Abstract
Translation initiation factor 1A stimulates 40S-binding of the eukaryotic initiation factor 2 (eIF2)/GTP/Met-tRNAiMet ternary complex (TC) and promotes scanning in vitro. eIF1A contains an OB-fold present in bacterial IF1 plus N- and C-terminal extensions. Truncating the C-terminus (ΔC) or mutating OB-fold residues (66–70) of eIF1A reduced general translation in vivo but increased GCN4 translation (Gcd− phenotype) in a manner suppressed by overexpressing TC. Consistent with this, both mutations diminished 40S-bound TC, eIF5 and eIF3 in vivo, and ΔC impaired TC recruitment in vitro. The assembly defects of the OB-fold mutation can be attributed to reduced 40S-binding of eIF1A, whereas ΔC impairs eIF1A function on the ribosome. A substitution in the C-terminal helix (98–101) also reduced 43S assembly in vivo. Rather than producing a Gcd− phenotype, however, 98–101 impairs GCN4 derepression in a manner consistent with defective scanning by reinitiating ribosomes. Indeed, 98–101 allows formation of aberrant 48S complexes in vitro and increases utilization of non-AUG codons in vivo. Thus, the OB-fold is crucial for ribosome-binding and the C-terminal domain of eIF1A has eukaryotic-specific functions in TC recruitment and scanning.
Keywords: eIF1A, eIF2, GCN4, scanning, translation
Introduction
Translation initiation begins with the binding of Met-tRNAiMet to the 40S ribosomal subunit in a ternary complex (TC) with eukaryotic initiation factor 2 (eIF2) and GTP. This reaction is stimulated in vitro by eIF1, eIF1A and eIF3 (Hinnebusch, 2000; Algire et al, 2002; Majumdar et al, 2003; Kolupaeva et al, 2005). The 43S preinitiation complex (PIC) thus formed binds to the 5′-end of mRNA and scans the leader until Met-tRNAiMet base-pairs with an AUG, triggering GTP hydrolysis by eIF2 in a reaction stimulated by eIF5 (Hershey and Merrick, 2000). Biochemical evidence indicates that mammalian eIF1 and eIF1A promote scanning and formation of a 48S complex at the start codon (Pestova et al, 1998). eIF2, eIF5, eIF1 and eIF3 have been implicated in AUG selection, as mutations in these factors permit increased initiation at a UUG codon in yeast (Donahue, 2000; He et al, 2003; Valasek et al, 2004). Consistent with this, eIF1 inhibits 48S assembly at near-cognate triplets or AUGs in a poor context in vitro (Pestova and Kolupaeva, 2002).
While biochemical studies have implicated eIF3, eIF1 and eIF1A in 40S-binding of TC, their relative importance in vivo and functions in 43S assembly are poorly understood. The TC in yeast cells is linked to eIF3, eIF5 and eIF1 in a multifactor complex (MFC) (Asano et al, 2000), and evidence is accumulating that these interactions promote cooperative binding of MFC components to the 40S ribosome in vivo (Asano et al, 2001; Valášek et al, 2002, 2003; Nielsen et al, 2004; Singh et al, 2004). We presented genetic evidence that eIF1A is required for wild-type (WT) binding of TC to 40S subunits in yeast, as deleting the C-terminal domain and adjacent 310 helix of eIF1A (the Δ108–153 mutation) increased translation of GCN4 mRNA in cells lacking protein kinase GCN2 (Olsen et al, 2003). GCN4 translation is normally induced in amino-acid-starved cells by phosphorylation of eIF2 (Hinnebusch, 1996). The ensuing reduction in TC concentration allows 40S subunits that translate the first upstream open-reading frame (uORF1) and resume scanning to bypass the three remaining uORFs and reinitiate downstream at GCN4. In nonstarved WT or gcn2Δ cells, by contrast, a high level of TC ensures that all 40S subunits scanning downstream from uORF1 quickly rebind TC, reinitiate at uORFs 2, 3 or 4 and dissociate from the mRNA without translating GCN4 (Hinnebusch, 1996). Importantly, the derepressed GCN4 translation in gcn2Δ cells (Gcd− phenotype) produced by the Δ108–153 mutation was suppressed by overexpressing the TC. This suggested that the rate of TC binding to 40S subunits scanning downstream from uORF1 was reduced by Δ108–153, allowing a fraction to bypass uORFs 2–4 and reinitiate at GCN4 instead (Olsen et al, 2003). However, this interpretation was not confirmed biochemically; nor was it determined whether Δ108–153 impairs 40S-binding of eIF1A or disrupts its biochemical function in 43S assembly.
eIF1A is related to bacterial IF1, and both proteins contain the β-barrel ‘OB'-fold (Sette et al, 1997; Battiste et al, 2000), while eIF1A also contains an α-helical domain and random-coil N- and C-terminal extensions (NTD and CTD) (Figure 1A). As IF1 binds to the A-site of the 30S subunit (Moazed et al, 1995; Carter et al, 2001), the OB-fold in eIF1A most likely binds to the A-site of 40S subunits in eukaryotes. To assign its functions to specific residues, we mutated conserved surface-exposed residues in the OB-fold, α-helical domains, NTD and CTD of yeast eIF1A, reasoning that such residues would mediate eIF1A interactions with the ribosome or other eIFs. From genetic and biochemical analysis of substitutions in the OB-fold and helical domain, and of the Δ108–153 mutation described previously, we conclude that the OB-fold plays a key role in ribosome-binding while the helical domain and CTD function in 43S assembly and scanning.
Figure 1.
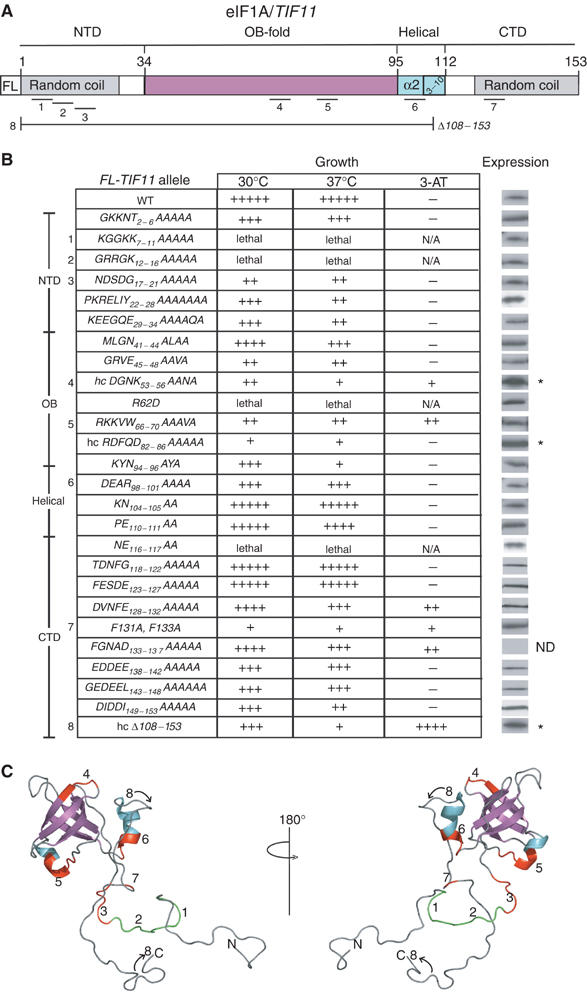
Phenotypes of eIF1A mutations. (A) Schematic representation of eIF1A domain organization, including the FLAG (FL) epitope, amino-acid positions (above) and locations of key mutations (1–8; underlined below). (B) Growth of gcn2Δ strains harboring the indicated mutant or WT FL-TIF11 alleles on SC medium lacking Leu (SC-L) at 30 or 37°C and on SC medium lacking leucine and histidine (SC–LH) and containing 10 mM 3-AT at 30°C. Relative growth is summarized on a scale from (−) to (+++++). The domain locations of the mutations are listed on the left along with designations of selected mutations 1–8 as indicated in panel A. The far-right column (Expression) contains results of Western analysis of WCEs from the same strains analyzed for growth phenotypes (except for the lethal alleles) using FLAG antibodies. Separate probing of the blots with antibodies against eIF2α ensured similar protein loading for each extract (not shown). Three FL-TIF11 alleles (*) were expressed on hc plasmids; ND, not detectable. Four additional lethal alleles were identified for which no protein products were detected: K64A, K67D, Q85A and HIRGK60−64AAAA (data not shown). (C) Predicted locations of residues altered by mutations 1–8 (described in panel A) in the 3D structure of eIF1A, designated red (viable alleles) or green (lethal alleles), as determined using MacPyMOL (DeLano, 2002).
