitrogen). Sense and antisense RNA probes were generated by in vitro transcription with the MaxiScript kit (Ambion) by using linearized plasmids in which the ORF was in sense or antisense orientation relative to the T7 transcription start site.
Crosses and Analysis of Gonadal Atrophy. All crosses were performed at 25°C. Crosses for the purpose of RNA collection were performed en masse. Crosses to measure the maternal effect on gonadal atrophy were carried out with single males and females. The single pairs were transferred every 1-2 days, and all F1 males from at least two vials per cross were counted. Gonadal atrophy in males can be determined by examination of testes. Atrophied testes assume a distinctly reduced morphology. In the 15 years since the original observation of hybrid dysgenesis between strain 9 and strain 160, the degree of sterility found in dysgenic F1 males and females has decreased somewhat. We confirmed by means of PCR that strain 9 had not been contaminated by Penelope copies from strain 160. Statistical analysis of gonadal atrophy was performed with sas in a logistic regression framework using the genmod procedure in which frequency data for gonadal atrophy was fit according to the model log(πi/1 - π) = μ + τi + βj..., where π equals the probability of atrophy for each testis (in this case, we count each of the two testes separately), μ is equal to the parameter representing the probability with no treatment, and τi and βj, etc., represent each treatment, which may be, for example, maternal cytoplasm from strain 9 or strain 160 or each of the chromosomes from strain 160 being present in the mother. The parameter for each treatment and its 95% confidence interval were then determined. This parameter estimate represents the estimated probability of atrophy for each testis under that particular treatment.
Results
TE siRNA Expression in Adults and Embryos. If lack of endogenous maternally inherited siRNA is associated with TE mobilization, the siRNA should be enriched in strain 160 relative to strain 9. Using Northern hybridization with probes for Paris, Helena, and Ulysses, we found no evidence that siRNA derived from these elements was enriched in strain 160 relative to strain 9 (data not shown). This result is subject to the limit of detectability of siRNA under our conditions, and we cannot rule out effects of siRNA due to these elements, especially if siRNA-mediated repression is not stoicheometric at low levels of siRNA.
In contrast, analysis with a probe covering the region of Penelope diagrammed in Fig. 1A yielded a clear distinction between strains 9 and 160. In extracts from whole adults, dsRNA molecules 21-25 nt were found in both sexes of strain 160 but not in strain 9 (Fig. 1B). This observation correlates with the presence of abundant functional copies of Penelope in strain 160 but only degraded copies in strain 9. We also examined Penelope siRNA expression in adult progeny of both the dysgenic (Fig. 1B, lane Dys) and nondysgenic (Fig. 1B, lane Ndys) cross. In the nondysgenic cross, adults of both sexes exhibited presumptive Penelope siRNA, whereas in the dysgenic cross, this species of RNA was absent from adult males but present in adult females. Evidence presented in the next section indicates that the presumptive siRNA observed in the adult females of the dysgenic cross derives from the X chromosome transmitted by the strain 160 males in the cross.
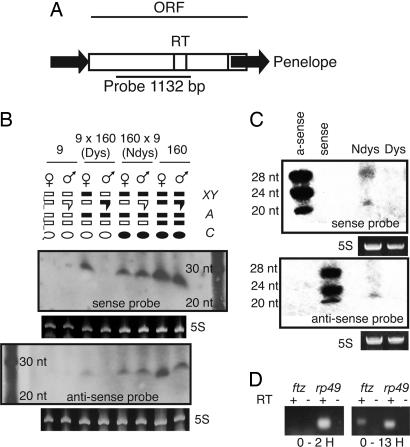
Fig. 1.
