Abstract
The characterization of Mycobacterium tuberculosis antigens inducing CD4+ T-cell responses could critically contribute to the development of subunit vaccines for M. tuberculosis. Here we performed computational analysis by using T-cell epitope prediction software (known as TEPITOPE) to predict promiscuous HLA-DR ligands in the products of the mce genes of M. tuberculosis. The analysis of the proliferative responses of CD4+ T cells from patients with pulmonary tuberculosis to selected peptides displaying promiscuous binding to HLA-DR in vitro led us to the identification of a peptide that induced proliferation of CD4+ cells from 50% of the tested subjects. This study demonstrates that a systematic computational approach can be used to identify T-cell epitopes in proteins expressed by an intracellular pathogen.
Mycobacterium tuberculosis survives phagocytosis and replicates within macrophages. Following infection with M. tuberculosis, both healthy subjects and patients with active tuberculosis develop T-cell responses against mycobacterial antigens. In mice CD4+ T cells (5, 22, 24, 32) and gamma interferon (IFN-γ) (8, 11), as well as IFN-γ receptor in humans (16, 23), were shown to be essential to the generation of a protective immune response to M. tuberculosis. Recently, depletion of CD4+ cells was shown to result in reactivation of latent M. tuberculosis infection in mice in spite of unaltered expression of IFN-γ, suggesting that CD4+ cells could have roles in addition to IFN-γ production in controlling M. tuberculosis infection (25). Therefore, the identification of antigens capable of eliciting CD4+ T-cell responses and the characterization of immunodominant T-cell epitopes are of primary importance for the development of subunit vaccines for tuberculosis. The role of CD8+ T cells in immunity to M. tuberculosis is less defined (10). CD8+ cytotoxic T cells capable of lysing infected macrophages as well as reducing the viability of intracellular mycobacteria through a granulysin-dependent mechanism were described (29). Moreover, epitopes from mycobacterial antigens capable of eliciting cytotoxic CD8+ T-cell responses were recently identified (17, 21, 6) as well as CD8+ T cells specific for mycobacterial antigens presented through an alternative major histocompatibility complex class I (MHC-I) processing pathway(s) (4, 19). MHC-II-restricted responses against M. tuberculosis have been more extensively characterized, and a number of M. tuberculosis antigens for CD4+ T cells have been identified to date. Recently, an immunodominant family of M. tuberculosis antigens recognized by T cells from healthy PPD (purified protein derivative)-reactive subjects was isolated (1). Expanding the knowledge of the human T-cell repertoire to peptide epitopes derived from M. tuberculosis antigens is also of potential interest for immunodiagnostic applications.
Activation of CD4+ T cells is dependent upon the presentation of peptides from disease-related protein antigens in the context of MHC-II molecules. The MHC genes are the most polymorphic present in the genome of every species analyzed. Most of the differences in the products of individual MHC-II alleles are localized in the peptide binding groove and determine the peptide binding properties of the different MHC molecules. In this study, the products of the mycobacterial cell entry (mce) genes were submitted to analysis by T-cell epitope prediction software (TEPITOPE), which enables the computational identification of promiscuous and allele-specific HLA-DR ligands (31). The mce1 gene was originally defined as an element conferring invasiveness to a nonpathogenic strain of Escherichia coli (2). Moreover, an mce-deficient Mycobacterium bovis BCG mutant exhibited reduced ability to invade nonphagocytic cells (9). The analysis of the complete genome of M. tuberculosis revealed the existence of four mce gene homologues very similarly organized in operons containing eight genes (7). Five peptides predicted by TEPITOPE as potential HLA-DR ligands and based on the sequence of the Mce2 protein were tested for induction of proliferation of CD4+ cells isolated from M. tuberculosis-infected subjects bearing 12 different aplotypes. This analysis led us to the identification of a peptide inducing CD4+ cell proliferation in 50% of the tested subjects, indicating that the application of the TEPITOPE software to mycobacterial antigens could lead to the identification of promiscuous epitopes eliciting MHC-II-restricted responses during infection with M. tuberculosis.
MATERIALS AND METHODS
Study subjects.