Results
Mutagenesis of conserved surface residues of yeast eIF1A
Using the 3D structure of human eIF1A (Battiste et al, 2000) and sequence alignments of eIF1A from different species, we constructed mutations of conserved surface-exposed residues in the OB-fold and α-helical domains of yeast eIF1A (Figure 1), in most cases substituting three to five contiguous residues with alanines. We also made consecutive blocks of Ala substitutions in the NTD and CTD. All mutations were generated in an FL-TIF11 allele on a single-copy plasmid, bearing the TIF11 promoter and coding sequences for FLAG (FL) epitope at the 5′-end of the ORF (Olsen et al, 2003). The mutant alleles were used to replace episomal WT TIF11 (TIF11+) in a tif11Δ gcn2Δ strain by plasmid-shuffling.
Eight of the clustered substitutions were lethal (Figure 1B and legend), preventing eviction of TIF11+ from the strain. Western analysis of whole-cell extracts (WCEs) from strains harboring both FL-tagged mutant alleles and untagged TIF11+ with FL antibodies showed that four of the lethal alleles (altering residues 60–64, 67 or 85) produced no detectable protein and were not studied further. The other four lethal alleles produced WT levels of protein (Figure 1B), implying that they disrupt an essential function of eIF1A. We compared the 40S-binding of WT and two of the FL-tagged mutants containing the lethal substitutions R62D or NE116−117AA in cells expressing untagged eIF1A using a chemical crosslinking assay described below. The OB-fold mutation R62D strongly reduced 40S-binding of eIF1A to only ∼17% of WT, whereas the CTD mutation NE116−117AA impaired 40S-binding by <30% (data not shown). A nonlethal OB-fold mutation discussed below decreased 40S-binding more so than did NE116−117AA, suggesting that the latter probably disrupts an essential function of eIF1A carried out on the ribosome.
Western analysis of the 22 nonlethal tif11 mutants after eviction of TIF11+ showed that all but three produced WT levels of eIF1A (Figure 1B). The three exceptional alleles, DGNK53–56AANA, RDFQD82–86AAAAA and Δ108–153, required overexpression to compensate for reduced steady-state levels, and when present on high-copy (hc) plasmids, produced protein levels ∼3-fold higher than WT. All subsequent analysis was conducted with the hc versions of these alleles.
The viable mutants containing only FL-tagged alleles were compared to the FL-TIF11+ strain for growth on complete medium (SC) and on SC lacking histidine and containing 3-aminotriazole (3-AT), an inhibitor of histidine biosynthesis. The gcn2Δ mutation in these strains blocks induction of GCN4 translation and attendant derepression of histidine biosynthetic enzymes regulated by GCN4. Mutations that derepress GCN4 translation independently of eIF2 phosphorylation suppress the 3-AT-sensitive phenotype (3-ATS) of gcn2Δ cells, producing a Gcd− phenotype. All but three of the 21 nonlethal substitutions produced a slow-growth phenotype (Slg−) on SC, and five of these conferred 3-AT-resistance (3-ATR) (Figure 1B). The Δ108–153 deletion produced both Slg− and 3-ATR phenotypes, as reported previously (Olsen et al, 2003).
The RKKVW66–70AAAVA and Δ108–153 mutations disrupt PIC assembly by different mechanisms
Among the new Gcd− alleles, RKKVW66−70AAAVA was of interest because it restored growth on 3-AT medium while strongly impairing growth on SC (Figure 2B). Indeed, RKKVW66−70AAAVA elicits greater depletion of polysomes and accumulation of 80S monosomes than does Δ108–153 (Figure 2A), indicating a strong defect in translation initiation. (Henceforth, we abbreviate RKKVW66−70AAAVA and Δ108–153 as 66–70 and ΔC, respectively.) Importantly, the 3-ATR/Gcd− phenotype of 66–70 was suppressed by overexpressing all three eIF2 subunits and tRNAiMet from an hc plasmid (hc-TC) (Figure 2B, cf. rows 4–5), as observed for ΔC (Olsen et al, 2003) (cf. rows 6–7). Hence, both 66–70 and ΔC appear to derepress GCN4 by reducing TC binding to scanning 40S ribosomes. Note that overexpressing TC also reduces the derepression of GCN4 that occurs when TC recruitment is impaired by eIF2 phosphorylation in starved GCN2+ cells (Dever et al, 1995). We quantified the Gcd− phenotype of the 66–70 mutant by assaying a GCN4-lacZ reporter containing all four uORFs, which is normally expressed at low levels in gcn2Δ cells owing to the absence of eIF2α phosphorylation. The 66–70 strain showed ∼5-fold greater GCN4-lacZ expression than did the TIF11+gcn2Δ strain (Figure 2C), whereas ΔC produced ∼8-fold derepression of GCN4-lacZ expression, in accordance with previous results (Olsen et al, 2003). Derepression of GCN4-lacZ in a GCN2 TIF11 strain treated with 3-AT was considerably higher (15-fold; data not shown), indicating that these mutations only partially suppress reinitiation at uORFs 2–4. As discussed below, this probably occurs because eIF1A mutations produce multiple initiation defects with offsetting effects on GCN4 translation.
Figure 2.
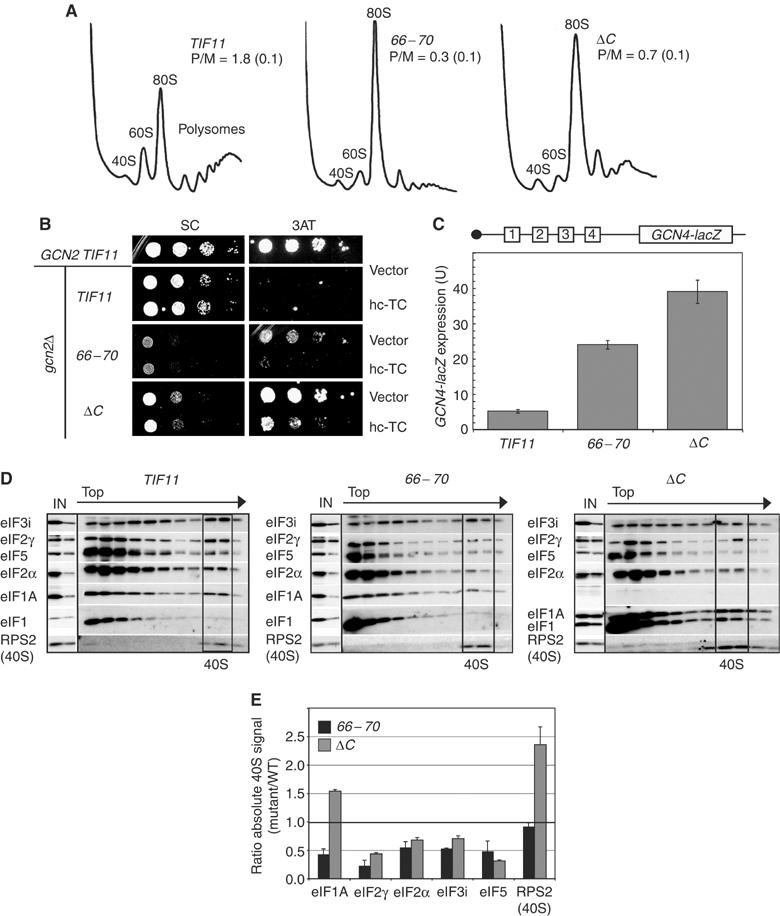
The 66–70 and ΔC mutations produce general reductions in translation initiation, derepressed translation of GCN4 and decreased 43S complex formation in vivo. (A) Polysome profiles of gcn2Δ strains harboring WT (H2999), 66–70 (H3580) or ΔC (H3002) alleles of TIF11-FL after culturing in SC-L medium to OD600≈1 and crosslinking cells with 1% (v/v) HCHO. WCEs were resolved by sedimentation through 4.5–45% sucrose gradients and gradients were scanned at A254. The polysome/monosome ratios (P/M, means±s.e., n=3) are indicated for each strain. (B) Slg− and Gcd− phenotypes were assayed by spotting serial 10-fold dilutions of the gcn2Δ strains described in (A) and isogenic GCN2 TIF11 strain H1642 (harboring YCplac111), on SC-L or SC-LH+10 mM 3-AT at 30°C. (C) Expression of the chromosomal GCN4-lacZ reporter depicted schematically was measured in the strains described in (A) grown in SC-L medium. β-Galactosidase activity in units (U) of nmol ONPG cleaved/min/mg was measured in WCEs from three three independent transformants and the mean and s.e. (n=6) values were plotted in the histogram. (D) 43S assembly was measured in the strains described in (A) after crosslinking cells with HCHO, as described in (A). WCEs were resolved by sedimentation through 7.5–30% sucrose gradients and gradient fractions were subjected to Western analysis using the indicated antibodies. In all, 1 and 0.2% aliquots of each input WCE (IN) were analyzed in parallel. The fractions containing free 40S subunits are boxed. (E) Initiation factor binding to 40S subunits in the experiment shown in (D), and in two replicate experiments, was quantified by calculating the ratio of the 40S signal in the mutant relative to the WT. The results are means±s.e. (n=3). The line at y=1.0 indicates the WT level of binding.