Putative Penelope siRNA is present in whole adults of strain 160, absent from strain 9, and maternally inherited in the nondysgenic cross. (A) Sequence organization of Penelope and location of the probe for siRNA. Black arrows indicate terminal repeats. RT, reverse transcriptase domain. (B) Northern analysis with 50 μg of total RNA from adults of strain 9 and 160, as well as dysgenic and nondysgenic progeny and sense and antisense orientations of the Penelope probe. 5S RNA is shown as a loading control, with a 10-bp RNA ladder shown on the side. Above the blots, we designate chromosomes as bars, with chromosomes from strain 160 in black and chromosomes from strain 9 in white. The pair of sex chromosomes, with the Y chromosome designated with a hooked bar, is shown above, and a representative pair of autosomes is shown below. Maternal cytoplasm is designated with an oval, with white and black indicating the presence or absence, respectively, of strain 160 chromosomes in the mother. Sense and antisense Penelope RNA between 20 and 30 nt is absent from strain 9 and present in strain 160. In the progeny, putative siRNA is present in males and females of the dysgenic (Dys) cross but only present in females of the nondysgenic (Ndys) crosses. (C) Northern analysis of 5 μg of enriched RNA from 0- to 2-h, nondysgenic and dysgenic embryos with 5S loading control. DNA oligos of 20, 24, and 28 nt with antisense and sense orientation to the Penelope probe were used as a polarity control. Small sense and antisense Penelope RNA is detectable in early embryos of the nondysgenic cross but not in those of the dysgenic cross. (D) RT-PCR for ftz expression in 0- to 2-h and 0- to 13-h nondysgenic embryos. Reverse transcriptase-negative controls (-) and rp49-positive controls (+) are shown. Lack of ftz expression in 0- to 2-h embryos indicates that genome-wide transcription has not yet been initiated in the nondysgenic embryos from C.
We also examined the pattern of siRNA presence in early embryos to test whether maternal loading from females correlates with the maintenance of TE repression (Fig. 1C). In D. melanogaster, genome-wide zygotic transcription begins at ≈2 h at 25°C (33). Therefore, gene expression in embryos younger than 2 h should primarily represent the RNA component of the embryo that is maternally loaded into the oocyte. Consistent with siRNA-mediated repression, we observed the presence of 21-25 nt dsRNA corresponding to Penelope in 0- to 2-h nondysgenic embryos but not in 0- to 2-h dysgenic embryos in which Penelope becomes mobilized.
Although genome-wide zygotic transcription begins at ≈2 h in D. melanogaster, this may not be true in D. virilis. If genome-wide zygotic transcription is initiated earlier in D. virilis, it is possible that the source of siRNA observed in nondysgenic embryos could result from transcribed Penelope elements present in the zygote. To rule out this possibility, we performed RT-PCR on the pair-rule gene fushi-tarazu (ftz) in nondysgenic embryos at 0-2 h as well as 0-13 h, using the maternally loaded ribosomal protein transcript rp49 as a control (Fig. 1D). The ftz gene is one of the earliest zygotic genes transcribed in embryos. The results confirm that ftz expression is absent in 0- to 2-h nondysgenic embryos, whereas rp49 RNA is present. This result indicates that the source of Penelope siRNA in the early embryo is likely of maternal, rather than zygotic, origin.
Penelope siRNA in Adult Dysgenic Females Is Due Primarily to the 160 X Chromosome. Dysgenic female adults exhibit Penelope siRNA expression, whereas males do not (Fig. 1B). This difference may reflect intrinsic differences in the biology of the sexes (for example, a sex-specific ability of females to mount an siRNA response), or it may result from the presence of the X chromosome from strain 160 in dysgenic females but not dysgenic males. To distinguish between these hypotheses, we examined strains with genotypes resembling progeny of the dysgenic and nondsysgenic crosses but with differences in the origin of the X chromosome. In Fig. 2A (Fig. 5, which is published as supporting information on the PNAS web site, provides a detailed diagram of the crosses carried out to generate the genotypes shown), the lanes labeled XX̂ are progeny of the cross XX̂/9 females with strain 160 males. The symbol XX̂ /9 denotes a strain carrying an attached-X chromosome [C(1)RM, w] with the autosomes from strain 9. This cross is reminiscent of the dysgenic cross in that females with strain 9 autosomes are mated with males of strain 160. However, because the attached-X chromosomes are inherited together, the sons inherit their X chromosome from their father instead of their mother. If Penelope siRNA expression is driven from the X chromosome from strain 160, then adult sons of this cross would exhibit Penelope siRNA expression and adult daughters would not. This result is observed for the XX̂ /9 female × 160 male cross (Fig. 2 A). If females possessed a sex-specific ability to mount an siRNA response, then the XX̂ result in Fig. 2 A would have been reversed.