Patients were recruited at Consorzio Antitubercolare “Istituto Villa Marelli,” Milan, Italy, and Azienda Ospedaliera “S. Maria degli Angeli,” Pordenone, Italy, and were typed for HLA-DR at the Tissue Typing Laboratory of S. Raffaele Hospital, Milan, Italy. All sera employed in Western blotting were from tuberculin-positive and human immunodeficiency virus-seronegative subjects. Individuals defined as M. tuberculosis positive were a group of patients with postprimary pulmonary tuberculosis who were sputum positive by at least two of the following criteria: direct observation, cultural isolation, and PCR (Amplicor MTB test; Roche), whereas M. tuberculosis-negative subjects had recovered from postprimary pulmonary tuberculosis and were free of symptoms as well as sputum negative as determined by direct observation, cultural isolation, and PCR for at least 6 months. Peripheral blood mononuclear cells (PBMCs) for immortalization of autologous antigen-presenting cells (APCs) and analysis of CD4+ T-cell response to mce2 peptides were from tuberculin-positive, human immunodeficiency virus-negative subjects with postprimary pulmonary tuberculosis (n = 22 [15 males and 7 females]; average age, 30.2 years) who had been treated with antituberculous chemotherapy for at least 1 month. Four PPD-negative healthy donors were used as negative controls.
T-cell epitope prediction.
The sequences of the Mce proteins were subjected to HLA-DR ligand prediction by the TEPITOPE software to identify promiscuous HLA-DR ligands (13). We used a version of TEPITOPE incorporating 25 virtual matrices covering most of the human HLA class II peptide binding specificity in the Caucasian population. We set the TEPITOPE prediction threshold at 3% (31) and picked peptide sequences predicted to bind the most common HLA-DR alleles (DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, and DRB1*1501). The peptides were chosen based on the predicted binding as well as the degree of similarity between the four mce members and were synthesized according to the sequence of the Mce2 protein. Peptide no. 4, which is present in two proteins and showed poor prediction of promiscuous binding, was used as a nonpromiscuous control.
Cloning, expression, and immunoblot of Mce2-Myc fusion protein with sera from M. tuberculosis-infected subjects.
The mce2 gene was obtained from E. coli (NM554 strain) transformed with the cosmid MTCY19H5 (kindly provided by S. Cole) by PCR amplification with the following primers: forward 5′-CCATGGTGCCAACGCTGGTGACG and reverse 5′-AAGCTTTCATTAATTCAGATCCTCTTCTGAGATGAGTTTTTGTTCTGGGTTGATCGTGTTCTCTCC containing the sequence (underlined) encoding the c-myc-derived epitope. The mce-myc fragment was cloned in pCR2.1 (Invitrogen, Groningen, The Netherlands) that was used to transform Top 10 E. coli. Purified plasmid DNA was digested with HindIII and NcoI and cloned in pSE420 (Invitrogen) under the control of the trp-lac (trc) promoter. Isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM was used to induce the resulting transformants. The Mce2-Myc protein was selectively detected, by immunoblotting with an anti-Myc monoclonal antibody (MAb) in the subcellular fraction containing membranes and inclusion bodies of IPTG-induced transformants, as a band migrating with an apparent molecular mass of 45 kDa (not shown). To immunoprecipitate the Mce2-Myc protein, IPTG-induced bacteria were lysed in 1% sodium dodecyl sulfate (SDS) at 95°C for 20 min. After centrifugation at 20,800 × g for 15 min, the supernatant was diluted to 1:10 with 1% NP-40 in Tris-EDTA buffer and was immunoprecipitated with 9E10 anti-Myc MAb (CRL-1279; American Type Culture Collection) followed by Sepharose-coupled protein G. The immunoprecipitates were resolved in SDS-12% polyacrylamide gel electrophoresis under reducing conditions and were blotted onto nitrocellulose membranes. Immunoblots were performed with sera from M. tuberculosis-infected patients that were revealed by horseradish peroxidase-conjugated antibodies specific for human immunoglobulin G (IgG) and anti-Myc MAb as a control that was revealed by horseradish peroxidase-conjugated antibodies specific for mouse IgG. The enhanced chemiluminescence detection system (Amersham) was used to reveal the binding. Positive sera were reactive with the recombinant Mce2-Myc protein in Western blotting at 1:100 and 1:1,000 dilutions and were negative on anti-Myc-immunoprecipitated lysates from noninduced bacteria at 1:100 dilution.
DR peptide binding assay.
The Mce2 peptides together with a control peptide derived from the sequence of Epithelial V-like antigen (12) were synthesized with a multiple peptide synthesizer (model 396; Advanced Chem Tech, Louisville, Ky.) using fluorenylmethoxycarbonyl chemistry and solid-phase synthesis. The ability of unlabeled Mce2-derived peptides to compete with a biotinylated indicator peptide was determined using an enzyme-linked immunosorbent assay (ELISA)-based competition assay, as described earlier (13). The following biotinylated indicator peptides were used: GFKA7 for DRB1*0101 (DR1) and DRB1*0701 (DR7), GIRA2YA4 for DRB1*1501 (DR2), IAYDA5 for DRB1*0301 (DR3), YPKFVKQNTLKA2 for DRB1*0401 (DR4), tetanus toxoid peptide 830-843 for DRB1*1101 (DR5), and GYRA6L for DRB1*0801 (DR8). The relative binding affinity of the predicted ligands for the different HLA-DR molecules was determined as the 50% inhibitory concentration (IC50) (i.e., the concentration of competitor peptide required to inhibit binding of the biotinylated indicator peptide by 50%) and was compared with the promiscuous HA307-319 peptide from influenza hemagglutinin.