The 3-ATR phenotypes of the other four Gcd− mutations described in Figure 1 were not suppressed by hc-TC (data not shown). Presumably, these mutations allow a fraction of the 40S subunits that have rebound TC to leaky-scan past uORF4 and reinitiate at GCN4. This class of mutations was not analyzed further.
To obtain biochemical support for our interpretation of the genetic results on the 66–70 and ΔC mutants, we measured binding of eIF2 to 40S PICs in WCEs of cells treated with formaldehyde. This treatment crosslinks factors to 40S ribosomes in vivo, minimizing dissociation of PICs during sedimentation through sucrose gradients without addition of heparin as a stabilizing agent (Nielsen et al, 2004). The crosslinks are reversed by heating prior to resolving the fractions by SDS–PAGE for Western analysis. Typical results of this assay are shown in Figure 2D and the results of replicate experiments are quantified in Figure 2E. (Henceforth, we use the term 40S-binding to describe the association of factors with either free 40S subunits or 43S/48S PICs, which cannot be distinguished in this assay.)
The 40S-binding of the 66–70 mutant protein was reduced to <50% of WT, whereas the ΔC protein bound at a greater than WT level, probably reflecting its overexpression (Figures 2D and E). Interestingly, 66–70 also reduced the 40S-binding of the TC components eIF2α and eIF2γ, eIF3 and eIF5 to ⩽50% of WT. Similar results were obtained for the ΔC mutant (Figures 2D and E). (We chose not to quantify the 40S-binding of eIF1 in these experiments because it is overexpressed in the mutants and it frequently does not show an obvious peak in the 40S fractions of WT cells.) The total amounts of eIF3i, eIF2γ and eIF5 over the entire gradient were somewhat lower in the 66–70 and ΔC mutants versus WT (Figure 2D), even though these factors were present at WT levels in the WCEs applied to the gradients (cf. ‘IN' lanes). Hence, it appears that the unbound factors are more susceptible to degradation during centrifugation in the mutant extracts. This phenomenon was described previously for mutations in the c/NIP1 subunit of eIF3 (Valasek et al, 2004). Despite the reductions in total levels of eIF3i, eIF2γ and eIF5 in the gradients, their distributions were altered such that the proportions in the 40S fractions were reduced with corresponding increases in fractions closer to the top of the gradient (Supplementary Figures S1 and S2). Thus, it appears that eIF1A is required for optimal 40S-binding of TC, eIF3 and eIF5, and that both 66–70 and ΔC impair these assembly reactions in vivo.
In accordance with the fact that hc-TC suppressed the Gcd− phenotypes of 66–70 and ΔC, hc-TC restored 40S-binding of eIF2α and eIF2γ to WT or higher levels in both mutants, and eIF3i binding in the 66–70 strain (cf. Figures 3A and B; quantification in Figure 3C). By contrast, hc-TC did not rescue 40S-binding of eIF5 in either mutant (Figure 3C). The large amounts of eIF2 subunits in the extracts containing hc-TC made it somewhat difficult to discern 40S-binding of eIF2 from trailing of eIF2 from the upper fractions into the 40S fractions. To confirm our conclusions, we resedimented the 40S fractions from gradients similar to those shown in Figures 3A and B and quantified the factors that cosedimented a second time with the 40S subunits. The results in Figure 3D confirm that 66–70 and ΔC impair 40S-binding of eIF2, eIF3 and eIF5, and also that hc-TC restores 40S-binding of eIF2, but not eIF5, by mass action. Hence, eIF1A promotes eIF5 recruitment independent of its effect on TC, and it enhances eIF3 recruitment by mechanisms both TC-dependent (evident in 66–70 cells) and TC-independent (evident in ΔC cells).
Figure 3.
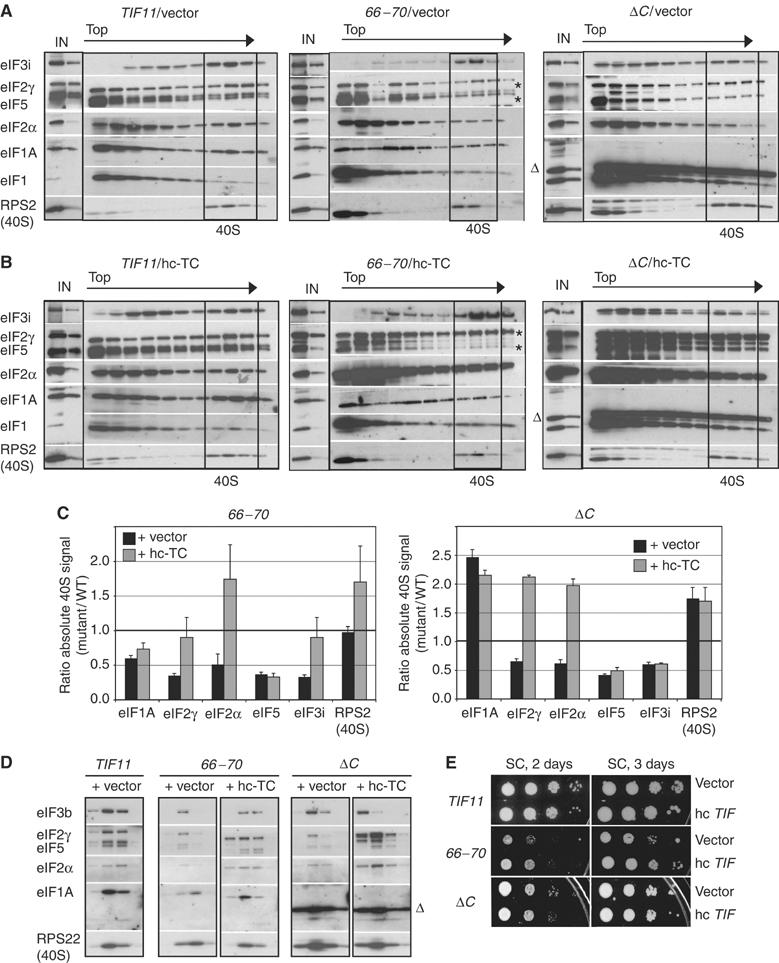
Overexpression of TC restores WT binding of TC, but not eIF5, in the 66–70 and ΔC mutants. (A, B) 43S assembly was measured in HCHO crosslinked cells of the gcn2Δ strains described in Figure 2A transformed with empty vector YEp24 (A) or p1780-IMT (hc-TC) (B), exactly as described in Figure 2. ‘Δ' indicates the ΔC product; ‘*' indicate that darker exposures of the indicated panels were presented. (C) The results in (A) and (B) were quantified as in Figure 2E, showing the means±s.e. from three experiments. (D) 40S fractions from gradients similar to (A) and (B) were subjected to resedimentation through a second sucrose gradient. Western analysis of gradient fractions containing 40S peaks in each strain is shown. (E) Overexpression of eIF5 (TIF5) partially suppresses the Slg− phenotype of the 66–70 mutant. Serial 10-fold dilutions of the gcn2Δ strains from Figure 2A transformed with empty vector YEp24 or hc TIF5 plasmid YEpTIF5-U were spotted on SC-U and grown at 30°C as indicated.
