Abstract
Chlamydia pneumoniae is a causative agent for many respiratory infections and has been associated with cardiovascular diseases in humans. The pathogenicity of C. pneumoniae is thought to depend on its ability to cause persistent infection and to evade host defense. Genome sequence analysis indicates that C. pneumoniae encodes a homologue of a chlamydial protease-like activity factor from C. trachomatis (CPAFct). We designated the C. pneumoniae homologue as CPAFcp. Recombinant CPAFcp was produced and found to degrade RFX5, a host transcription factor required for major histocompatibility complex (MHC) antigen expression. The degradation was inhibitable by lactacystin, an irreversible proteasome inhibitor. Furthermore, CPAFcp was secreted into host cytosol by C. pneumoniae organisms. Depletion of the C. pneumoniae-secreted CPAFcp with specific antibodies completely ablated the RFX5 degradation activity in the infected cells, suggesting that CPAFcp is necessary for the degradation of host transcription factors required for MHC antigen expression during C. pneumoniae infection. These observations have revealed a unique molecular mechanism for C. pneumoniae to evade host adaptive immunity that may aid in its persistence.
Chlamydia is an obligate intracellular pathogen that has to replicate within a cytoplasmic vacuole of eukaryotic cells (13). There are two major chlamydial species that cause human diseases (12, ). The Chlamydia trachomatis species is a leading cause of trachoma and sexually transmitted diseases (12), while the C. pneumoniae species causes various respiratory infections (11). Although the C. pneumoniae-induced respiratory infections are often asymptomatic, its association with atherosclerosis has attracted the attention of many investigators (10). C. pneumoniae organisms have been detected in a large proportion of atherotic plaques but not in nonatherotic cardiovascular tissues (20, 25). In cell culture, C. pneumoniae infection was able to transform macrophages into foam cells (15), a hallmark of atherosclerosis. Several groups including ourselves have demonstrated that respiratory infection with C. pneumoniae organisms can greatly enhance atherosclerotic lesion development in animal models (3, 6, 14, 18). More importantly, antibiotic treatment of the infected animals can prevent the C. pneumoniae exacerbation of atherosclerosis (5, 19). Despite the important role of C. pneumoniae infection in atherosclerosis, the mechanism of the C. pneumoniae atherogenicity is still not clear.
It is thought that the continuous release of inflammatory cytokines by persistently infected cells may play a major role in chlamydial pathogenesis (2, 22). One of the hallmarks of C. pneumoniae infection is persistence (1, 7, 9). The question is how C. pneumoniae is able to successfully maintain the persistence in its hosts. We have previously demonstrated that C. trachomatis organisms can escape from host immune detection by secreting a proteolytically active molecule (designated CPAFct for chlamydial protease-like activity factor from C. trachomatis) into host cell cytosol (29, 30, 31). CPAFct can selectively degrade host transcription factors, including RFX5. RFX5 is a critical component of the RFX transcription complexes that are required for major histocompatibility complex (MHC) antigen expression (26, 27, 31). Mutations in RFX5 can lead to deficiency in MHC antigen expression (24). It is not surprising that C. trachomatis has targeted RFX5 for evading host adaptive immunity (31). In the present study, we show that C. pneumoniae organisms produce a CPAF homologue designated CPAFcp. Although there is only 48% amino acid sequence identity between CPAFct and CPAFcp (http://violet.berkeley.edu:4231/orf/CTD_858_v_CPN.html), CPAFcp possesses a similar proteolytic activity for degrading RFX5 as CPAFct does, suggesting that C. pneumoniae may utilize a similar strategy for evading host defense.
MATERIALS AND METHODS
Cell-free degradation assay.