Immortalization of autologous APCs and analysis of CD4+ T-cell response to Mce2 peptides.
To immortalize autologous APCs, PBMCs isolated on a Ficoll density gradient were resuspended in RPMI 1640 medium (Gibco-BRL, Gaithersburg, Md.) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 50 U of penicillin-streptomycin/ml, 50 nM β-mercaptoethanol, 10% heat-inactivated fetal calf serum, 30% culture supernatant of the Epstein-Barr virus (EBV)-producing cell line B95.8 (generous gift of Paola Panina-Bordignon), and cyclosporine A (Novartis Pharma, Basel, Switzerland) at 300 to 600 ng/ml. After 3 days of culture, the supernatant was replaced with normal medium and cells were cultivated for 2 weeks. After an appropriate period of culture, cells were frozen. To analyze CD4+ T-cell responses to Mce2 peptides, PBMCs were resuspended at 106 cells/ml in complete RPMI 1640 medium supplemented with 10% human AB+ serum and 10 μg of whole-cell lysate of M. tuberculosis H37Rv (kindly provided by John T. Belisle, Colorado State University, Fort Collins, Colo.)/ml and cultivated for 3 days. Stimulated cells were expanded in medium with interleukin 2 (20 U/ml) to propagate T cells for stimulation with EBV-immortalized autologous B cells (27). Then 4 × 105 CD4+ T cells and 2 × 105 autologous, irradiated (6,000 rads) B cells were distributed in triplicate in 96-well plates and were stimulated with the various peptides at 1 and 10 μM concentrations. After 72 h, cell proliferation was assessed by [3H]thymidine incorporation and scintillation counting. The proliferative response was expressed as stimulation index (SI), the ratio between counts per minute of triplicate wells from CD4+ cells cultured with peptide and counts per minute for cells cultured without peptide. Cells displaying an SI/PPD-negative SI ratio of >2.5 were calculated to be significantly stimulated by a paired Student t test (P < 0.05).
RESULTS
Prediction of HLA-DR-restricted epitopes in Mce proteins and binding of selected Mce2 peptides to soluble HLA-DR.
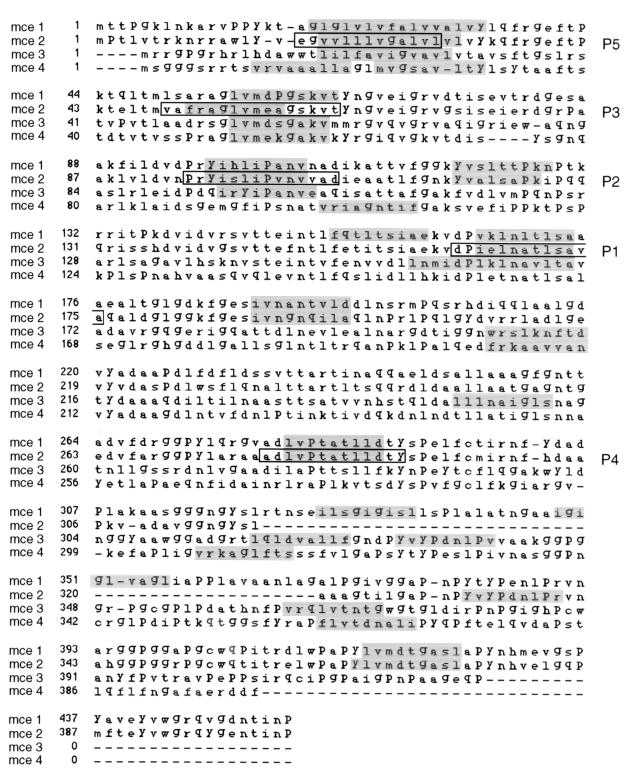
The identification of allele-specific motifs in antigenic peptides together with the structural definition of HLA-peptide complexes provided the basis for the development of methods to predict MHC-II-binding peptides (15, 30). TEPITOPE was applied to the four members of the mce family and led to the identification of peptides predicted to bind to HLA-DR (shaded sequences in Fig. 1). We synthesized four peptides with strong prediction for promiscuous binding to HLA-DR that were identified in the same region of at least three members of the mce family. A fourth peptide (P4 in Fig. 1) with poor prediction of promiscuous binding was synthesized as a nonpromiscuous control. To the nonamer selected by TEPITOPE as the core HLA-binding motif, two amino acids were added at both ends (peptides 1 and 4) or two at the N terminus and three at the C terminus (peptide 2) based on the sequence homology between the four mce members. These extensions of the nonamer increase the efficiency of in vitro peptide presentation to CD4+ T cells. Peptide 3 was synthesized as a 16-mer to include the four nonamers selected by TEPITOPE in the Mce sequences with the addition of two amino acids at the N terminus of the Mce2 nonamer for the reason mentioned above. The increased lengths of predicted epitopes in the regions corresponding to peptide 5 were due to the presence of more than one hypothetical HLA-binding frame. The synthesized 13-mer was chosen in the Mce2 sequence with the inclusion at the N terminus of glutamic acid and glycine to limit the presence of nonpolar residues. The five peptides were tested for binding to soluble HLA-DR molecules using an ELISA-based competition assay (13). Table 1 shows the relative binding capacity (IC50 in micromolars) for each peptide, calculated as the concentration of competitor peptide required to inhibit 50% of the binding of an allele-specific biotinylated peptide (indicator peptide). The lowest IC50s with the different aplotypes were obtained with peptides 2 and 3.