The fact that hc-TC did not restore WT 40S-binding of eIF5 in both mutants fits with our finding that the Slg− phenotypes of these mutants were not suppressed by hc-TC (Figure 2B), indicating a rate-limiting defect distinct from TC recruitment. Consistent with this, overexpressing eIF5 partially suppressed the Slg− phenotype of 66–70 (Figure 3E) and increased 40S-binding of eIF5, but did not alter 40S-binding of other factors or of the mutant eIF1A itself (Supplementary Figure S3). Thus, reduced 40S-binding of eIF5 is partly responsible for the strong initiation defect in 66–70 cells. Overexpressing eIF5 did not reduce the Slg− phenotype of the ΔC mutant, indicating an additional rate-limiting defect in this strain. As shown below, ΔC impairs ribosomal scanning and AUG recognition.
The 40S-binding of eIF1A was reduced by 66–70 to ∼50% of the WT level (Figures 2D and E). From the crystal structure of the IF1–30S complex (Carter et al, 2001), it can be predicted that this mutation substitutes OB-fold residues that interact with the A-site of the 40S subunit. Hence, the defects conferred by 66–70 could result from diminished ribosome binding of the mutant eIF1A. Supporting this idea, introducing 66–70 on an hc plasmid partly suppressed its Slg− and 3-ATR/Gcd− phenotypes (Figure 4A) and produced a greater than WT level of 40S-binding by the mutant eIF1A protein (Figures 4B and C) Thus, the 40S-binding defect of the 66–70 protein can be corrected by mass action. The hc 66–70 allele also increased 40S-binding of eIF2, eIF3 and eIF5 compared to that seen with sc 66–70, albeit not to WT levels (Figures 4B and C). The residual 43S assembly defects and Slg− phenotype (Figure 4A) in cells with hc 66–70 suggest that eIF1A function in PIC assembly is impaired by this mutation apart from its effects on 40S-binding of eIF1A itself.
Figure 4.
Overexpressing the 66–70 mutant diminishes its Slg− and Gcd− phenotypes, and the mutant is defective for 40S-binding in vitro. (A) Serial dilutions of gcn2Δ strains carrying the WT (H2999) or 66–70-tif11-FL alleles on sc or hc plasmids (strains H3580 and H3581, respectively), and H1642 transformed with YCplac111, were spotted on SC-L and SC-LH+10 mM 3-AT and grown at 30°C. (B, C) Overexpressing the 66–70 mutant partially suppresses its 43S assembly defect. 43S assembly was measured in HCHO crosslinked cells of the strains described in (A) and quantified as described in Figure 2. (D) The 66–70 mutation reduces the affinity of eIF1A for 40S subunits in vitro. Dissociation of preformed 40S/eIF1A-TAMRA complex in the presence of increasing concentrations of unlabeled WT or mutant eIF1A and 1 μM eIF1 was measured by the decrease in fluorescence anisotropy. (E) Kd values (nM) were calculated from the curves in (D) and reported as means±s.e. (F) Rate of TC binding to 40S subunits is reduced by ΔC in vitro. Binding of TC to 40S subunits as a function of time (in reactions also containing eIF1, model mRNA and 1 μM WT or mutant eIF1A) was measured as the fraction of [35S]-Met-tRNAiMet associated with the 40S in a native gel assay. The apparent TC-binding defect in the 66–70 mutant is suppressed by increasing its concentration to 10 μM.
To determine whether 66–70 impairs direct contact between eIF1A and the 40S subunit, we analyzed its effect on binding of fluorescently tagged eIF1A to purified 40S ribosomes. Tetramethylrhodamine (TAMRA)-labeled WT eIF1A was prebound to 40S subunits and unlabeled mutant or WT eIF1A was added to compete for binding of labeled eIF1A. The decrease in fluorescence anisotropy as a function of the concentration of mutant proteins allowed us to calculate the Kd for each mutant (Maag and Lorsch, 2003). As shown in Figures 4D and E, the 66–70 mutant has a Kd ∼90-fold greater than that of WT eIF1A, whereas the ΔC mutant binds to 40S subunits with WT or higher affinity. These data indicate that 66–70, but not ΔC, reduces the intrinsic affinity of eIF1A for the 40S ribosome.
We also asked whether the ΔC and 66–70 mutations impair the rate of PIC assembly in vitro. Preformed TC containing [35S]-Met-tRNAiMet was incubated with 40S subunits, eIF1, a model mRNA, and either mutant or WT eIF1A, and the fraction of labeled [35S]-Met-tRNAiMet bound to 40S subunits was measured with a native gel assay (Algire et al, 2002). As shown in Figure 4F, ΔC greatly reduced the rate of TC binding to 40S subunits in this assay. The ΔC mutant retains some activity, as the rate of TC binding was ⩾2-fold higher than in the absence of eIF1A and reached the same end point given by WT protein after 20 min (data not shown). The 66–70 protein also showed a strong defect in TC binding; however, this defect was suppressed when using a concentration of 66–70 protein large enough to overcome its 40S-binding defect (10 μM) (Figure 4F). These findings support the idea that ΔC decreases the ability of 40S-bound eIF1A to stimulate TC recruitment, whereas 66–70 primarily impairs 40S-association of the mutant protein.
TC overexpression suppresses the Slg− and 43S assembly defects in the helical domain DEAR98–101AAAA mutant
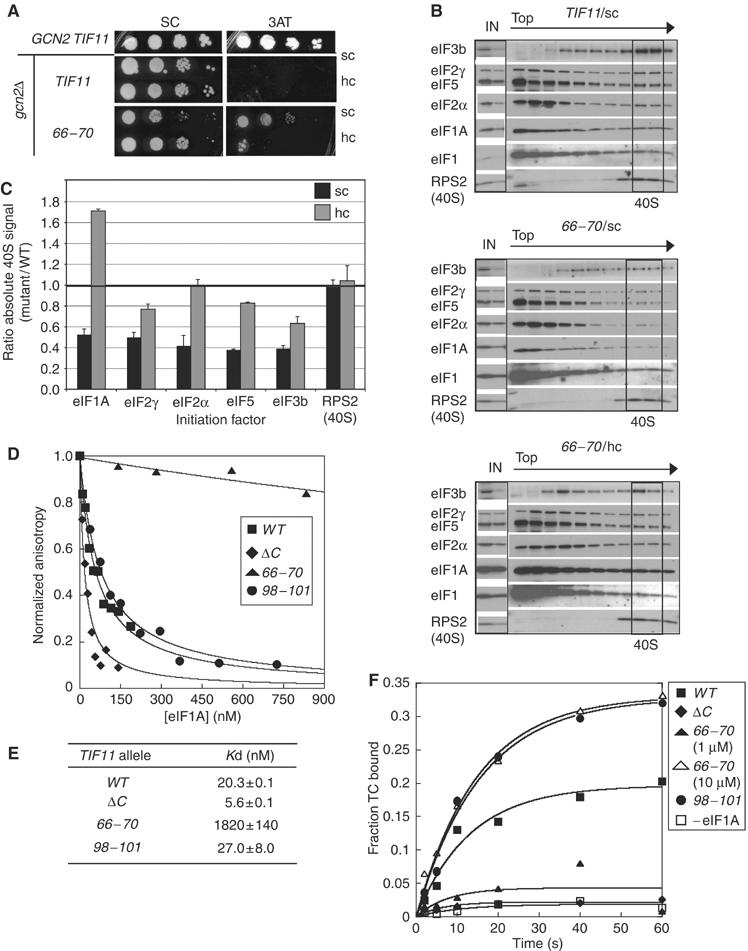
Unlike the 66–70 and ΔC mutations, the DEAR98−101AAAA substitution in the helical domain (Figure 1C, #6; abbreviated below as 98–101) produced a Slg− phenotype that is partially suppressed by hc TC (Figure 5A). Consistent with this, hc-TC decreased polysome runoff and accumulation of 80S monosomes in this mutant (Figure 5C; cf. last two profiles). In vivo analysis of 43S complexes revealed >50% reductions in 40S-bound eIF2 subunits and also reductions in 40S-binding of eIF3 (by ∼40%), eIF5 (by 80%) and eIF1A itself (by 55%), suggesting that 98–101 causes a broad defect in 43S assembly (Figures 6A and B, cf. TIF11/vector and 98–101/vector, and 6C). (As described above, there were reduced levels of eIF2, eIF3 and eIF1A in fractions at the top of the gradient, but these reductions were not evident in the input WCEs, suggesting that they arise from degradation of unbound factors in the mutant extract during centrifugation.) Consistent with the partial suppression of the growth defect in 98–101 cells by hc-TC, the 40S-binding defects of eIF2, eIF3 and eIF5, and the mutant eIF1A were diminished by hc-TC in this strain (Figures 6A and B, cf. 98–101/vector and 98–101/hc-TC). The fact that eIF3 and eIF5 recruitment was not fully rescued by hc-TC in the 98–101 mutant indicates that this mutation also impairs 40S-binding of these factors by a TC-independent mechanism.