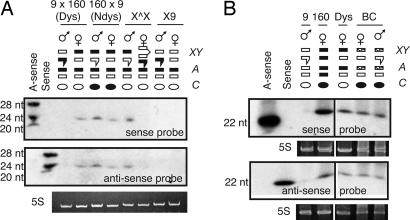
Fig. 2.
Putative siRNA is driven from the X chromosome of strain 160. (A) Northern analysis of 5 μg of enriched RNA per lane (with 5S loading control) shown for dysgenic (Dys) and nondysgenic (Ndys) adults and for adult progeny of the XX̂ and X9 crosses. Membrane was stripped and reprobed. Expression of Penelope siRNA in adults correlates with the presence of the X chromosome from strain 160. (B) Northern analysis of total RNA from the progeny of a backcross (dysgenic females × 9 males, lane labeled BC) containing a random mixture of recombinant X chromosomes (shown with stipled bars). Penelope siRNA is found in males and females. Cytoplasm in backcross adults is designated black, indicating the presence of 160 chromosomes in the mother.
The lanes labeled X9 in Fig. 2 A refer to the progeny of a cross in which females of strain 9 were mated with males of a strain designated X9 that carry the X chromosome of strain 9 but the autosomes of strain 160. In this case, neither daughters nor sons of the cross possess the X chromosome from strain 160. Fig. 2 A shows that neither adult daughters nor adult sons of the X9 cross express Penelope siRNA, which is also consistent with the X chromosome from strain 160 mediating the expression of siRNA in the adult females of the dysgenic cross.
Finally, Fig. 2 shows the pattern of siRNA expression in adult progeny of a backcross (Fig. 2B, lane BC) of semifertile dysgenic females with strain 9 males. In this case, the X chromosomes should include random recombinants of the strain 160 X and the strain 9 X chromosomes, and, accordingly, progeny of both sexes should exhibit Penelope siRNA. As before, these results are consistent with a model in which Penelope siRNA in adult flies is driven from one or more loci present in the X chromosome of strain 160.
The Strain 160 X Chromosome Is Necessary for Maternal Transmission of Penelope siRNA. Becausee the primary source of Penelope siRNA in whole adults is the 160 X chromosome, we determined whether this expression is limited to either the germ line or the soma. Fig. 3 (see Fig. 6, which is published as supporting information on the PNAS web site, for crossing schemes) shows that Penelope siRNA expression in 0- to 2-h embryos depends on the presence of the 160 X chromosome in the mother. Dysgenic embryos (Fig. 3, lane Dys) or embryos whose mother lacks the 160 X chromosome (Fig. 3, lane A) do not exhibit Penelope siRNA, but embryos whose mother does carry the 160 X chromosome (Fig. 3, lane B) do show this siRNA. Furthermore, adult females from strain 9 or those lacking the 160 X chromosome (Fig. 3, lane C) do not express the Penelope siRNA in ovaries or soma, whereas adult females possessing the 160 X chromosome (Fig. 3, lane D) exhibit this siRNA in ovaries and soma.