FIG. 1.
Sequence alignment of Mce proteins. Shading indicates the peptides selected by TEPITOPE as ligands for the most common HLA alleles. The boxed peptides identified by the indicated numbers were synthesized and used in the HLA-DR-binding assay as well as the CD4+ T-cell proliferation assay.
TABLE 1.
HLA-DR-binding assay of Mce2-derived peptidesa
| Peptide | IC50 (μM) for:
|
||||||
|---|---|---|---|---|---|---|---|
| DRB1*0101 (DR1) | DRB1*1501 (DR2) | DRB1*0301 (DR3) | DRB1*0401 (DR4) | DRB1*1101 (DR5) | DRB1*0701 (DR7) | DRB1*0801 (DR8) | |
| 1 (aa 163–175) | 30 | 15 | 0.45 | 0.8 | 2 | 0.025 | 100 |
| 2 (aa 95–108) | 0.02 | 0.008 | 8 | 2 | 0.4 | 0.025 | 0.15 |
| 3 (aa 49–64) | 0.8 | 1 | 100 | 8 | 1 | 0.4 | 10 |
| 4 (aa 278–290) | >100 | >100 | 25 | >100 | >100 | 0.6 | 0.2 |
| 5 (aa 17–29) | 20 | 1 | 30 | >100 | 18 | 0.07 | 100 |
| HA307–319 | 0.12 | 4 | 8 | 1.5 | 0.5 | 0.3 | 4 |
Synthesized Mce2-derived peptides are listed. Peptides were tested by ELISA for the competition of binding to HLA-DR of an indicator peptide and were compared to a promiscuous HLA-DR ligand from influenza hemagglutinin (HA307–319). The HLA-DR binding is expressed as the micromolar concentration of the competitor peptide able to inhibit the binding of the biotinylated indicator peptide by 50% (IC50 in micromolars). aa, amino acids.
IgG responses to Mce2 in M. tuberculosis-infected individuals.
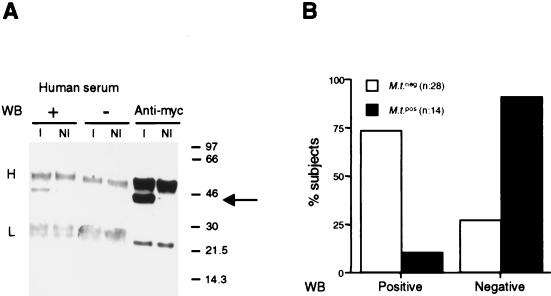
The Mce2 protein was expressed in E. coli transformants upon induction with IPTG as a fusion protein with a c-Myc epitope tag. To detect human Mce2-specific IgGs, bacteria were lysed and the lysates were subjected to immunoprecipitation with anti-Myc MAb. The immunoprecipitates were resolved by SDS-12% polyacrylamide gel electrophoresis and were transferred onto nitrocellulose membranes. Figure 2A shows the selective detection by a positive serum of the 45-kDa band corresponding to the Mce2-Myc protein in the anti-Myc immunoprecipitate from the lysate of IPTG-induced transformants. Mce2-specific IgGs were detected predominantly in treated individuals who recovered from pulmonary tuberculosis and who were negative for M. tuberculosis by cultural analysis (73% of tested subjects). In contrast, patients with active infections were mostly negative in this assay (90% of tested subjects) (Fig. 2B). Thus, the detection by immunoblotting of IgGs recognizing Mce2-Myc correlated with the recovery from M. tuberculosis infection following pharmacological treatment.
