Abstract
Nuclear envelope (NE) formation during cell division in multicellular organisms is a central yet poorly understood biological process. We report that the conserved nucleoporin Nup155 has an essential function in NE formation in Caenorhabditis elegans embryos and in Xenopus laevis egg extracts. In vivo depletion of Nup155 led to failure of nuclear lamina formation and defects in chromosome segregation at anaphase. Nup155 depletion inhibited accumulation of nucleoporins at the nuclear periphery, including those recruited to chromatin early in NE formation. Electron microscopy analysis revealed that Nup155 is also required for the formation of a continuous nuclear membrane in vivo and in vitro. Time-course experiments indicated that Nup155 is recruited to chromatin at the time of NE sealing, suggesting that nuclear pore complex assembly has to progress to a relatively late stage before NE membrane assembly occurs.
Keywords: Caenorhabditis elegans, NPC, nuclear envelope, Nup155, Xenopus laevis
Introduction
The nuclear envelope (NE) of eukaryotes serves as an important barrier between the cell nucleus and the cytoplasm, preventing free diffusion of macromolecules between the two compartments and separating the nucleic acid and protein synthesis machineries (Gant and Wilson, 1997; Vasu and Forbes, 2001). The NE consists of an outer (ONM) and an inner nuclear membrane (INM), which are joined at nuclear pore complexes (NPCs) that form aqueous channels in the NE. The ONM is continuous with the endoplasmic reticulum, whereas the INM is characterized by multiple INM-specific proteins including MAN1, emerin and lamin B receptor. The nuclear lamins, intermediate filament-type proteins, form a network adjacent to the INM. Interactions between lamins and INM proteins are important for localizing the INM proteins. In addition, the lamins and several INM proteins interact directly with chromatin and/or with chromatin-associated proteins such as BAF and HP1 (Gruenbaum et al, 2005).
When cells of multicellular organisms enter mitosis, the NE breaks down. This involves disassembly of the nuclear lamina by phosphorylation and solubilization of lamins while NE membranes and their integral proteins are partially or entirely absorbed into the endoplasmic reticulum (Mattaj, 2004 and references therein). Most nucleoporins are soluble during mitosis, although some are integral membrane proteins. During anaphase or telophase, depending on the cell type, the NE starts to reform. In vitro models have been developed to recapitulate NE formation, and in particular Xenopus laevis egg extracts provide a useful biochemical system to study the individual steps of NE assembly (Lohka and Masui, 1983; Gant and Wilson, 1997). Initially, chromatin decondenses and membranes and some early nucleoporins associate with chromatin. Fusion of membranes and recruitment of additional nucleoporins lead to a complete enclosure of chromatin by an NE containing NPCs, followed by nuclear growth and complete decondensation of chromatin.
NPCs are large structures consisting of multiple copies of approximately 30 different nucleoporins (Rout et al, 2000; Vasu and Forbes, 2001; Cronshaw et al, 2002; Suntharalingam and Wente, 2003). To analyse NE formation in vivo, we previously identified 20 Caenorhabditis elegans nucleoporins and showed that a subset is required for normal nuclear appearance, suggesting an important role in NE formation (Galy et al, 2003). This was unexpected, since in vitro studies had shown that a closed NE can form in the absence of NPCs (Macaulay and Forbes, 1996; Harel et al, 2003; Walther et al, 2003). One of the nucleoporins whose depletion from C. elegans disrupted nuclear assembly was the highly conserved Nup155.
Nup155 was first identified in rodent cells and shown to be localized symmetrically to both the nucleoplasmic and cytoplasmic faces of the NPC (Radu et al, 1993). Vertebrate Nup155 has been reported to interact with both Nup35 (Nup35 is also known as Nup53 due to its homology with yeast Nup53p) and the mRNA export factor Gle1, but the function of these interactions is unknown (Rayala et al, 2004; Hawryluk-Gara et al, 2005). Drosophila Nup155 is an essential gene (Gigliotti et al, 1998; Kiger et al, 1999), while Saccharomyces cerevisiae contains two genes with high homology to vertebrate Nup155, NUP170 and NUP157. Individually, neither is essential but mutant yeast lacking both Nup155 homologues are inviable (Aitchison et al, 1995).