The cell-free degradation assay was performed as previously described (29). A cytosolic extract (CE) of either chlamydia-infected or normal HeLa cells was made with a buffer consisting of 1% NP-40 and 150 mM NaCl in 50 mM Tris (pH 8.0) plus a protease inhibitor cocktail. The CE thus prepared were used as the source of enzymes. To generate fusion proteins for the cell-free assay, C. pneumoniae AR39 DNA sequences coding for CPAFcp or CPAFcp fragments were cloned into a pGEX vector (Pharmacia) and expressed as fusion proteins with glutathione S-transferase (GST) as the fusion partner. The fusion proteins were purified with glutathione-conjugated agarose beads as described in the manufacturer”s manual (Pharmacia). The degradation activity of the purified protein was measured in the cell-free assay. The following procedure was used to prepare nuclear extracts (NEs) as substrate (containing RFX5) for the cell-free assay. Normal HeLa cells were homogenized to break cytoplasmic membranes and the residual pellets were repeatedly washed with the NP-40 buffer as described above to remove cytosol or membrane proteins as much as possible. The final washed nuclear pellets were extracted with a buffer consisting of 0.5 M NaCl and 1% Triton X-100 in 20 mM Tris (pH 8.0). To prepare the purified RFX5 as substrate, the human RFX5 gene from pREP-4/RFX5 plasmid (kindly provided by Peter J. van den Elsen [21]) was cloned into the pGEX vector, and the fusion protein GST-RFX5 was expressed and purified to homogeneity by using glutathione-conjugated agrose beads as previously described (31). The purified GST-RFX5 was used as a substrate in the cell-free degradation assays.
Western blot.
The Western blot assay was carried out as previously described (28). Samples from the various cell-free degradation assays were directly subjected to sodium dodecyl sulfate-polyacrylamide gel separation and Western blot analysis. Samples from infection dose titration experiments were obtained as follows: HeLa cells were infected with C. pneumoniae AR39 strain at various multiplicities of infection (MOIs) as indicated in the legend to Fig. 2 in the presence of 2 μg of cycloheximide/ml. At 48 h after infection, the cell culture was washed and replenished with fresh growth medium without cycloheximide in order to allow host cells to recover their ability to synthesize new proteins. After an additional 24 h of culture, the cell samples were harvested for sodium dodecyl sulfate-polyacrylamide gel separation and Western blot analysis. Mouse antibodies were used to detect CPAFcp C terminus (antiserum was generated by immunizing mice with a GST fusion protein containing the C-terminal half of the CPAFcp [data not shown]). Rabbit antibodies were used to detect RFX5 (Rockland Immunochemicals, Gilbertsville, Pa.).
FIG. 2.
Correlation of CPAFcp secretion by C. pneumoniae with host transcription factor degradation in C. pneumoniae-infected cells. (A) An immunofluorescence staining approach was used to identify CPAFcp in C. pneumoniae-infected HeLa cells. HeLa monolayer cells were infected with C. pneumoniae at a low MOI so that only a small portion of cells was infected. The infection was allowed for 72 h in the presence of 2 μg of cycloheximide/ml. The processed monolayer cells were costained with Hoechst 32258 for DNA (blue), anti-AR39 organism rabbit antiserum (green), and anti-CPAFcp mouse antiserum (red). Images were acquired individually for each stain in gray (top row), and the single-color images were merged in frame into the triple-color image (bottom). Note that the anti-CPAFcp antibody only stained the cytosol (red) of the cells harboring C. pneumoniae inclusions (green). (B) A Western blot assay was used to compare the levels of CPAFcp and RFX5 in HeLa cells alone or HeLa cells infected with C. pneumoniae at various MOIs as indicated in the figure. HeLa cells were infected and cultured as described in Materials and Methods. Cell lysates from 2 × 105 cells were loaded into each lane. After transfer onto nitrocellular membrane, CPAFcp and RFX5 were detected with the corresponding antibodies. Note the inverse correlation between the levels of CPAFcp and RFX5.
Immunofluorescence staining assay.
Immunoflorescence detection of CPAFcp in C. pneumoniae-infected cells was carried out as previously described (29, 32). Briefly, HeLa cell monolayer was infected with C. pneumoniae AR39 for 72 h. The monolayer, after fixation with paraformaldehyde (Sigma, St. Louis, Mo.) and permeabilization with Saponin (Sigma), was costained with Hoechst 32258 (blue), a rabbit anti-AR39 antiserum (raised with purified AR39 elementary bodies [data not shown]; probed with a Cy2-conjugated goat anti-rabbit immunoglobulin G [IgG]), and a mouse anti-CPAFcp antiserum (probed with a Cy3-conjugated goat anti-mouse IgG). Images were acquired individually for each stain by using a Hamamatsu digital camera connected to an AX70 Olympus microscope, and the single-color images were merged in frame into the triple-color image by using the software SimplePCI.
Immunoprecipitation assay.