FIG. 2.
(A) Western blots (WB) with human positive (+) and negative (−) sera as well as with anti-Myc MAb of anti-Myc immunoprecipitates from lysates of noninduced (NI) and IPTG-induced (I) E. coli transformants expressing the Mce2-Myc fusion protein. Heavy (H) and light (L) chains of the immunoprecipitating anti-Myc MAb as well as the Mce2-Myc protein (arrow) are indicated. Numbers on the right are molecular sizes in kilodaltons. (B) Relative distributions of M. tuberculosis-positive (black bars) and -negative (white bars) subjects with respect to IgG reactivity to Mce2-Myc protein in immunoblot.
Proliferative responses of CD4+ T cells to predicted immunogenic peptides.
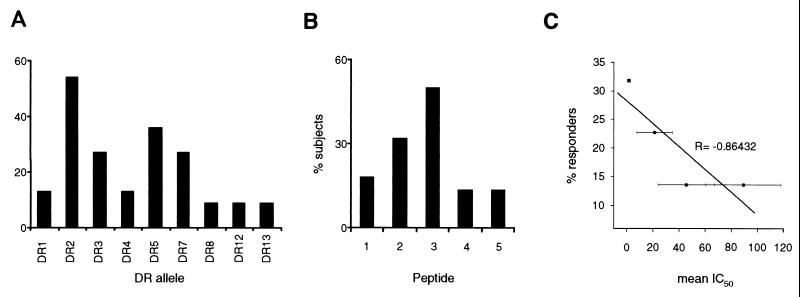
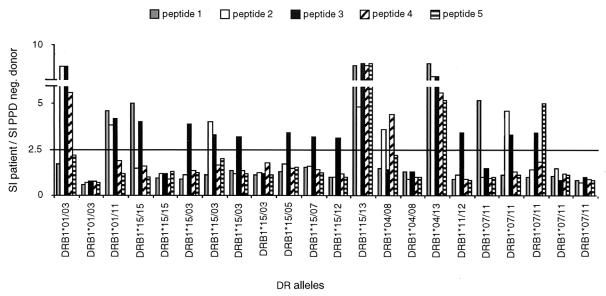
The HLA-DRs of PPD-negative healthy donors and M. tuberculosis-infected subjects were identified by molecular typing of the HLA-DRB locus. The relative distributions of the various alleles in the tested population are shown in Fig. 3A. T cells contained in the PBMC fractions from healthy donors (n = 4) and M. tuberculosis-infected subjects (n = 22) were stimulated with whole-cell lysates of H37Rv mycobacteria and propagated in interleukin 2. Stimulation of H37Rv-specific CD4+ T cells with Mce2-derived peptides was performed using previously EBV-immortalized autologous B cells as APCs. Proliferative responses displaying an SI/PPD-negative SI ratio of >2.5 were considered specific for a given peptide (P < 0.05 and <0.001). Peptides 2 and 3 were the best inducers of proliferative responses with different alleles (Fig. 3B and 4), whereas no significant proliferations were observed when CD4+ cells were stimulated with a control peptide. The percentages of proliferative responses were negatively correlated with the mean IC50s detected in the HLA-DR-binding assay (Fig. 3C). Peptide 3, which induced 50% of proliferative responses with a mean IC50 of 17.3, was excluded from such an analysis because of the increased length (16 amino acids) with respect to the other four peptides. The extended length was determined by the inclusion of contiguous homologous Mce regions to incorporate the slightly shifted nonamer in the Mce2 sequence and the predicted nonamers in the other Mce members (Fig. 1). This should allow the detection of more anti-Mce CD4+ T-cell responses (see discussion). The proliferative responses could be correlated to the binding of the peptide in vitro to a particular HLA-DR apart from DR13, which was not employed in such an assay. The efficiency of peptide 3 in eliciting CD4+ proliferative responses suggests that it could be a promiscuous epitope generated by the natural processing of mce proteins.
FIG. 3.
(A) Relative representations of HLA-DR alleles in subjects tested for CD4+ T-cell proliferative responses. (B) Percentages of subjects responding to the different Mce2-derived peptides. (C) Correlation between percentages of proliferative responses obtained in the CD4+ T-cell stimulation assay and mean IC50s detected in the HLA-DR-binding assay with peptides 1, 2, 4, and 5.
FIG. 4.
Proliferative responses (expressed as ratio of SI of patient/SI of PPD-negative donor) to Mce2-derived peptides of typed patients with pulmonary tuberculosis. A paired Student t test was used to determine the P value that was <0.05 or <0.001 for cellular responses above the marked value of 2.5.