We show here that Nup155 is required for both NE membrane fusion and NPC assembly. Using a combination of in vivo and in vitro analyses, we find that depletion of Nup155 prevents accumulation of several nucleoporins, including those recruited to chromatin early during nuclear assembly like Nup107 and Pom121, at the NE. In the absence of Nup155, nuclear membranes dock to chromatin but do not fuse to form a closed NE. Surprisingly, Nup155 is recruited relatively late during NE formation, suggesting that Nup155 defines an essential late step in NE assembly subsequent to Nup107 and Pom121 recruitment.
Results
Analysis of nuclear morphology upon Nup155 depletion in vivo
Our previous analysis of nucleoporins in C. elegans by differential interference contrast (DIC) microscopy demonstrated that Nup155 is required for normal nuclear appearance (Galy et al, 2003). To address the role of Nup155 in NE formation in detail, we initially depleted Nup155 from embryos coexpressing fluorescent β-tubulin and lamin and monitored the embryos by time-lapse confocal microscopy. In control embryos, the pronuclei were clearly visible by DIC and fluorescence microscopy. Centrosomes were closely associated with the pronuclear envelopes and a mitotic spindle formed (Figure 1A, left panels, time points −4:00–0:40). The NE reformed rapidly after sister chromatid separation and lamin was imported and assembled to a lamina (Figure 1A, left panels, 3:20–9:00). In contrast, in Nup155 RNAi embryos, pronuclei were not visible either by DIC microscopy or by lamin staining and centrosomes separated prematurely (Figure 1A, right panels, −4:00 to −1:00) (n=3/3 embryos), presumably due to a lack of anchoring to the pronuclear envelopes. Following mitosis, the embryos failed to assemble a visible nucleus or nuclear lamina, suggesting a severe defect in NE formation (Figure 1A, right panels, 3:20–9:00).
Figure 1.
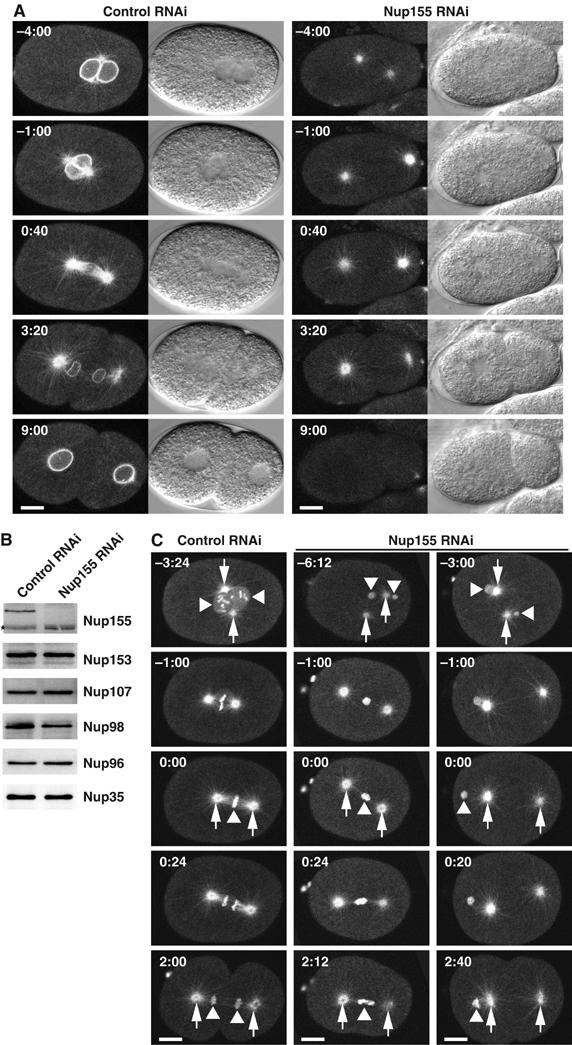
Depletion of Nup155 from C. elegans embryos affects nuclear appearance. (A) Embryos expressing GFP-β-tubulin and YFP-lamin from control (left panels) or Nup155-depleted worms (right panels) were observed by time-lapse microscopy. In this and other figures, images are shown with indication of time (minutes:seconds) relative to anaphase onset and with embryos anterior to the left. In the Nup155 RNAi embryo, neither pronuclei nor nuclei were visible. See Supplementary video 4. (B) Western blot analysis of embryonic lysates from mock-depleted (left lane) or Nup155-depleted worms (right lane) with antisera against various nucleoporins. The asterisk marks a protein that crossreacts with the Nup155 antibody, and demonstrates equal gel loading. (C) Control (left) or Nup155-depleted (middle and right) embryos expressing GFP-β-tubulin and GFP-histone H2B were observed by time-lapse microscopy. In selected panels, centrosomes are indicated with arrows whereas triangles point to chromatin. See Supplementary video 5. Scale bars, 10 μm.