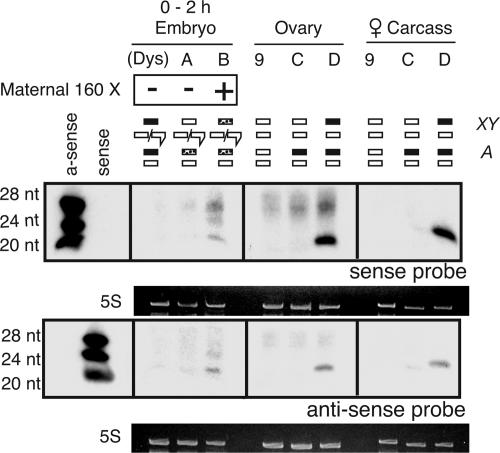
Fig. 3.
The X chromosome is the source of Penelope siRNA within the female germ line. In 0- to 2-h embryos, Penelope siRNA is found only if mothers possess the 160 X chromosome. In females, the X chromosome drives the expression of Penelope siRNA in the germ line and soma. Females that lack this chromosome do not demonstrate Penelope siRNA in either tissue. Five micrograms of enriched RNA was used per lane.
Confirmation That Repression of Dysgenesis in Males Results from a Maternal Effect. Because dysgenic and nondysgenic females are genetically identical and differ only in whether strain 9 or strain 160 was the mother, the repression of hybrid dysgenesis in females is due to a maternal effect. However, it is a formal possibility that the mechanism of TE mobilization in the germ line of males is different from that in females. Nondysgenic sons possess the X chromosome from strain 160, whereas dysgenic sons lack this chromosome. It could be that maintenance of repression in the male germ line is zygotic and due to the presence of the X chromosome from strain 160. Specifically, because the X chromosome drives Penelope siRNA expression in adults, it may also drive repressive siRNA expression in the early male germ line. If this possibility were the case, then mobilization of TEs in dysgenic males may result from absence of the “protective” X chromosome from strain 160 rather than absence of maternally transmitted siRNA.
To determine whether TE mobilization in the male germ line is due to the absence of maternal repression, we compared the level of dysgenesis in genetically identical males, all possessing the 160 X chromosome but differing in the genotype of the mother (Fig. 4; see Fig. 7, which is published as supporting information on the PNAS web site, for crosses). Nondysgenic males from the 160 female × 9 male cross and male progeny from the XX̂ /9 female × 160 male cross are genetically identical. In particular, both types of males possess an X chromosome from strain 160. However, maternal inheritance differs between these males, because the mothers in the nondysgenic cross 160 female × 9 male are from strain 160, whereas mothers of the XX̂ /9 female × 160 male cross possess an attached-X chromosome in a strain 9 background. For both types of males, the degree of sterility was determined by counting atrophied testes as shown in Fig. 4A, and the probability of gonadal atrophy was determined. Nondysgenic males showed nearly complete repression of testicular atrophy, whereas the probability of atrophy for each of the testes of identical males whose mothers lack chromosomes from strain 160 is nearly 40% (Fig. 4B). This result is similar to currently observed levels of gonadal atrophy in the dysgenic cross (see below). Thus, hybrid dysgenesis in males correlates with the absence of a maternal factor rather than the lack of the X chromosome from strain 160 in the zygote.
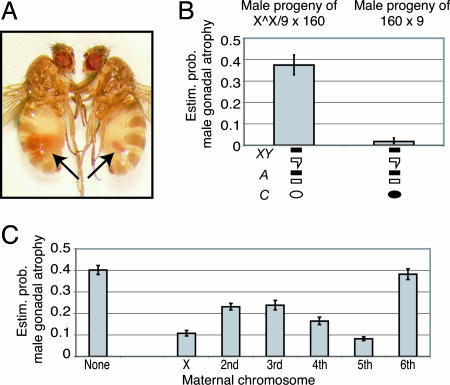
Fig. 4.