DISCUSSION
Several lines of evidence indicate that activation of M. tuberculosis-specific CD4+ T cells is critical for the control of mycobacterial infection. Analysis of human sera in immunoblots with recombinant Mce2-Myc protein detected Mce2-specific IgGs predominantly in subjects treated for pulmonary tuberculosis who were negative for M. tuberculosis isolation. Immunoglobulin isotype switching generally involves the recruitment of CD4+ helper T cells (28). Since the immunoblot assay likely detects also IgGs induced by other members of the Mce family, this result suggests that the Mce2 protein and/or other Mce members elicit an MHC-II-restricted response.
HLA-DR constitutes the dominant isotype of human MHC-II (33). TEPITOPE incorporates 25 virtual matrices covering the majority of human HLA-DR specificities and enables the systematic prediction of peptide ligands for a broad range of HLA-binding specificities (10). It was successfully employed to identify HLA-DR ligands derived from tumors (20, 31) and endogenous proteins involved in autoimmune diseases (14). We investigated whether promiscuous HLA-DR ligands could be identified in the sequences of the Mce protein family. We used a stringent threshold setting, which was proven to be effective in predicting up to 80% of an in vitro-selected peptide repertoire (31). The HLA-DR-binding assay with the five selected peptides corresponding to the Mce2 sequence confirmed the validity of such an approach. Indeed, peptides 2 and 3, which displayed the lowest IC50s, were predicted to be the most promiscuous ligands. Peptide 5 demonstrated high IC50s with most MHCs in spite of the promiscuous binding prediction, and this might be due to the high hydrophobicity of such a peptide. Nevertheless, we employed peptide 5 in the CD4+ T-cell proliferation assay, because it was shown that hydrophobic peptides could represent promiscuous T-cell epitopes and could be efficiently presented in vitro (26).
In vivo antigen processing and HLA class II binding are complex multistep processes that can be influenced by unpredictable mechanisms, i.e., the susceptibility to proteolysis of a given antigen, the specificity of the proteolytic enzymes involved during processing, and the stability of the generated peptides. We assessed the capacity of the selected peptides to be presented in vivo in the course of M. tuberculosis infection by testing the proliferation of CD4+ T cells from patients with pulmonary tuberculosis upon stimulation with autologous APCs loaded with the Mce2 peptides as well as a control, unrelated peptide. Such an analysis revealed that the percentage of proliferative responses was inversely correlated to the mean IC50 obtained in the HLA-binding assay. Peptide 3 displayed a significant increase in the percentage of proliferative responses with respect to the expected value by the correlation between mean IC50s and percentages of T-cell responses obtained with the other four peptides. This could be due to both the extended length with inclusion of more potential epitopes from the four Mce proteins and/or some preferential presentation of nonamers contained in peptide 3 to CD4+ cells in vivo. Two DRB1*04/08 patients did not respond to peptide 3 in the antigen presentation assay, whereas no restriction to particular aplotypes was evident in responses to peptide 2. Indeed, both DR4 and DR8 were not predicted to be optimal ligands for peptide 3 in the HLA-DR-binding assay, and peptide 2 displayed the most promiscuous pattern in such an assay.
The Mce proteins were predicted to bear signal sequences or hydrophobic stretches at the N terminus, suggesting that they could be either secreted or surface exposed (7), consistent with the proposed role of the Mce1 protein in invasion of host cells (2). The results presented in this study indicate that Mce proteins are immunogenic. Moreover, a promiscuous T-cell epitope could be identified by TEPITOPE in a region of homology of the four Mce proteins. Subunit vaccines consisting of mycobacterial protein antigens represent a potential safe and specific tool for the prevention of tuberculosis. A potential limit of subunit vaccines is limited persistence in vivo, resulting in the inability to induce strong primary responses with long-lived memory (18). However, it was recently shown that in mice previously immunized with BCG, memory CD4+ T cells could be boosted by a protein antigen (3). Since BCG is widely administered to children, immunization with BCG could be beneficial for obtaining more specific and protective secondary responses with subunit vaccines. The identification of promiscuous binding to HLA is an ideal prerequisite for the design of subunit vaccines. Since the complete genome of M. tuberculosis is available, TEPITOPE could constitute a valuable tool for the identification of immunogenic peptides from M. tuberculosis proteins selectively implicated in the various pathogenetic aspects of tuberculosis.
Acknowledgments
This work was supported by grant PPI 1502 from Istituto Superiore di Sanità, Progetto Nazionale Tubercolosi.