To investigate the efficiency and specificity of the knockdown of Nup155, we analysed extracts from Nup155 RNAi embryos by Western blotting. Compared to control-depleted extracts, Nup155 RNAi reduced the level of Nup155 protein below the detection limit without affecting five other nucleoporins significantly (Figure 1B). Thus, the phenotypes caused by Nup155 RNAi can likely be ascribed entirely to depletion of Nup155.
The observation that pronuclear appearance was strongly disturbed by Nup155 depletion prompted us to examine chromatin in the affected embryos using a strain expressing GFP fusions of β-tubulin and histone H2B. In control embryos, chromosomes could be monitored from condensation in prophase through congression at the metaphase plate to segregation during anaphase (Figure 1C, left column). In Nup155-depleted embryos, the pronuclei were dramatically reduced in size, consistent with not being detectable by DIC microscopy, and individual chromosomes could not be recognized (Figure 1C, middle and right columns). In most embryos (n=6/9 embryos), the pronuclei did not meet and the weaker association between pronuclei and centrosomes discussed above (Figure 1A) was confirmed. Two classes of mitotic defect were observed. In some embryos, chromatin was positioned between the two centrosomes, and microtubules connecting the centrosomes to chromatin were visible (n=6/9 embryos). These embryos displayed a strong anaphase segregation phenotype, with chromatin trapped between the centrosomes even after formation of the cleavage furrow (Figure 1C, middle column, and data not shown). In the second class, no chromatin was detected between the centrosomes, and a mitotic spindle was not formed (n=3/9 embryos) (Figure 1C, right column). The presence of two polar bodies in all Nup155 RNAi embryos indicated that meiosis was not significantly impaired, and no defects in oogenesis were detected (Figure 1C, and data not shown).
Recruitment of nucleoporins to the NE in Nup155-depleted embryos
The observation that nuclear appearance was severely affected by the depletion of Nup155 led us to investigate the fate of individual nucleoporins and NPC formation. Nup155-depleted embryos were fixed and analysed with antibodies specific for Nup35, Nup96 and Nup153 as well as with mAb414, which in C. elegans predominantly recognizes Nup96, Nup98 and Nup358 (Galy et al, 2003). While all four antibodies gave rise to clear nuclear rim staining in control embryos (Figure 2A and B), the patterns were very different in Nup155 RNAi embryos. Nup153 still associated with chromatin but no longer accumulated at the nuclear periphery (Figure 2A). Nup96 showed a variable pattern with both chromatin staining and some clustering on the surface of chromatin. The nuclear accumulation of Nup96 was clearly diminished and cytoplasmic foci were observed (Figure 2B). Most dramatically, Nup35 was completely absent from the chromatin of Nup155 RNAi embryos (Figure 2B). In all embryos, mAb414 staining was highly irregular with prominent cytoplasmic foci in which Nup96 was colocalized (Figure 2B).
Figure 2.