The contribution of maternal 160 chromosomes to the repression of gonadal atrophy. (A) Wild-type testis (left) and atrophied testis (right). (B) Genetically identical progeny of crosses with or without an attached-X chromosome (XX̂) provide evidence that hybrid dysgenesis in males results from lack of maternally transmitted repression. Shown is the estimated probability of gonadal atrophy in genetically identical males that differ only in respect to the cytoplasm of the mother. Males that possess the X chromosome from strain 160 but whose mothers lack chromosomes from strain 160 show high levels of gonadal atrophy (n = 211). Males whose mothers possess 160 chromosomes do not (n = 228). (C) Maternal effects of individual strain 160 chromosomes on suppression of gonadal dysgenesis; the lower the bar, the greater the level of repression. The largest effects are due to the X chromosome and chromosome V.
The 160 X Chromosome Is Sufficient But Not Necessary for Suppression of Dysgenesis. Because the X chromosome from strain 160 drives the expression of Penelope siRNA in whole adults, we determined whether this chromosome is sufficient by itself and when placed into a strain 9 genetic background to maternally suppress hybrid dysgenesis. Chromosomes from strain 160 were made heterozygous in the genetic background of strain 9, either alone or in combination with other chromosomes from strain 160, by means of two generations of crossing with strain 9 (see Fig. 8, which is published as supporting information on the PNAS web site, for the crossing schemes and sample sizes). Females of each genotype were then singly mated with males of strain 160 to determine the degree to which each female genotype could maternally transmit factors suppressing dysgenesis. The mother's genotype was assessed post hoc by examining the progeny for the presence of recessive markers on all five autosomes.
Fig. 4C shows that a maternal X chromosome from strain 160 contributes substantially to suppression of sterility in the male offspring. The estimated probability of gonadal atrophy is reduced from 40% to 11% when a maternal X chromosome alone is provided. The second, third, and fourth chromosomes exhibit a weaker maternal effect on sterility, although the fifth chromosome has a similar effect on suppression as the X chromosome (estimated 8% gonadal atrophy). Interestingly, the tiny sixth chromosome, which is homologous to the fourth chromosome of D. melanogaster, has a negligible effect on the suppression of dysgenesis. This chromosome in strain 160 lacks copies of Penelope, whereas all other chromosomes from strain 160 possess copies of Penelope (28).
The data in Fig. 4C also indicate that, although the X chromosome and autosomes 2-5 all contribute to maternal repression, no individual chromosome is sufficient to maximally repress hybrid dysgenesis. Rather, the chromosomes act cumulatively (Fig. 8), and, when the mother's genotype includes all six chromosomes from strain 160, complete repression is observed (proportion of atrophied testes, 0 of 248 males). Indeed, strong repression is observed when the mother's genotype includes chromosomes 2-6 from strain 160 but the X chromosome from strain 9 (proportion of atrophied testes, 1% of 368 males). These data imply that the X chromosome from strain 160, as a source of Penelope siRNA, has a strong repressing effect, but that other chromosomes from strain 160, which also carry Penelope, also are capable of mediating repression.
Discussion
We have demonstrated that small (21-25 nt) sense and antisense RNA species homologous to Penelope can be identified in strain 160, and, in females, these species are found both within the germ line and soma. We estimate the size of the small RNA molecules to be ≈23 nt, which is similar to the size found for most TE-derived siRNAs found in D. melanogaster (26). These species of endogenous RNA derive from a locus (or loci) on the X chromosome and are transmitted to offspring through the female, but not male, germ line. Additionally, the X chromosome provides substantial repression of dysgenesis in the offspring when present in the mother. Thus, Penelope siRNA complexes may function as an important maternal factor that mediates repression in nondysgenic embryos.
The X-linked locus (or loci) that functions as the source of siRNA molecules in the female germ line is not known, but it presumably generates double-stranded Penelope species that are processed by the RNAi machinery. We have shown definitively that this presumptive Penelope siRNA is maternally, but not paternally, transmitted. We have no direct evidence that these Penelope siRNAs account for the strong suppression of dysgenesis observed for the X chromosome, although the circumstantial evidence supports this hypothesis.