We thank Katharina Fleischhauer (Dibit-HSR, Milan, Italy) for HLA typing, Maria Guttinger (Dibit-HSR, Milan, Italy) and Paola Panina-Bordignon (Roche Milano Ricerche, Milan, Italy) for skillful advice on the T-cell proliferation assay, Massimiliano Tattanelli (Università di Milano) for help in cloning the Mce2-Myc protein, Luca Scorrano (Dana Farber Cancer Institute, Boston, Mass.) for linear regression graphics, Stewart T. Cole and Karin Eiglmeier (Institut Pasteur, Paris, France) for the MTCY19H5 cosmid, and John T. Belisle (Colorado State University) for H37Rv whole-cell lysate (NIH, NIAID Contract N01 AI-75320).
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Alderson, M. R., T. Bement, C. H. Day, L. Zhu, D. Molesh, Y. A. Skeiky, R. Coler, D. M. Lewinsohn, S. G. Reed, and D. C. Dillon. 2000. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4+ T cells. J. Exp. Med. 191: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, S., G. Bomfim, R. Knights, T. Huima-Byron, and L. W. Riley. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261: 1454–1457. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, J. V., A. A. Frank, M. A. Keen, J. T. Bellisle, and I. M. Orme. 2001. Boosting vaccine for tuberculosis. Infect. Immun. 69: 2714–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canaday, D. H., C. Ziebold, E. H. Noss, K. A. Chervenak, C. V. Harding, and W. H. Boom. 1999. Activation of human CD8+ αβ TCR+ cells by Mycobacterium tuberculosis via an alternate class I MHC antigen-processing pathway. J. Immunol. 162: 372–379. [PubMed] [Google Scholar]
- 5.Caruso, A. M., N. Serbina, E. Klein, K. Triebold, B. R. Bloom, and J. L. Flynn. 1999. Mice deficient in CD4 T cells have only transiently diminished levels of IFN-γ, yet succumb to tuberculosis. J. Immunol. 162: 5407–5416. [PubMed] [Google Scholar]
- 6.Cho, S., V. Mehra, S. Thoma-Uszynski, S. Stenger, N. Serbina, R. J. Mazzaccaro, J. L. Flynn, P. F. Barnes, S. Southwood, E. Celis, B. R. Bloom, R. L. Modlin, and A. Sette. 2000. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc. Natl. Acad. Sci. USA 97: 12210–12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, A. M., D. K. Dalton, T. A. Stewart, J. P. Griffin, D. G. Russell, and I. M. Orme. 1993. Disseminated tuberculosis in interferon γ gene-disrupted mice. J. Exp. Med. 178: 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flesselles, B., N. N. Anand, J. Remani, S. M. Loosmore, and M. H. Klein. 1999. Disruption of the mycobacterial cell entry gene of Mycobacterium bovis BCG results in a mutant that exhibits a reduced invasiveness for epithelial cells. FEMS Microbiol. Lett. 177: 237–242. [DOI] [PubMed] [Google Scholar]
- 10.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19: 93–129. [DOI] [PubMed] [Google Scholar]
- 11.Flynn, J. L., J. Chan, K. J. Triebold, D. K. Dalton, T. A. Stewart, and B. R. Bloom. 1993. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttinger, M., F. Sutti, M. Panigada, S. Porcellini, B. Merati, M. Mariani, T. Teesalu, G. G. Consalez, and F. Grassi. 1998. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J. Cell Biol. 141: 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer, J., E. Bono, F. Gallazzi, C. Belunis, Z. Nagy, and F. Sinigaglia. 1994. Precise prediction of major histocompatibility complex class II-peptide interaction based on peptide side chain scanning. J. Exp. Med. 180: 2353–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer, J., T. Sturniolo, and F. Sinigaglia. 1997. HLA class II peptide binding specificity and autoimmunity. Adv. Immunol. 66: 67–100. [DOI] [PubMed] [Google Scholar]
- 15.Hammer, J., P. Valsasnini, K. Tolba, D. Bolin, J. Higelin, B. Takacs, and F. Sinigaglia. 1993. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell 74: 197–203. [DOI] [PubMed] [Google Scholar]
- 16.Jouanguy, E., F. Altare, S. Lamhamedi, P. Revy, J. F. Emile, M. Newport, M. Levin, S. Blanche, E. Seboun, A. Fischer, and J. L. Casanova. 1996. Interferon-γ-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N. Engl J. Med. 335: 1956–1961. [DOI] [PubMed] [Google Scholar]