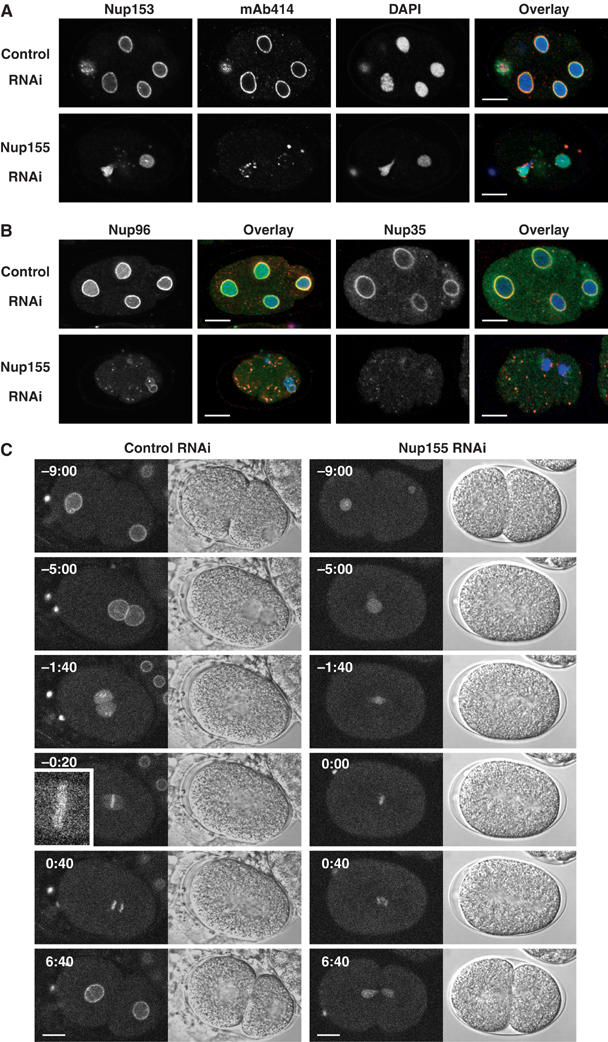
C. elegans Nup155 is required for NPC assembly. (A) Control (upper panels) or Nup155-depleted (lower panels) embryos were fixed and analysed using anti-Nup153 antibodies (green in overlay) or mAb414 (red in overlay). Chromatin was visualized with DAPI (blue in overlay). (B) Control (upper panels) or Nup155-depleted (lower panels) embryos were analysed with anti-Nup96 (left two columns, green in overlay) or anti-Nup35 (right two columns, green in overlay) antibodies together with mAb414 and DAPI (red and blue in overlay, respectively). (C) Still images from time-lapse recordings of YFP-Nup107-expressing control embryos (left panels) or Nup155-depleted embryos (right panels). The inset in left panel shows a magnification of YFP-Nup107 localization to the metaphase plate (−0:20). See Supplementary video 6. Scale bars, 10 μm.
The Nup107–160 complex has been found to be important for NPC formation and has been suggested to function in nucleation of NPC assembly (Walther et al, 2003). We therefore investigated the effect of Nup155 depletion on Nup107 localization, to determine whether Nup155 acts upstream or downstream of the Nup107–160 complex. We generated a C. elegans strain that expresses fluorescent YFP-tagged Nup107. YFP-Nup107 localized to both the periphery and interior of pronuclei (Figure 2C, left panels, −9:00 to −5:00). At pronuclear envelope breakdown, the rim staining was lost and Nup107 accumulated on the condensing chromosomes (Figure 2C, left panels, −1:40). In metaphase, YFP-Nup107 localized along the chromosomes (Figure 2C, left panels, −0:20). (Note that Nup107 is present on kinetochores in mammalian cells (Belgareh et al, 2001) and that C. elegans chromosomes are holocentric (Moore et al, 1999). This staining is therefore likely to correspond to kinetochores.) Upon exit from mitosis, YFP-Nup107 reaccumulated at the nuclear periphery (Figure 2C, left panels, 6:40). When Nup155 was depleted by RNAi, YFP-Nup107 staining at the pronuclear rim was abolished and instead a diffuse localization in the pronuclei was seen (Figure 2C, right panels, −9:00 to −5:00). Association of YFP-Nup107 to the missegregating chromatin persisted throughout mitosis and peripheral accumulation after mitosis failed (Figure 2C, right panels, 0:00–6:40). In conclusion, our analyses of fixed and living embryos demonstrate that all nucleoporins tested required Nup155 for accumulation at the nuclear rim. To determine whether this function is conserved, we next analysed in vitro nuclear assembly in Xenopus egg extracts.
Characterization of Xenopus Nup155
Polyclonal antibodies raised against recombinant full-length Xenopus Nup155 detected a single band of ∼150 kDa in a cytosolic fraction of Xenopus egg extracts whereas preimmune serum did not (Figure 3A). The serum also recognized human Nup155 by Western blot (data not shown) and showed a prominent nuclear rim staining in Xenopus Xl177 and human HeLa cells by immunofluorescence (Supplementary Figure S1).
Figure 3.