The mechanism or mechanisms by which the other chromosomes mediate their maternal effect is not known. All chromosomes that possess Penelope elements contribute cumulatively to maternal repression (Figs. 4 and 8). There are a number of possibilities. The autosomes may contribute siRNA molecules at a level below the limit of detection. By way of precedent, RNAi silencing has been observed in Caenorhabditis elegans in the absence of detectable levels of siRNA (34). Alternatively, sequence divergence between autosomal copies of Penelope in strain 160 may preclude their detection by our probe, or they may originate primarily from regions of Penelope not included in the probe. Independently of any siRNA effects, maternal repression of Penelope may be mediated by means that are reminiscent of the “poison subunit” effect on P element transposition caused by certain internally deleted elements designated KP (35, 36). Finally, the hybrid dysgenesis system in D. virilis is unique in that multiple TEs become mobilized, and, although Penelope appears to be the primary cause of the comobilization, the other TEs may also contribute (28). The failure of the 160 X chromosome to completely suppress dysgenesis and the cumulative effects of the autosomes may simply reflect the complex nature of TE repression and mobilization.
Several aspects of the dysgenic syndrome observed here in D. virilis differ from those for the P element observed in D. melanogaster. First, it appears that repression is maintained when a sufficient number of chromosomes from strain 160 are present in the mother, even if these chromosomes were inherited paternally. Our crossing scheme used to measure the maternal effect of different chromosomes on suppression (Fig. 8) entailed backcrossing hybrid males to strain 9 females. Despite the backcross to strain 9, females that are heterozygous for all chromosomes from strains 9 and 160 still transmit repression to their offspring. This effect is not observed with P elements in D. melanogaster. In this system, when a mother is mated with a male possessing P elements, her ability to transmit repression to her progeny depends, in part, on the genotype of her own mother (8, 16), and the maternal transmission of repression depends on the continued presence of P copies in the genome of the mother (37). Over time, when transmitted maternally, the level of repression accumulates. The delay in the accumulation of maternal repression is also observed in the case of the I element when repression is mediated by a transgene (13). Here, by contrast, females are capable of transmitting repression as long as they possess a sufficient number of chromosomes from strain 160, even if their mothers were originally from strain 9. Nonetheless, although these results show that a grandmother contribution is not essential to the repression of dysgenesis, it can certainly modulate it (28).
A second aspect that differs between hybrid dysgenesis in D. melanogaster and that in D. virilis is the comobilization of unrelated TEs. When first observed, it was difficult to imagine how multiple unrelated elements (class I LTR and non-LTR elements as well as class II DNA elements) could be mobilized by similar mechanisms (27). However, it is becoming increasingly clear that the RNAi machinery regulates a diverse array of genetic parasites. For example, in nematodes, a DNA transposon is regulated by the RNAi machinery (20), whereas, in flies, a likely component of the RNAi machinery, spn-E, plays a role in repressing LTR and non-LTR elements in ovaries and testes (21). In an analysis of the small RNA profile during D. melanogaster development, small species of RNA derived from LTR and non-LTR elements as well as DNA elements were all found (26). The cause of comobilization in D. virilis is not known, but it appears that Penelope plays a primary role. Many viruses have evolved the ability to antagonize the RNAi machinery (38-40). Furthermore, it has been demonstrated in plants that these suppressors can cause developmental defects by interfering with the proper function of microRNAs important in development (41, 42). It is therefore possible that Penelope possesses factors that suppress RNA silencing, and the comobilization of other elements may be a pleiotropic consequence of a compromised RNAi machinery. It is tempting to speculate that the sterility is also a pleiotropic consequence of a compromised RNAi machinery, especially in light of evidence that components of the RNAi machinery, such as piwi (43, 44), also play a role in germ-cell maintenance
Although the D. virilis syndrome of hybrid dysgenesis differs in several aspects from the P and I element system, the most significant similarity is that, in all three cases, the female germ line maintains TEs in a repressed state, whereas the male germ line does not. In D. melanogaster, this observation also extends to lacZ reporter constructs that only undergo transsilencing in the germ line when inherited maternally (45). From an evolutionary perspective, it may be that, because females invest substantial resources in the production of oocytes, natural selection favors additional investment in maintaining genetic parasites in a silent state. Interestingly, microarray experimental data (46) imply that, among the core components of the RNAi machinery, seven of nine differentially expressed components are up-regulated in ovaries relative to testes (Fig. 9, which is published as supporting information on the PNAS web site). Furthermore, if silencing is mediated by protein-RNA complexes, males that produce and transmit these complexes may be at a disadvantage to the extent that success in fertilization requires the production of large numbers of energetically inexpensive sperm (47).