The immunoprecipitation assays were carried out as previously described (28, 31). A mouse antiserum raised with the C-terminal fragment of CPAFcp was used for depleting CPAFcp and a control antiserum from mouse similarly immunized with an unrelated GST fusion protein for mock depletion. Then, 5 μl of each antiserum was conjugated to protein G-agarose beads, and the antibody-bead complexes were used to absorb 20 μl of cytosol extracts of AR39-infected HeLa cells (AR39-CE) in a total volume of 60 μl for 1 h at room temperature. After a second absorption, 15 μl of the final remaining supernatant was compared with 5 μl of control AR39-CE (without absorption) for their ability to degrade RFX5 in a cell-free assay.
RESULTS
A recombinant chlamydial protease-like activity factor cloned from the C. pneumoniae genome is sufficient for degrading host transcription factor RFX5.
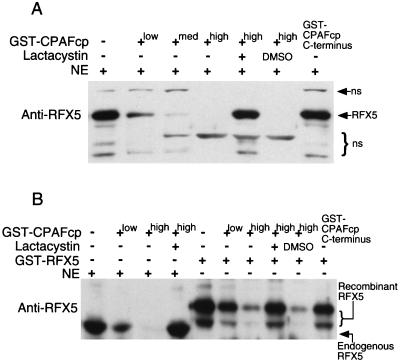
Sequence homology searching (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was used to identify a conserved hypothetical open reading frame (cpn1016; http://violet.berkeley.edu:4231/cpn/p1016.html) in the C. pneumoniae genome (16, 23) that encodes a homologue of CPAFct (29). The C. pneumoniae homologue is designated CPAFcp. There is 48% amino acid sequence identity between CPAFct and CPAFcp (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi#7388442). We expressed CPAFcp as a fusion protein with GST as the fusion partner and tested the purified GST-CPAFcp fusion protein for its ability to degrade host transcription factor RFX5 in a cell-free degradation assay (Fig. 1). The GST-CPAFcp readily degraded RFX5 in a HeLa cell NE, and the degradation activity was inhibited by lactacystin but not the solvent dimethyl sulfoxide alone (Fig. 1A). A GST fusion protein containing just the C terminus of CPAFcp did not degrade RFX5. These observations demonstrated that CPAFcp possesses a proteolytic activity similar to that of CPAFct (29). To exclude the possible contribution of other components in the HeLa NEs to the CPAFcp degradation activity, we used a recombinant RFX5 purified from a bacterial expression system as the substrate to further evaluate the CPAFcp degradation activity in a cell-free assay (Fig. 1B). CPAFcp effectively degraded the recombinant RFX5, and the degradation was also inhibited by lactacystin, suggesting that the same enzymatic activity is responsible for degrading both the endogenous and recombinant RFX5. Again, a fusion protein containing the C terminus of CPAFcp did not degrade the recombinant RFX5, suggesting that bacterial contaminants did not contribute to the degradation activity. Together, these observations demonstrate that CPAFcp alone is sufficient for degrading RFX5.
FIG. 1.
Degradation of RFX5 by recombinant CPAFcp. CPAFcp was expressed as a fusion protein with GST as the fusion partner and the purified GST-CPAFcp was tested for its ability to degrade either the endogenous RFX5 in HeLa cell NE (A) or a recombinant human RFX5 (GST-RFX5) purified from a bacterial expression system (B). The degradation was carried out in a cell-free assay as described in Materials and Methods. CPAFcp was used at final concentrations of 0.05 (low), 0.2 (med), or 0.6 (high) μM, while the proteasome inhibitor lactacystin was used at 200 μM. A fusion protein containing GST and just the COOH-terminal portion of CPAFcp (GST-CPAFcp C terminus) failed to degrade either the endogenous or the recombinant RFX5 even at a final concentration of 2 μM. DMSO, dimethyl sulfoxide (a solvent used for dissolving lactacystin); ns, nonspecific binding.
Correlation of CPAFcp secretion by C. pneumoniae organisms with host transcription factor degradation in C. pneumoniae-infected cells.
Although we have demonstrated that the recombinant CPAFcp purified from a bacterial expression system is sufficient for degrading RFX5, it is not known whether CPAFcp is actually produced by C. pneumoniae organisms and whether the C. pneumoniae-synthesized CPAFcp is functional. We first used a CPAFcp-specific antibody to detect endogenous CPAFcp in C. pneumoniae-infected HeLa cells (Fig. 2A). CPAFcp was found predominantly in the cytosol of the infected cells, suggesting that C. pneumoniae organisms not only produce CPAFcp but also secrete the CPAFcp into host cell cytoplasm to allow CPAFcp to access host proteins. The fact that CPAFcp is only detected in the infected cells but not in the adjacent uninfected cells suggests that CPAFcp is restricted to the infected cells only. To correlate CPAFcp production with the host transcription factor degradation activity in the infected cells, we used a Western blot assay to compare the levels of CPAFcp and transcription factor RFX5 in HeLa cells alone or HeLa cells infected with C. pneumoniae at various MOIs (Fig. 2B). The level of CPAFcp produced by C. pneumoniae increased, while the level of the host transcription factor RFX5 decreased in an infection dose-dependent manner. The inverse relationship between CPAFcp and RFX5 suggests that CPAFcp may be responsible for the disappearance of RFX5.