- 17.Lalvani, A., R. Brookes, R. J. Wilkinson, A. S. Malin, A. A. Pathan, P. Andersen, H. Dockrell, G. Pasvol, and A. V. Hill. 1998. Human cytolytic and interferon γ-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95: 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letvin, N. L., B. R. Bloom, and S. L. Hoffman. 2001. Prospects for vaccines to protect against AIDS, tuberculosis, and malaria. JAMA 285: 606–611. [DOI] [PubMed] [Google Scholar]
- 19.Lewinsohn, D. M., M. R. Alderson, A. L. Briden, S. R. Riddell, S. G. Reed, and K. H. Grabstein. 1998. Characterization of human CD8+ T cells reactive with Mycobacterium tuberculosis-infected antigen-presenting cells. J. Exp. Med. 187: 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manici, S., T. Sturniolo, M. A. Imro, J. Hammer, F. Sinigaglia, C. Noppen, G. Spagnoli, B. Mazzi, M. Bellone, P. Dellabona, and M. P. Protti. 1999. Melanoma cells present a MAGE-3 epitope to CD4+ cytotoxic T cells in association with histocompatibility leukocyte antigen DR11. J. Exp. Med. 189: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohagheghpour, N., D. Gammon, L. M. Kawamura, A. van Vollenhoven, C. J. Benike, and E. G. Engleman. 1998. CTL response to Mycobacterium tuberculosis: identification of an immunogenic epitope in the 19-kDa lipoprotein. J. Immunol. 161: 2400–2406. [PubMed] [Google Scholar]
- 22.Muller, I., S. P. Cobbold, H. Waldmann, and S. H. Kaufmann. 1987. Impaired resistance to Mycobacterium tuberculosis infection after selective in vivo depletion of L3T4+ and Lyt-2+ T cells. Infect. Immun. 55: 2037–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newport, M. J., C. M. Huxley, S. Huston, C. M. Hawrylowicz, B. A. Oostra, R. Williamson, and M. Levin. 1996. A mutation in the interferon-γ-receptor gene and susceptibility to mycobacterial infection. N. Engl. J. Med. 335: 1941–1949. [DOI] [PubMed] [Google Scholar]
- 24.Orme, I. M. 1988. Characteristics and specificity of acquired immunologic memory to Mycobacterium tuberculosis infection. J. Immunol. 140: 3589–3593. [PubMed] [Google Scholar]
- 25.Scanga, C. A., V. P. Mohan, K. Yu, H. Joseph, K. Tanaka, J. Chan, and J. L. Flynn. 2000. Depletion of CD4+ T cells causes reactivation of murine persistent tuberculosis despite continued expression of interferon γ and nitric oxide synthase 2. J. Exp. Med. 192: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinigaglia, F., M. Guttinger, J. Kilgus, D. M. Doran, H. Matile, H. Etlinger, A. Trzeciak, D. Gillessen, and J. R. Pink. 1988. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature 336: 778–780. [DOI] [PubMed] [Google Scholar]
- 27.Sinigaglia, F., P. Romagnoli, M. Guttinger, B. Takacs, and J. R. Pink. 1991. Selection of T cell epitopes and vaccine engineering. Methods Enzymol. 203: 370–386. [DOI] [PubMed] [Google Scholar]
- 28.Snapper, C. M., and F. D. Finkelman. 1993. Immunoglobulin class switching, p. 837–863. In W. E. Paul (ed.), Fundamental immunology, 3rd ed. Raven Press Ltd., New York, N.Y.
- 29.Stenger, S., D. A. Hanson, R. Teitelbaum, P. Dewan, K. R. Niazi, C. J. Froelich, T. Ganz, S. Thoma-Uszynski, A. Melian, C. Bogdan, S. A. Porcelli, B. R. Bloom, A. M. Krensky, and R. L. Modlin. 1998. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282: 121–125. [DOI] [PubMed] [Google Scholar]
- 30.Stern, L. J., J. H. Brown, T. S. Jardetzky, J. C. Gorga, R. G. Urban, J. L. Strominger, and D. C. Wiley. 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368: 215–221. [DOI] [PubMed] [Google Scholar]
- 31.Sturniolo, T., E. Bono, J. Ding, L. Raddrizzani, O. Tuereci, U. Sahin, M. Braxenthaler, F. Gallazzi, M. P. Protti, F. Sinigaglia, and J. Hammer. 1999. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 17: 555–561. [DOI] [PubMed] [Google Scholar]
- 32.Tascon, R. E., E. Stavropoulos, K. V. Lukacs, and M. J. Colston. 1998. Protection against Mycobacterium tuberculosis infection by CD8+ T cells requires the production of gamma interferon. Infect. Immun. 66: 830–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji, K., M. Aizawa, and T. Sasazuki. 1992. HLA 1991. Proceedings of the Eleventh International Histocompatibility Workshop and Conference. Oxford University Press, New York, N.Y.