Supplementary Material
Acknowledgments
We thank Elena Lozovsky, Yun Tao, James Birchler, Lena Hileman, Josh Chang Mell, and Jorge Vieira for discussions and advice. We also thank Lisa Megna and other members of the Thomas Cline laboratory for providing the attached-X stock. This work was supported by National Institutes of Health Grants GM068465 and GM60035.
Author contributions: J.P.B. and D.L.H. designed research; J.P.B. performed research; J.P.B. analyzed data; and J.P.B. and D.L.H. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: RNAi, RNA interference; siRNA, small interfering RNA; TE, transposable element.
References
- 1.Hickey, D. A. (1982) Genetics 101, 519-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green, M. M. (1988) in Banbury Report, eds. Lambert, M. E., McDonald, J. F. & Weinstein, I. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 41-50.
- 3.Charlesworth, B. & Langley, C. H. (1989) Annu. Rev. Genet. 23, 251-287. [DOI] [PubMed] [Google Scholar]
- 4.Montgomery, E., Charlesworth, B. & Langley, C. H. (1987) Genet. Res. 49, 31-41. [DOI] [PubMed] [Google Scholar]
- 5.Langley, C. H., Montgomery, E., Hudson, R., Kaplan, N. & Charlesworth, B. (1988) Genet. Res. 52, 223-235. [DOI] [PubMed] [Google Scholar]
- 6.Nuzhdin, S. V., Pasyukova, E. G. & Mackay, T. F. C. (1996) Proc. R. Soc. London B 263, 823-831. [DOI] [PubMed] [Google Scholar]
- 7.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 8.Engels, W. R. (1979) Genet. Res. 33, 219-236. [DOI] [PubMed] [Google Scholar]
- 9.Kidwell, M. G. (1981) Genetics 98, 275-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bingham, P. M., Kidwell, M. G. & Rubin, G. M. (1982) Cell 29, 995-1004. [DOI] [PubMed] [Google Scholar]
- 11.Bucheton, A., Paro, R., Sang, H. M., Pelisson, A. & Finnegan, D. J. (1984) Cell 38, 153-163. [DOI] [PubMed] [Google Scholar]
- 12.Yannopoulos, G., Stamatis, N., Monastirioti, M., Hatzopoulos, P. & Louis, C. (1987) Cell 49, 487-495. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, S., Gassama, M. P. & Heidmann, T. (1999) Nat. Genet. 21, 209-212. [DOI] [PubMed] [Google Scholar]
- 14.Marin, L., Lehmann, M., Nouaud, D., Izaabel, H., Anxolabehere, D. & Ronsseray, S. (2000) Genetics 155, 1841-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart, J. R., Haley, K. J., Swedzinski, D., Lockner, S., Kocian, P. E., Merriman, P. J. & Simmons, M. J. (2002) Genetics 162, 1641-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronsseray, S., Lemaitre, B. & Coen, D. (1993) Mol. Gen. Genet. 241, 115-123. [DOI] [PubMed] [Google Scholar]
- 17.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir, S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99, 123-132. [DOI] [PubMed] [Google Scholar]
- 20.Sijen, T. & Plasterk, R. H. A. (2003) Nature 426, 310-314. [DOI] [PubMed] [Google Scholar]
- 21.Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M. & Gvozdev, V. A. (2001) Curr. Biol. 11, 1017-1027. [DOI] [PubMed] [Google Scholar]
- 22.Reiss, D., Josse, T., Anxolabehere, D. & Ronsseray, S. (2004) Mol. Genet. Genomics 272, 336-343. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 24.Zilberman, D., Cao, X. F. & Jacobsen, S. E. (2003) Science 299, 716-719. [DOI] [PubMed] [Google Scholar]