CPAFcp is required for the degradation of RFX5 in C. pneumoniae-infected cells.
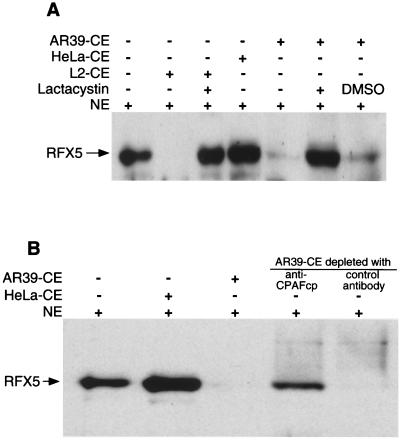
After correlating C. pneumoniae secretion of CPAFcp with the degradation of RFX5 in C. pneumoniae-infected cells, we next measured the RFX5 degradation activity in the cytosol of C. pneumoniae-infected cells by using a cell-free degradation assay (Fig. 3A). A CE from C. trachomatis LGV2 strain-infected cells (L2-CE) was used as positive control since L2-CE has been previously shown to contain the RFX5 degradation activity (29). A CE from uninfected HeLa cells (HeLa-CE) was used as negative control. The CE from C. pneumoniae AR39 strain-infected cells (AR39-CE) completely degraded RFX5, whereas the negative control HeLa-CE failed to do so. The RFX5 degradation activity in AR39-CE was inhibited by lactacystin, suggesting that AR39-CE possesses a proteolytic activity similar to that of the recombinant CPAFcp (see Fig. 1).
FIG. 3.
CPAFcp is necessary for degradation of the host transcription factor RFX5. (A) CEs of HeLa cells alone (HeLa-CE) or HeLa cells infected with C. pneumoniae AR39 strain (AR39-CE) or infected with C. trachomatis LGV2 strain (L2-CE) were used as the sources of enzymes for degrading RFX5 in HeLa cell NE in a cell-free degradation assay as described in the Fig. 1 legend. Equivalent amounts of CEs were used for each reaction. RFX5 was detected with a rabbit anti-RFX5 antiserum. Note that both AR39 and L2 CEs degraded RFX5, but HeLa-CE failed to do so. (B) AR39-CE was absorbed with an anti-CPAFcp or control antibody as described in Materials and Methods, and both the intact AR39-CE and the final remaining supernatants after antibody absorption were measured for their ability to degrade RFX5 in a HeLa cell NE by using the cell-free degradation assay. Note that AR39-CE depleted with the anti-CPAFcp antibody lost most of its RFX5 degradation activity, whereas the AR39-CE depleted with the control antibody maintained the same level of degradation activity as the intact AR39-CE.
To directly assess whether CPAFcp is responsible for the RFX5 degradation activity in the cytosol of infected cells, we used a CPAFcp-specific antibody to perform a depletion experiment (Fig. 3). AR39-CE but not HeLa-CE degraded RFX5. More importantly, the AR39-CE supernatant after being absorbed with a CPAFcp-specific antibody conjugated to agarose beads could no longer degrade RFX5, while the AR39-CE supernatant similarly absorbed with a control antibody still maintained the RFX5 degradation activity. This result demonstrates that CPAFcp is necessary for the RFX5 degradation activity in the C. pneumoniae-infected cell cytosol.
DISCUSSION
Evasion of host defense is likely advantageous for chlamydia to survive for long periods of time in its host. We have previously identified various immune evasion strategies utilized by C. trachomatis (4, 29, 30, 31) and have now shown that similar mechanisms are used by C. pneumoniae. Our results demonstrate that CPAFcp by itself is sufficient for degrading the host transcription factor RFX5 in a cell-free degradation assay with a purified human RFX5 as substrate. We have also shown that CPAFcp is required for the RFX5 degradation activity in the C. pneumoniae-infected cells by using an antibody depletion experiment. There is no system for genetic transformation of chlamydia, so definitive gene knockout experiments are not possible. However, antibody depletion of CPAFcp and blocking CPAFcp function with specific protease inhibitors clearly indicate the biological function of CPAFcp. These experiments together have provided the first line of evidence demonstrating that C. pneumoniae has indeed evolved specific strategies for evading host adaptive immunity.