Figure 5.
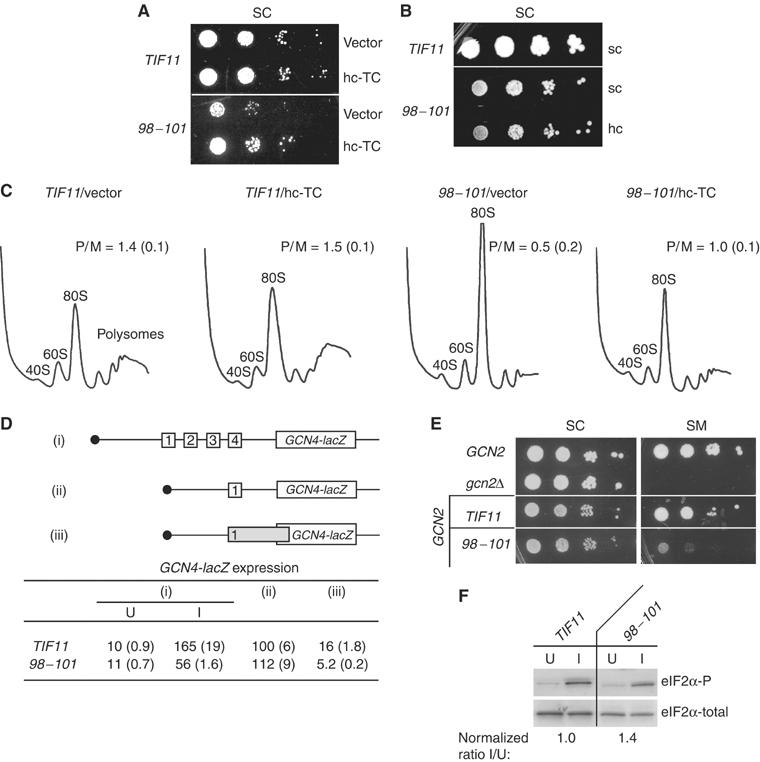
The 98–101 mutation confers a general initiation defect partially suppressed by hc-TC and also a Gcn− phenotype. (A) Serial dilutions of GCN2 strains H3583 and H3584 transformed with vector YEp24 or hc-TC plasmid p1780-IMT were spotted on SC-U and grown at 30°C for 2 days. (B) Serial dilutions of GCN2 strains containing sc WT TIF11-FL (H3583) or tif11-98–101-FL on an sc (H3584) or hc (H3585) plasmid were spotted on SC-L and grown at 30°C for 3 days. (C) Polysome profiles of the TIF11 and 98–101 transformants described in (A) were determined as described in Figure 2A. (D) The Gcn− phenotype of the 98–101 allele is not caused by impaired reinitiation or by leaky-scanning of uORF1. The GCN2 strains H3583 and H3584 harboring plasmids p180, pM199 or pM226, containing GCN4-lacZ reporters depicted schematically in (i), (ii) and (iii), respectively, were assayed for β-galactosidase activity after culturing in the absence (U) or presence (I) of 0.5 μg/ml SM for reporter (i), or in the absence of SM for reporters (ii) and (iii). Units of specific activity are means±s.e. (n=6). (E) The 98–101 mutation confers a Gcn− phenotype. GCN2 strains H3583 and H3584, together with H1642 harboring YCplac111 and H2999, were spotted on SC-L, and SC lacking Leu, Ile and Val (SC-LIV)+1 μg/ml SM, and grown at 30°C for 3–4 days. (F) The 98–101 mutation does not reduce eIF2α phosphorylation. The GCN2 strains H3583 and H3584 were grown to OD600≈1 in SC (U, uninducing) or SC-IV (I, inducing) medium and 0.5 μg/ml SM was added to the latter for 3 h at 30°C. WCEs were subjected to Western analysis using antibodies specific for eIF2α phosphorylated on Ser-51 (eIF2α-P; upper panel) and antibodies against total eIF2α lower panel), and the mean ratios of eIF2α-P/eIF2α were calculated and normalized to the WT ratio in each medium from two independent experiments.
Figure 6.
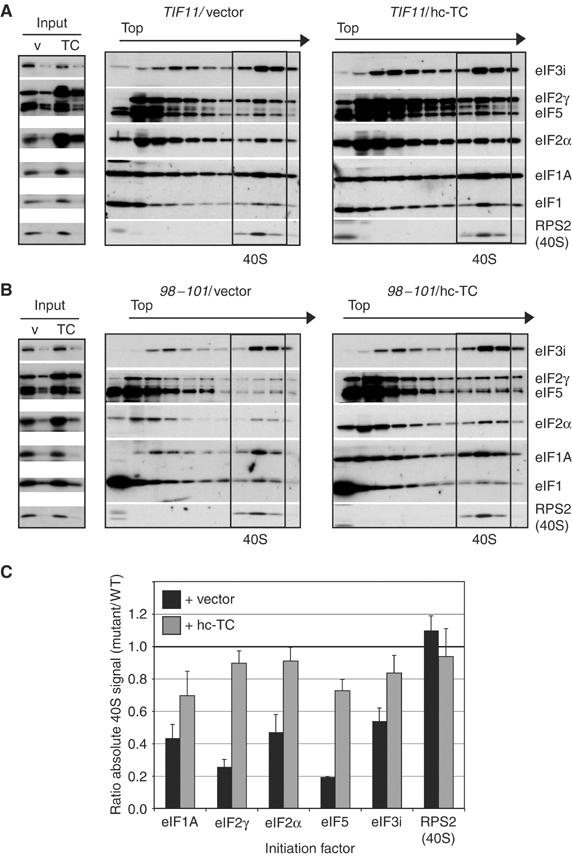
The 98–101 allele confers a defect in 43S assembly that is rescued by TC overexpression. (A–C) 43S assembly was measured in HCHO crosslinked cells of strains H3583 and H3584 bearing empty vector or hc-TC plasmid p1780-IMT, as described in Figure 2. The results quantified in panel C are means±s.e., n=3.
Although 98–101 reduces 40S-binding of eIF1A in vivo by ∼60% (Figure 6C), analysis of recombinant 98–101 protein in vitro revealed no reduction in its intrinsic affinity for the 40S subunit (Figures 4D and E). This suggests that the impaired 40S-binding of the 98–101 protein in vivo results from defective interactions with other eIFs that promote PIC assembly, rather than loss of a direct contact between eIF1A and the 40S subunit. This interpretation is supported by the fact that hc-TC increased binding of the 98–101 protein in parallel with that of eIF3 and eIF5 (Figures 6B and C). Overexpression of the 98–101 protein from an hc plasmid fully suppressed its 40S-binding defect, giving 240±70% of WT binding (data not shown), but did not diminish the growth defect (Figure 5B) or 40S-binding defects of eIF2, eIF3 or eIF5 produced by this mutation (increases of <20% compared to sc 98–101). Together, our findings suggest that 98–101 impairs a biochemical function of the helical domain required for WT PIC assembly in vivo rather than disrupting a critical contact between eIF1A and the ribosome.