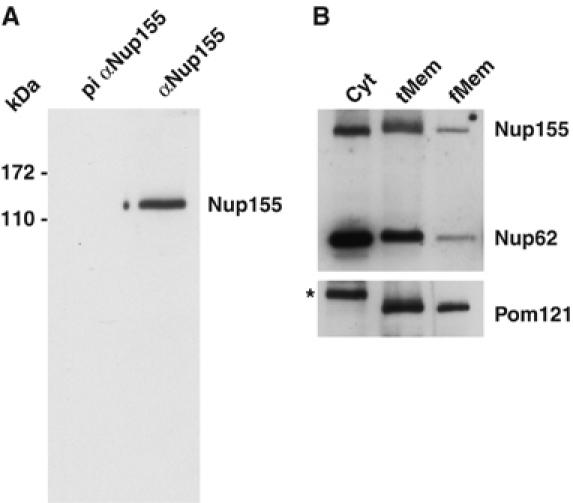
Biochemical characterization of Nup155 in Xenopus egg extract. (A) The cytosolic fraction of Xenopus egg extracts was analysed by Western blotting with preimmune (pi αNup155) or anti-Nup155 sera (αNup155). Molecular weight markers are indicated on the left. (B) Equal amounts of protein of the cytosol (Cyt), total membrane (tMem) and floated membrane (fMem) fractions of Xenopus egg extract were probed for the antigens Nup155, Nup62 and Pom121 on a Western blot. The asterisk in the cytosol lane indicates a crossreactivity of the Pom121 antibody.
Nup155 was also present in the total membrane fraction prepared from Xenopus eggs (Figure 3B, lane tMem). We investigated whether Nup155 could be removed from the crude membrane fraction by high-salt washes, and found that some Nup155 remained membrane-associated, similar to the behaviour of a control soluble nucleoporin, Nup62 (data not shown). To examine the possibility that Nup155 might be pelleted with membranes as part of a large soluble protein complex, we floated the membranes through a sucrose step gradient (Wilson and Newport, 1988). The lightest membrane fraction was collected and the total (tMem) and floated (fMem) membrane fractions were analysed by Western blot (Figure 3B). Nup155 and Nup62 behaved similarly and were significantly reduced after floating the membranes, whereas the transmembrane nucleoporin Pom121 fractionated with the membranes (Figure 3B). In the subsequent depletion experiments, membranes were routinely purified via flotation.
Nup155 depletion leads to a block in NPC assembly
Mock- and Nup155-immunodepleted cytosolic fractions of Xenopus egg extracts were prepared. Floated membranes were incubated together with cytosol, sperm chromatin, an ATP regeneration system and glycogen to reconstitute nuclei in vitro (Lohka and Masui, 1983; Gant and Wilson, 1997). After two rounds of depletion, >95% of Nup155 was removed (Figure 4B). The stringent depletion conditions needed to reduce Nup155 to this level were required to observe the defects in NE formation presented below, but resulted in the formation of small, irregularly shaped, nuclei after mock depletion.
Figure 4.
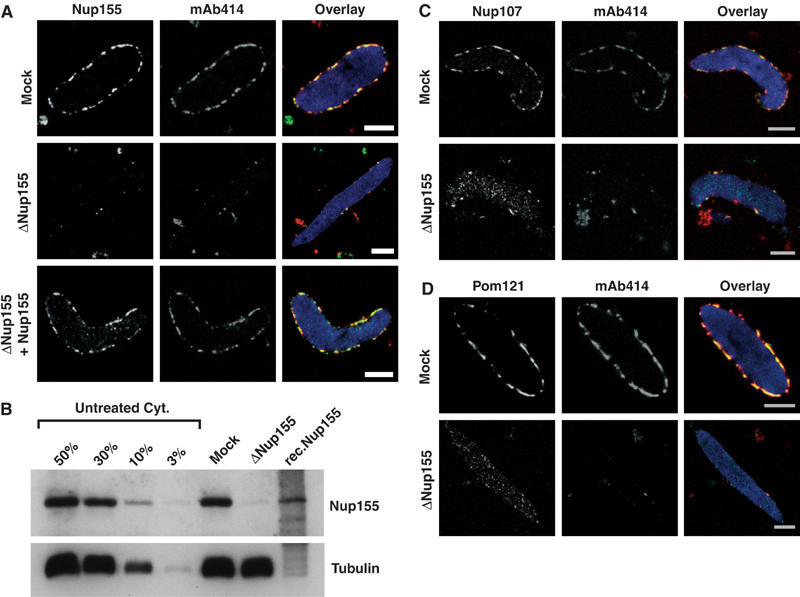
Immunodepletion of Nup155 inhibits NPC formation in vitro. (A) Nup155 was immunodepleted from cytosol and nuclei were assembled in vitro. Nucleoporin localization was analysed by immunofluorescence in mock-depleted (Mock) and Nup155-depleted reactions (ΔNup155), or after readdition of recombinant Nup155 (ΔNup155+Nup155). Nup155, mAb414 and overlay (Nup155 (green), mAb414 (red) and DAPI in blue) are costainings of the same nuclei. Scale bars, 5 μm. (B) Amounts of Nup155 in mock-depleted (Mock) and Nup155-depleted (ΔNup155) cytosol was compared by Western blotting. Equal volumes were loaded while varying amounts of untreated cytosol (Cyt) were loaded for comparison. A 0.6 μl portion of cytosol (25 μg total protein) corresponds to 50% of the sample volume loaded of mock- and Nup155-depleted cytosol. Tubulin serves as a loading control. (C, D) As in panel A, but co-immunodetection of Nup107 (C) or Pom121 (D) with mAb414 antigens.