Although there is only 48% amino acid sequence identity between CPAFct and CPAFcp (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi#7388442), the proteolytic activity of both CPAFs is conserved, suggesting that degradation of host transcription factors required for MHC antigen expression may be essential for chlamydial survival in its hosts. Our studies on CPAFct (29) and CPAFcp (Fig. 2A) have shown that CPAF is mainly secreted into host cell cytosol without an obvious presence in the organisms themselves, suggesting that the main purpose for chlamydial synthesis of CPAF is to use CPAF to manipulate host cells. However, secretion of chlamydial proteins into host cell cytosol is itself a danger for the chlamydial organisms since microbial products present in the host cell cytosol can be processed and presented to host T lymphocytes. These lymphocytes can potentially detect and attack the infected cells (8). Then, why does chlamydia secrete CPAF at all? We speculate that the intimate interactions between chlamydia and host cells may inevitably require chlamydia to actively secrete factors into host cells. Efforts are under way to identify other chlamydial proteins that are secreted into host cells.
Although we still do not know how CPAF works precisely, the identification of CPAFcp has provided an additional tool for us to further delineate the mechanism of CPAF functions. It is known that C. trachomatis and C. pneumoniae have different tissue tropism in vivo and different growth requirements in vitro. It is likely that besides common targets such as RFX5, CPAFct and CPAFcp may also have their unique substrates. Identification of these unique substrates may reveal new functions of CPAFs and provide additional information regarding the mechanisms of CPAF function. Efforts to search for additional CPAF substrates are under way.
Acknowledgments
This work was supported in part by grants (to G.Z.) from the National Institutes of Health (R01 HL64883-01 and R01 AI47997-02).
We thank Grant McClarty for help with the manuscript.
Editor: E. I. Tuomanen
REFERENCES
- 1.Airenne, S., H. M. Surcel, H. Alakarppa, K. Laitinen, J. Paavonen, P. Saikku, and A. Laurila. 1999. Chlamydia pneumoniae infection in human monocytes. Infect. Immun. 67: 1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, L. A., E. Blessing, M. Rosenfeld, T. Lin, and C. Kuo. 2000. Mouse models of C. pneumoniae infection and atherosclerosis. J. Infect. Dis. 181(Suppl. 3): S508–S513. [DOI] [PubMed] [Google Scholar]
- 4.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong, I. W. 2000. Antibiotics effects in a rabbit model of Chlamydia pneumoniae-induced atherosclerosis. J. Infect. Dis. 181(Suppl. 3): S514–S518. [DOI] [PubMed] [Google Scholar]
- 6.Fong, I. W., T. Quinn, E. Blessing, C. Kuo, R. Malinverni, M. Lauer, S. Mawhorter, K. Bachmaier, M. Rosenfeld, C. Taylor, and G. Zhong. 2000. Collaborative multidisciplinary workshop report: what questions regarding the role of Chlamydia pneumoniae in atherosclerosis and cardiovascular disease need to be addressed utilizing animal models? J. Infect. Dis. 181(Suppl. 3): S519–S520. [DOI] [PubMed] [Google Scholar]
- 7.Gabriel, A. S., H. Gnarpe, J. Gnarpe, H. Hallander, O. Nyquist, and A. Martinsson. 1998. The prevalence of chronic Chlamydia pneumoniae infection as detected by polymerase chain reaction in pharyngeal samples from patients with ischaemic heart disease. Eur. Heart J. 19: 1321–1327. [DOI] [PubMed] [Google Scholar]
- 8.Germain, R. N. 1994. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell 76: 287–299. [DOI] [PubMed] [Google Scholar]
- 9.Gieffers, J., H. Fullgraf, J. Jahn, M. Klinger, K. Dalhoff, H. A. Katus, W. Solbach, and M. Maass. 2001. Chlamydia pneumoniae infection in circulating human monocytes is refractory to antibiotic treatment. Circulation 103: 351–356. [DOI] [PubMed] [Google Scholar]
- 10.Grayston, J. T. 2000. Background and current knowledge of Chlamydia pneumoniae and atherosclerosis. J. Infect. Dis. 181(Suppl. 3): S402–S410. [DOI] [PubMed] [Google Scholar]