The 98–101 mutation increases, rather than decreases, the rate of TC binding to 40S subunits in the reconstituted system containing purified eIF1, TC and mutant eIF1A (Figure 4F). Thus, 98–101 does not disrupt an inherent function of eIF1A in promoting TC recruitment in the manner observed for the ΔC mutation. To reconcile this conclusion with our finding that TC recruitment is impaired in 98–101 cells, and that hc-TC suppresses this defect (Figure 6), we suggest that the deficits in eIF5 and eIF3 recruitment produced by 98–101 in vivo decrease TC recruitment indirectly because of coupled 40S-binding by these factors in the context of the MFC. Overexpressing TC compensates for these defects in eIF5 and eIF3 recruitment and thereby restores 40S-binding of all three MFC components through mass action and cooperative binding to the 40S subunit.
Evidence that the 98–101 and ΔC mutations produce defects in scanning and AUG selection
Even though the 98–101 mutation reduces TC recruitment in vivo, it does not elicit a Gcd− phenotype (Figure 1B). This suggests that the mutation produces a postassembly defect that suppresses the derepression of GCN4 translation expected from impaired TC recruitment. A similar phenomenon has been described for mutations in eIF5 that produce multiple defects in the initiation pathway by disrupting the MFC (Nielsen et al, 2004; Singh et al, 2005). Supporting this interpretation, 98–101 produces a Gcn− phenotype in a GCN2+ strain, decreasing GCN4-lacZ expression to ∼30% of WT in response to Ile/Val starvation (Figure 5D, construct i) and conferring sensitivity to an inhibitor of isoleucine/valine biosynthesis (sulfometuron methyl (SM)) (Figure 5E). The Gcn− phenotype of the 98–101 mutant does not arise from diminished eIF2α phosphorylation in the starved cells (Figure 5F).
One explanation for a Gcn− phenotype in cells containing ample phosphorylated eIF2α is that ribosomes leaky-scan through the uORF1 AUG and initiate first at uORFs 2, 3 or 4 instead. Following translation of these uORFs, the ribosomes dissociate from the mRNA and fail to reach the GCN4 start codon. To address this possibility, we assayed a GCN4-lacZ reporter containing only an elongated version of uORF1 that overlaps the GCN4 start codon (Figure 5D, construct iii). Expression of this construct is normally very low because nearly all 40S ribosomes translate uORF1 but fail to reinitiate at GCN4, as elongating uORF1 destroys its ability to retain 40S subunits that resume scanning after termination (Grant et al, 1994). Leaky-scanning of the elongated uORF1 in response to the 98–101 mutation would be expected to increase expression of this reporter; however, we observed decreased reporter expression instead (∼30% of WT; Figure 5D, iii). Thus, 98–101 leads to lower, rather than higher, leaky-scanning through uORF1.
Another possible explanation for the Gcn− phenotype of 98–101 is that it impairs the ability of 40S subunits to resume scanning and reinitiate at GCN4 following translation of uORF1. This mechanism can also be eliminated because 98–101 does not decrease expression of the GCN4-lacZ reporter containing only WT uORF1 (Figure 5D, construct ii). This last finding, that eliminating uORFs 2–4 suppresses the deleterious effect of 98–101 on GCN4 translation, strongly suggests that the Gcn− phenotype results from the inability of rescanning 40S subunits to bypass uORFs 2–4 even when TC recruitment is diminished by eIF2α phosphorylation. A Gcn− phenotype with the same characteristics was observed previously when the distance between uORFs 1 and 4 was expanded to provide increased time for rebinding of TC before the rescanning 40S subunits reach uORF4 (Abastado et al, 1991). Hence, 98–101 may mimic the effect of increased uORF1–4 separation by reducing the rate of 40S scanning between uORFs 1 and 4. A reduced efficiency of scanning could also explain the decrease in leaky-scanning past elongated uORF1 produced by 98–101 (Figure 5D, iii), as a structural impediment to scanning was shown previously to reduce leaky-scanning through an AUG codon (Kozak, 1990).
We sought next to obtain biochemical evidence that the 98–101 mutation decreases the efficiency of ribosomal scanning. Mammalian eIF1A participates with eIF1 in allowing the 43S complex to scan from the 5′-end of mRNA and form a stable 48S complex at the start codon in reactions containing purified ribosomes, eIFs and β-globin mRNA. Inhibition of primer extension by reverse transcriptase is used to map the leading edge of the ribosome in the 48S complexes (toe-printing) (Pestova et al, 1998). We first demonstrated that yeast eIF1A can substitute for mammalian eIF1A in such reactions, and then examined the effects of eIF1A mutations on this activity.
With all human factors except eIF1A present, a significant proportion of 48S complexes were formed with the AUG codon in the P-site, yielding toe-prints 16 nucleotides (nt) downstream of the start codon (complex II) (Figure 7A). A sizable fraction of aberrant complexes also occurred with toe-prints 16 nt from a near-cognate GUG codon. Smaller proportions of aberrant complexes near the 5′-terminus (complex I) or with toe-prints 7 nt upstream of complex II (designated complex III) also formed in the absence of eIF1A. Supplementation with WT yeast or human eIF1A eliminated or strongly suppressed formation of complex I and the GUG complex, while yeast eIF1A also suppressed complex III (Figure 7A). Thus, WT yeast eIF1A functions efficiently in this assay to promote nearly exclusive formation of 48S PICs at the AUG codon.
Figure 7.
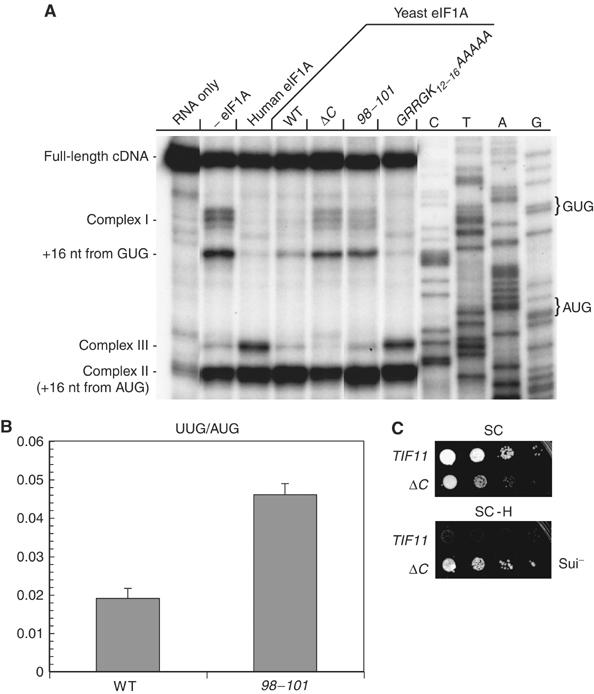
C-terminal mutations in eIF1A cause scanning/AUG recognition defects in vitro and in vivo. (A) Toe-printing analysis of 48S complexes on β-globin mRNA in reaction mixtures containing 40S subunits and mammalian translation initiation factors in the absence or presence of eIF1A, as indicated. See text for explanation of different complexes labeled on the left. (B) The 98–101 mutation increases utilization of a UUG start codon in vivo. Strains H3583 and H3584 carrying plasmids YCp50-HIS4-AUG-lacZ or YCp50-HIS4-UUG-lacZ were grown in SC-U medium and assayed for β-galactosidase activity. The specific activities±s.e. (n=6) determined for the YCp50-HIS4-UUG-lacZ versus YCp50-HIS4-AUG-lacZ transformants were measured in three independent experiments and used to calculate the mean ratios±s.e. (n=3) shown in the histogram. (C) The ΔC mutation suppresses the His− phenotype conferred by the his4-303 allele. Serial 10-fold dilutions of strains H3583 and H3586 were spotted on SC lacking leucine and histidine and grown at 30°C for 2 days (SC, upper panel) or 14 days (SC-H, lower panel).
Interestingly, the 98–101 and ΔC proteins allowed formation of complex I and the GUG complex at levels ∼50% of that observed in the absence of eIF1A, whereas a different yeast mutant (GRRGK12−16AAAAA) showed WT inhibition of these aberrant complexes. The latter finding is significant because the GRRGK12−16AAAAA mutant showed a lower affinity for 40S ribosomes in the binding assays of Figures 4D and E than did the 98–101 mutant (data not shown). Hence, it is unlikely that the mutant phenotypes of 98–101 (or ΔC) in the toe-print assays arise from reduced 40S-binding of mutant eIF1A proteins. We interpret these results to indicate that 98–101, and ΔC decrease the propensity of the 43S complex to scan the mRNA, increasing the stability of aberrant 48S complexes upstream of the AUG start codon (Battiste et al, 2000).