Nup155 localized to the nuclear rim of nuclei assembled in mock-depleted extract (Figure 4A, first row, left). This staining was undetectable on nuclei assembled after Nup155 depletion (Figure 4A, second row, left). mAb414, which recognizes four vertebrate nucleoporins (Nup62, Nup153, Nup214 and Nup358), also showed rim staining of mock-depleted nuclei that was lost upon Nup155 depletion. Probing for Nup107 and Pom121 led to a similar observation with regard to the nuclear rim, but uniform chromatin staining was still visible in these cases (Figure 4C and D). Thus, none of the nucleoporins investigated by immunofluorescence localized to the site of NE formation in the absence of Nup155 in vitro, similar to the situation in C. elegans Nup155 RNAi embryos.
Full-length recombinant Xenopus Nup155 was purified under native conditions to investigate the specificity of the depletion phenotype. Small amounts of Nup155 (roughly 10% of that shown in Figure 4B) were sufficient to restore rim staining of Nup155, mAb414, (Figure 4A, last row) Nup107 and Pom121 (data not shown), demonstrating that the loss of these proteins from NPCs was specifically due to Nup155 depletion. Recombinant Nup155 was produced in very small amounts, but, by both Western blotting (Figure 4B) and mass spectrometric analysis (Supplementary Figure S2), appeared to be mostly full length.
Nup155 is required for nuclear membrane fusion
The observation that Nup155 is essential for NPC formation and for the accumulation of the integral membrane protein Pom121 on chromatin led us to address the fate of the nuclear membranes in the absence of Nup155. We depleted Nup155 from C. elegans embryos expressing the INM protein MAN1 fused to GFP. In control embryos, GFP-MAN1 marked the NE of the pronuclei, while after NE breakdown, GFP-MAN1 accumulated in the endoplasmic reticulum surrounding the mitotic spindle and the centrosomes (Figure 5A, left panels, −6:40–0:40; see Poteryaev et al (2005) for a description of the endoplasmic reticulum during mitosis in C. elegans embryos). Roughly 1 min after anaphase onset, GFP-MAN1 was recruited to the surface of the chromatin, followed rapidly by complete chromatin enclosure and nuclear growth (Figure 5A, left panels, 2:40–8:00, and Supplementary video 7). In Nup155-depleted embryos, the pronuclear GFP-MAN1 staining pattern was irregular and disappeared earlier than in control embryos (Figure 5A, right panels, −6:40 to −3:40) (n=3/3 embryos). During mitosis, GFP-MAN1 stained the centrosomes, which, as observed above, separated prematurely (Figure 5A, right panels, −3:40–0:40). Although GFP-MAN1 was recruited to chromatin at the end of mitosis, the distribution was not as homogenous as in the control embryos and NE morphology was clearly abnormal (Figure 5A, right panels, 2:40–8:00). These results suggested that NE formation might be defective in the absence of Nup155.
Figure 5.