- 25.Wu-Scharf, D., Jeong, B. R., Zhang, C. M. & Cerutti, H. (2000) Science 290, 1159-1162. [DOI] [PubMed] [Google Scholar]
- 26.Aravin, A. A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J. & Tuschl, T. (2003) Dev. Cell 5, 337-350. [DOI] [PubMed] [Google Scholar]
- 27.Petrov, D. A., Schutzman, J. L., Hartl, D. L. & Lozovskaya, E. R. (1995) Proc. Natl. Acad. Sci. USA 92, 8050-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vieira, J., Vieira, C. P., Hartl, D. L. & Lozovskaya, E. R. (1998) Genet. Res. 71, 109-117. [DOI] [PubMed] [Google Scholar]
- 29.Lozovskaya, E. R., Scheinker, V. S. & Evgenev, M. B. (1990) Genetics 126, 619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arkhipova, I. R., Pyatkov, K. I., Meselson, M. & Evgen'ev, M. B. (2003) Nat. Genet. 33, 123-124. [DOI] [PubMed] [Google Scholar]
- 31.Evgenev, M. B., Zelentsova, H., Shostak, N., Kozitsina, M., Barskyi, V., Lankenau, D. H. & Corces, V. G. (1997) Proc. Natl. Acad. Sci. USA 94, 196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyozin, G. T., Makarova, K. S., Velikodvorskaja, V. V., Zelentsova, H. S., Khechumian, R. R., Kidwell, M. G., Koonin, E. V. & Evgen'ev, M. B. (2001) J. Mol. Evol. 52, 445-456. [DOI] [PubMed] [Google Scholar]
- 33.Foe, V. E., Odell, G. M. & Edgar, B. A. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Martineze Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 1, pp. 149-300. [Google Scholar]
- 34.Grishok, A., Sinskey, J. L. & Sharp, P. A. (2005) Genes Dev. 19, 683-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black, D. M., Jackson, M. S., Kidwell, M. G. & Dover, G. A. (1987) EMBO J. 6, 4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrews, J. D. & Gloor, G. B. (1995) Genetics 141, 587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sved, J. A. (1987) Genetics 115, 121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llave, C., Kasschau, K. D. & Carrington, J. C. (2000) Proc. Natl. Acad. Sci. USA 97, 13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vance, V. & Vaucheret, H. (2001) Science 292, 2277-2280. [DOI] [PubMed] [Google Scholar]
- 40.Li, H. W., Li, W. X. & Ding, S. W. (2002) Science 296, 1319-1321. [DOI] [PubMed] [Google Scholar]
- 41.Kasschau, K. D., Xie, Z. X., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A. & Carrington, J. C. (2003) Dev. Cell 4, 205-217. [DOI] [PubMed] [Google Scholar]
- 42.Chapman, E. J., Prokhnevsky, A. I., Gopinath, K., Dolja, V. V. & Carrington, J. C. (2004) Genes Dev. 18, 1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D. & Lin, H. F. (1998) Genes Dev. 12, 3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox, D. N., Chao, A. & Lin, H. F. (2000) Development (Cambridge, U.K.) 127, 503-514. [DOI] [PubMed] [Google Scholar]
- 45.Ronsseray, S., Boivin, A. & Anxolabehere, D. (2001) Genetics 159, 1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parisi, M., Nuttall, R., Naiman, D., Bouffard, G., Malley, J., Andrews, J., Eastman, S. & Oliver, B. (2003) Science 299, 697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, G. A. (1982) J. Theor. Biol. 96, 281-294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.