Finally, we asked whether the 98–101 or ΔC mutations lead to increased utilization of a near-cognate start codon in vivo. This defect can be detected by increased initiation at a UUG triplet in the 5′-end of the HIS4 ORF, allowing expression of the his4-303 allele lacking the normal AUG start codon. The resulting histidine auxotrophy indicates a Sui− (suppressor of initiation codon mutation) phenotype (Donahue, 2000). The ΔC mutation strongly suppressed the His− phenotype of his4-303, conferring a robust Sui− phenotype (Figure 7C). Furthermore, we found that ΔC is lethal in the presence of a dominant Sui− allele of TIF5 encoding a hyperactive form of eIF5 that likely produces a Sui− phenotype by increasing GTP hydrolysis at UUG codons (Huang et al, 1997). The synthetic lethality of the SUI5 tif11-ΔC combination may result from an intolerable level of initiation at near-cognate start codons. Although the 98–101 mutation did not suppress the His− phenotype associated with his4-303 (data not shown), it produced a 2.4-fold increase in translation of a his4-lacZ reporter containing UUG versus AUG as the start codon (Figure 7B). Hence, we believe that 98–101 also confers a Sui− phenotype, but weaker than that of ΔC.
Discussion
Our finding that the ΔC and 66–70 mutations derepress GCN4 translation in a manner suppressed by TC overexpression provides genetic evidence that eIF1A promotes TC recruitment in vivo. Consistent with this, we showed that both mutations impair binding of TC, eIF3 and eIF5 to 40S subunits in vivo, and that the defects in TC recruitment can be overcome through mass action by overexpressing the TC. Both mutations also conferred reduced rates of TC binding to 40S subunits in a reconstituted in vitro assay, although the defect produced by 66–70 was suppressed by using elevated levels of the mutant protein to compensate for its defect in 40S-binding. In agreement with this last result, the Slg− and Gcd− phenotypes and defects in PIC assembly produced by 66–70 in vivo were diminished by overexpressing the mutant protein. Thus, the TC recruitment defect and attendant derepression of GCN4 conferred by 66–70 arise largely from its defect in 40S-binding. This conclusion fits with the prediction from the 30S–IF1 complex (Carter et al, 2001) that residues 66–70 in the OB-fold of eIF1A are close to the A-site of the 40S subunit. The ΔC mutation, by contrast, impairs 43S assembly even though the mutant eIF1A is bound to 40S subunits at WT or greater levels. Hence, the C-terminal extension or adjacent 310 helix eliminated by ΔC appears to have an important biochemical function in TC recruitment.
Whereas overexpressing TC reduced the Gcd− phenotypes and TC recruitment defects in the 66–70 and ΔC mutants, it did not suppress their Slg− phenotypes nor restore 40S-binding of eIF5 or eIF3 (in the ΔC mutant) to WT levels. By contrast, overexpressing eIF5 rescued 40S-binding of eIF5 and diminished the growth defect in 66–70 cells. These results indicate that eIF1A promotes 40S-binding of eIF5 and eIF3 independent of its role in TC recruitment. Hence, we conclude that eIF1A independently promotes the 40S-binding of multiple constituents of the yeast MFC. As no direct physical interactions between eIF1A and MFC components have been described, the molecular basis for the stimulatory effects of eIF1A on MFC recruitment remains to be defined.
The 98–101 substitution in the α-helical domain was the only mutation that conferred a growth defect suppressible by hc-TC, and the mutation also impaired 40S-binding binding of TC, eIF3 and eIF5 in vivo in a manner largely corrected by overexpressing TC. The 98–101 mutation impairs 40S-binding of eIF1A itself in vivo, and while this defect was corrected by overexpressing the mutant protein, the Slg− phenotype and reductions in PIC assembly were not simultaneously suppressed. Hence, we conclude that the helical domain of eIF1A has an important function in promoting recruitment of the MFC to the 40S subunit.
The recombinant 98–101 protein exhibits WT affinity for 40S ribosomes in vitro, indicating that reduced 40S-binding by this mutant in vivo does not reflect loss of a contact between eIF1A and the ribosome. Similarly, 98–101 did not reduce the rate of TC binding to 40S subunits in the reconstituted assay, suggesting that the intrinsic function of eIF1A in TC recruitment is not impaired by this mutation. To reconcile these in vitro and in vivo findings, we suggest that 98–101 indirectly impairs TC recruitment in vivo by reducing 40S-binding of eIF3 and eIF5, thereby disrupting the coupled association of factors in the MFC with the ribosome. We further suggest that 40S-binding of eIF1A is coupled to that of the MFC, so that the 98–101 mutation indirectly impairs 40S-binding of eIF1A by diminishing recruitment of MFC components. Indeed, we recently obtained evidence that depleting eIF5 from cells impairs eIF1A recruitment in vivo (A Jivotovskaya, K Nielsen and AGH, unpublished observations). We suggest that overexpressing TC most likely restores 40S-binding of the MFC in the 98–101 mutant by driving the assembly of a TC–eIF5 binary complex on the ribosome by mass action. This in turn would stimulate recruitment of eIF3, as interaction with eIF2 increases the affinity of eIF5 for eIF3 (Singh et al, 2004).
Apart from its role in PIC assembly, we found that the C-terminal domain of eIF1A also functions in scanning and AUG recognition. Toe-print analysis of 48S assembly in a reconstituted mammalian system revealed that both the 98–101 and ΔC mutations increase formation of aberrant complex I and the 48S complexes at an upstream GUG on β-globin mRNA. As suggested previously (Battiste et al, 2000), formation of these aberrant complexes could result from a decreased rate of ribosomal scanning. Consistent with these findings, the 98–101 and ΔC mutations appeared to increase utilization of non-AUG triplets as start codons on HIS4 mRNA in vivo (Sui− phenotype).
A slower rate of scanning can also explain why the defect in TC recruitment conferred by 98–101 in vivo does not produce a Gcd− phenotype. In fact, this mutant displays a Gcn− phenotype in GCN2 cells that can be attributed to the failure of 40S ribosomes rescanning downstream from uORF1 to bypass uORFs 2–4 even though TC levels are reduced by eIF2α phosphorylation. This type of Gcn− defect was observed previously when the distance between uORFs 1 and 4 was expanded, increasing the time required for scanning 40S subunits to reach uORF4 after terminating at uORF1. This alteration compensates for the delay in TC binding elicited by eIF2α phosphorylation and ensures that nearly all 40S subunits rebind TC before reaching uORF4 and, hence, fail to reinitiate at GCN4 (Abastado et al, 1991). Thus, we suggest that the 98–101 mutation mimics the effect of lengthening the uORF1–4 interval and produces a Gcn− phenotype by decreasing the rate of scanning.
It might be expected that the ΔC mutation would also confer a Gcn− phenotype, given that it produced a scanning defect in the toe-print assay comparable to that of 98–101. In fact, we found that ΔC does confer sensitivity to SM and a modest (20%) reduction in GCN4-lacZ expression in a GCN2+ strain under starvation conditions, without lowering eIF2α phosphorylation (data not shown). Perhaps the defect in TC binding produced by ΔC outweighs the scanning defect and suppresses the expected Gcn− phenotype. In 98–101 cells, by contrast, the scanning defect would outweigh the deficit in TC binding and suppress the expected Gcd− phenotype.
Support for this interpretation came recently from the analysis of mutations in the eIF5-CTD that destabilize the MFC and produce Gcd− phenotypes at 30°C but Gcn− phenotypes at a higher temperature (36°C) where the mutations have a stronger effect on initiation. It was proposed that impaired TC recruitment at 30°C produces the Gcd− phenotype but more severe disruption of the MFC at 36°C elicits a scanning defect (leaky-scanning of uORF1 in this case) that outweighs the effect of reduced TC recruitment on GCN4 translation (Singh et al, 2005). We suggest that the ΔC and 98–101 mutations can be likened to these eIF5-CTD mutations at 30 and 36°C, respectively, in terms of their relative effects on TC recruitment versus scanning. Singh et al (2005) also showed that certain eIF5-CTD mutants display a Gcd− phenotype only when eIF1 is overexpressed, apparently because eIF1 overexpression exacerbates a moderate defect in TC recruitment to the point where it outweighs the scanning defects produced by these eIF5 mutations, thus derepressing GCN4. Consistent with this, we found that eIF1 overexpression reveals a Gcd− phenotype in the 98–101 mutant and exacerbates the Gcd− phenotype of ΔC (data not shown).