- 11.Grayston, J. T., M. B. Aldous, A. Easton, S. P. Wang, C. C. Kuo, L. A. Campbell, and J. Altman. 1993. Evidence that Chlamydia pneumoniae causes pneumonia and bronchitis. J. Infect. Dis. 168: 1231–1235. [DOI] [PubMed] [Google Scholar]
- 12.Grayston, J. T., and S. Wang. 1975. New knowledge of chlamydiae and the diseases they cause. J. Infect. Dis. 132: 87–105. [DOI] [PubMed] [Google Scholar]
- 13.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5: 288–293. [DOI] [PubMed] [Google Scholar]
- 14.Hu, H., G. N. Pierce, and G. Zhong. 1999. The atherogenic effects of chlamydia are dependent on serum cholesterol and specific to Chlamydia pneumoniae. J. Clin. Investig. 103: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalayoglu, M., V. Indrawati, R. P. Morrison, S. G. Morrison, Y. Yuan, and G. I. Byrne. 2000. Chlamydial virulence determinants in atherogenesis: the role of chlamydial lipopolysaccharide and heat shock protein 60 in macrophage-lipoprotein interactions. J. Infect. Dis. 181(Suppl. 3): S483–S489. [DOI] [PubMed] [Google Scholar]
- 16.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21: 385–389. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, L., H. Hu, H. Ji, A. D. Murdin, G. N. Pierce, and G. Zhong. 2000. Chlamydia pneumoniae infection significantly exacerbates aortic atherosclerosis in an LDLR−/− mouse model within six months. Mol. Cell. Biochem. 215: 123–128. [DOI] [PubMed] [Google Scholar]
- 19.Muhlestein, J. B. 2000. Chlamydia pneumoniae-induced atherosclerosis in a rabbit model. J. Infect. Dis. 181(Suppl. 3): S505–S507. [DOI] [PubMed] [Google Scholar]
- 20.Ouchi, K., B. Fujii, S. Kudo, M. Shirai, K. Yamashita, T. Gondo, T. Ishihara, H. Ito, and T. Nakazawa. 2000. Chlamydia pneumoniae in atherosclerotic and nonatherosclerotic tissue. J. Infect. Dis. 181(Suppl. 3): S441–S443. [DOI] [PubMed] [Google Scholar]
- 21.Peijnenburg, A., M. J. Van Eggermond, S. J. Gobin, R. Van den Berg, B. C. Godthelp, J. M. Vossen, and P. J. Van den Elsen. 1999. Discoordinate expression of invariant chain and MHC class II genes in class II transactivator-transfected fibroblasts defective for RFX5. J. Immunol. 163: 794–801. [PubMed] [Google Scholar]
- 22.Rasmussen, S. J., L. Eckmann, A. J. Quayle, L. Shen, Y. X. Zhang, D. J. Anderson, J. Fierer, R. S. Stephens, and M. F. Kagnoff. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Investig. 99: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steimle, V., B. Durand, E. Barras, M. Zufferey, M. R. Hadam, B. Mach, and W. Reith. 1995. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome). Genes Dev. 9: 1021–1032. [DOI] [PubMed] [Google Scholar]
- 25.Taylor-Robinson, D., and B. J. Thomas. 2000. Chlamydia pneumoniae in atherosclerotic tissue. J. Infect. Dis. 181(Suppl. 3): S437–S440. [DOI] [PubMed] [Google Scholar]
- 26.van den Elsen, P. J., S. J. Gobin, M. C. van Eggermond, and A. Peijnenburg. 1998. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48: 208–221. [DOI] [PubMed] [Google Scholar]
- 27.van den Elsen, P. J., A. Peijnenburg, M. C. van Eggermond, and S. J. Gobin. 1998. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol. Today 19: 308–312. [DOI] [PubMed] [Google Scholar]
- 28.Zhong, G., F. Castellino, P. Romagnoli, and R. N. Germain. 1996. Evidence that binding site occupancy is necessary and sufficient for effective major histocompatibility complex (MHC) class II transport through the secretory pathway redefines the primary function of class II-associated invariant chain peptides (CLIP). J. Exp. Med. 184: 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189: 1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J. Exp. Med. 191: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong, G., C. Reis e Sousa, and R. N. Germain. 1997. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc. Natl. Acad. Sci. USA 94: 13856–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]