Our results show that the α-helical domain and random-coil CTD of eIF1A, regions not present in bacterial IF1, stimulate binding of TC and other MFC components to the 40S subunit and promote efficient scanning and AUG selection. Residues 66–70 in the OB-fold, by contrast, likely provide a critical 40S–eIF1A interaction. It makes sense that functions of eIF1A involved in MFC recruitment and scanning depend on domains not present in IF1, as eIF2, eIF3 and eIF5 are absent in bacteria, and there are significant differences in Met-tRNAiMet recruitment and start codon recognition between eukaryotes and prokaryotes.
Materials and methods
Yeast strains and plasmid construction
The plasmids and yeast strains involving the 66–70, 98–101 and ΔC mutations are listed in Tables I and II. All other plasmids and yeast strains for the remaining TIF11 mutations are listed in the Supplementary data along with details of their construction, and the biochemical and genetic methods employed.
Table 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| YCplac111 | sc LEU2 cloning vector | Gietz and Sugino (1988) |
| YEplac181 | hc LEU2 cloning vector | Gietz and Sugino (1988) |
| p3392 | sc URA3 TIF11 in YCplac33 | Olsen et al (2003) |
| p3499 | sc LEU2 TIF11-FL in YCplac111 | Olsen et al (2003) |
| p3604 | hc LEU2 tif11-Δ108–153-FL in YEplac181 | Olsen et al (2003) |
| p4399 | sc LEU2 tif11-RKKVW66–70AAAVA-FL in YCplac111 | This study |
| p4443 | hc LEU2 tif11-RKKVW66–70AAAVA-FL in YEplac181 | This study |
| p4398 | sc LEU2 tif11-DEAR98–101AAAA-FL in YCplac111 | This study |
| p4444 | hc LEU2 tif11-DEAR98–101AAAA-FL in YEplac181 | This study |
| p1780-IMT | hc URA3 SUI2, GCD11, SUI3, IMT4 in YEp24 | Asano et al (1999) |
| YEp24 | hc URA3 cloning vector | Parent et al (1985) |
| YEpTIF5-U | hc URA3 TIF5-FL in YEplac195 | Valasek et al (2004) |
| YEplac195 | hc URA3 | Gietz and Sugino (1988) |
| p4385 | hc TRP1 SUI2, GCD11, SUI3, IMT4 in YEp24, ura3∷TRP1:kanR | This study |
| YEplac112 | hc TRP1 | Gietz and Sugino (1988) |
| p180 | sc URA3 GCN4-lacZ with WT leader | Hinnebusch (1985) |
| PM199 | sc URA3 GCN4-lacZ with uORF1 only | Grant et al (1994) |
| PM226 | sc URA3 GCN4-lacZ with uORF1 extending into GCN4 | Grant et al (1994) |
| p367 | sc URA3 AUG-HIS4-lacZ | Donahue and Cigan (1988) |
| p391 | sc URA3 AUU-HIS4-lacZ | Donahue and Cigan (1988) |
Table 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| H1642 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 | |
| <GCN4-lacZ, TRP1> | Dever et al (1992) | |
| H2809 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 gcn2Δ tif11Δ | |
| <GCN4-lacZ, TRP1>p3392 (sc TIF11, URA3) | Choi et al (2000) | |
| H2999 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 gcn2Δ tif11Δ | |
| <GCN4-lacZ, TRP1>p3499 (sc TIF11-FL, LEU2) | Olsen et al (2003) | |
| H3002 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 gcn2Δ tif11Δ | |
| <GCN4-lacZ, TRP1>p3604 (hc tif11-Δ108–153-FL, LEU2) | Olsen et al (2003) | |
| H3580 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 gcn2Δ tif11Δ | |
| <GCN4-lacZ, TRP1>p4399 (sc tif11-RKKVW66–70AAAVA-FL, LEU2) | This study | |
| H3581 | MATa ura3–52 leu2–3 leu2–112 trp1Δ63 gcn2Δ tif11Δ | |
| <GCN4-lacZ, TRP1>p4443 (hc tif11-RKKVW66–70AAAVA-FL, LEU2) | This study | |
| H3582 | MATa ura3–52 trp1Δ63 leu2–3, leu2–112 | |
| his4–303 (ATT) tif11Δ p3392 (sc TIF11, URA3) | This study | |
| H3583 | MATa ura3–52 trp1Δ63 leu2–3, leu2–112 | |
| his4–303 (ATT) tif11Δp3499 (sc TIF11-FL, LEU2) | This study | |
| H3584 | MATa ura3–52 trp1Δ63 leu2–3, leu2–112 | |
| his4–303 (ATT) tif11Δp4398 (sc tif11-DEAR98–101AAAA-FL, LEU2) | This study | |
| H3585 | MATa ura3–52 trp1Δ63 leu2–3, leu2–112 | |
| his4–303 (ATT) tif11Δ p4444 (hc tif11-DEAR98–101AAAA-FL, LEU2) | This study | |
| H3586 | MATa ura3–52 trp1Δ63 leu2–3, leu2–112 | |
| his4–303 (ATT) tif11Δ p3604 (hc tif11-Δ108–153-FL, LEU2) | This study |
Supplementary Material
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Acknowledgments
We thank Tom Dever and members of our laboratories for helpful suggestions.
References
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG (1991) Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, Hinnebusch AG, Lorsch JR (2002) Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG (2000) A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bɛ, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 18: 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valášek L, Donahue TF, Hinnebusch AG (2001) Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J 20: 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JB, Pestova TV, Hellen CUT, Wagner G (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell 5: 109–119 [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE (2000) Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol 20: 7183–7191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR (1997) ‘Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13: 647–653 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific. http://www.pymol.org [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TD, Hinnebusch AG (1992) Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596 [DOI] [PubMed] [Google Scholar]
- Dever TE, Yang W, Åström S, Byström AS, Hinnebusch AG (1995) Modulation of tRNAiMet, eIF-2 and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNAiMet ternary complexes. Mol Cell Biol 15: 6351–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T (2000) Genetic approaches to translation initiation in Saccharomyces cerevisiae. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB and Mathews MB (eds), pp 487–502. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Donahue TF, Cigan AM (1988) Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol 8: 2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Grant CM, Miller PF, Hinnebusch AG (1994) Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol Cell Biol 14: 2616–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K (2003) The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 23: 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) Pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB and Mathews MB (eds), pp 33–88. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hinnebusch AG (1985) A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (1996) Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In Translational Control, Hershey JWB, Mathews MB and Sonenberg N (eds), pp 199–244. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hinnebusch AG (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB and Mathews MB (eds), pp 185–243. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Huang H, Yoon H, Hannig EM, Donahue TF (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 11: 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV (2005) Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1990) Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA 87: 8301–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Maag D, Lorsch JR (2003) Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol 330: 917–924 [DOI] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U (2003) Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J Biol Chem 278: 6580–6587 [DOI] [PubMed] [Google Scholar]
- Moazed D, Samaha RR, Gualerzi C, Noller HF (1995) Specific protection of 16S rRNA by translational initiation factors. J Mol Biol 248: 207–210 [DOI] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG (1991) Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 11: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG (2004) Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J 23: 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG (2003) Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent SA, Fenimore CM, Bostian KA (1985) Vector systems for the expression, analysis and cloning of DNA sequences in S.cerevisiae. Yeast 1: 83–138 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol 18: 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M, van Tilborg P, Spurio R, Kaptein R, Paci M, Gualerzi CO, Boelens R (1997) The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J 16: 1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence CW (1974) Methods of Yeast Genetics, pp 61–64. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Singh CR, Curtis C, Yamamoto Y, Hall NS, Kruse DS, He H, Hannig EM, Asano K (2005) Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol Cell Biol 25: 5480–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Yamamoto Y, Asano K (2004) Physical association of eukaryotic initiation factor (eIF) 5 carboxyl-terminal domain with the lysine-rich eIF2beta segment strongly enhances its binding to eIF3. J Biol Chem 279: 49644–49655 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG (2003) The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Nielsen KH, Hinnebusch AG (2002) Direct eIF2–eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J 21: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG (2004) Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3