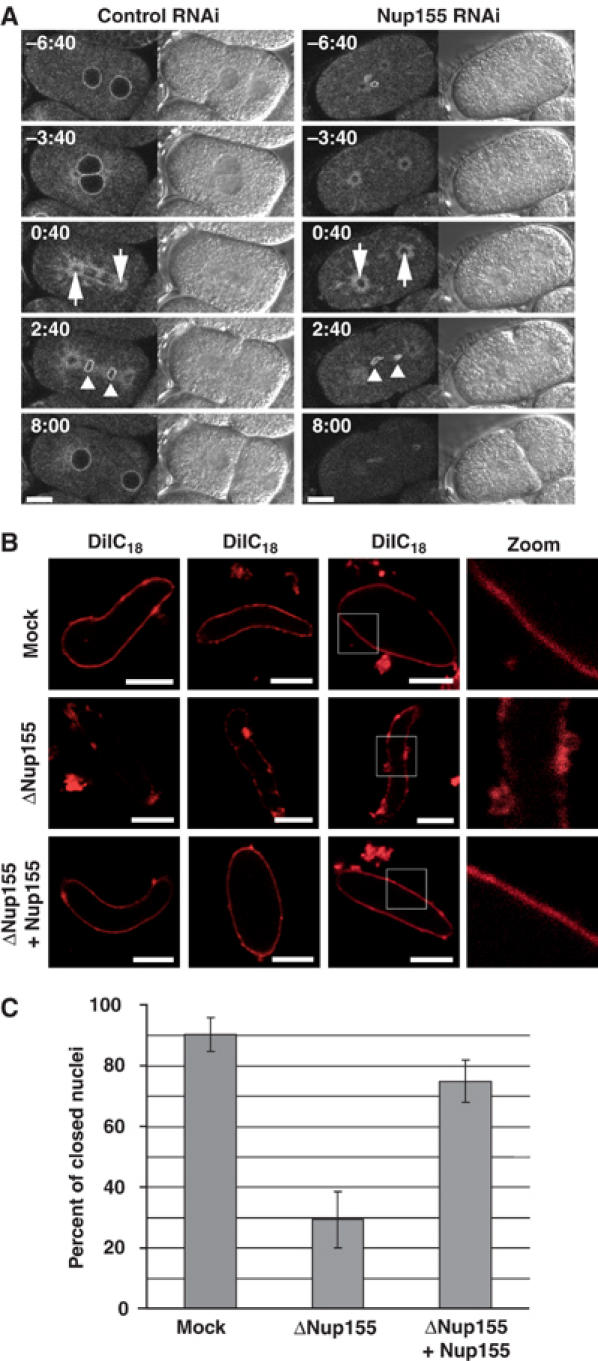
Membrane defects upon Nup155 depletion. (A) Control (left panels) or Nup155-depleted (right panels) embryos expressing GFP-MAN1 were observed by time-lapse microscopy. Centrosomes are indicated with arrows at anaphase (0:40) while triangles point to GFP-MAN1 association with chromatin during telophase (2:40). See Supplementary video 7. Scale bars, 10 μm. (B) Nuclei assembled in vitro in mock- or Nup155-depleted extracts, or in depleted extracts after addition of recombinant Nup155, were labelled with the membrane dye DiIC18 before fixation in 2% paraformaldehyde and 0.5% glutaraldehyde and analysed by confocal microscopy. Left and right image sets were derived from independent experiments. The two left panels are individually adjusted in signal intensity to illustrate the nature of the membranes best possible. Equal exposure times were chosen for right images. (C) Quantitation of closed NEs from panel B. In three independent experiments, 100 nuclei from each reaction were examined. The average of the three independent experiments is shown; error bars represent the total variation over the experiments.
To test this in the Xenopus in vitro system, we stained membranes with the lipophilic dye DiIC18 after nuclear assembly and before glutaraldehyde fixation (Figure 5B). By confocal microscopy, nuclear membranes were smooth and continuous in the mock-depleted reactions, but in the absence of Nup155, membrane vesicles associated with chromatin but did not form a continuous membrane (Figure 5B, see also Figure 6). In some cases, few vesicles were associated with the chromatin, and in others, membranes did associate but they did not fuse to form a smooth NE (Figure 5B). These phenotypes were also seen by electron microscopy (see below). The chromatin templates in Nup155-depleted extracts remained condensed, further indicating that closed NEs did not form (Figure 5B). We quantified this phenotype by scanning through 100 nuclei in each of three independent experiments by confocal microscopy. On average, 90% of the nuclei in the mock reactions had a continuous, closed NE. This number was reduced to 29% in Nup155-depleted extracts and could be restored to 75% by the addition of recombinant Nup155 (Figure 5C).
Figure 6.
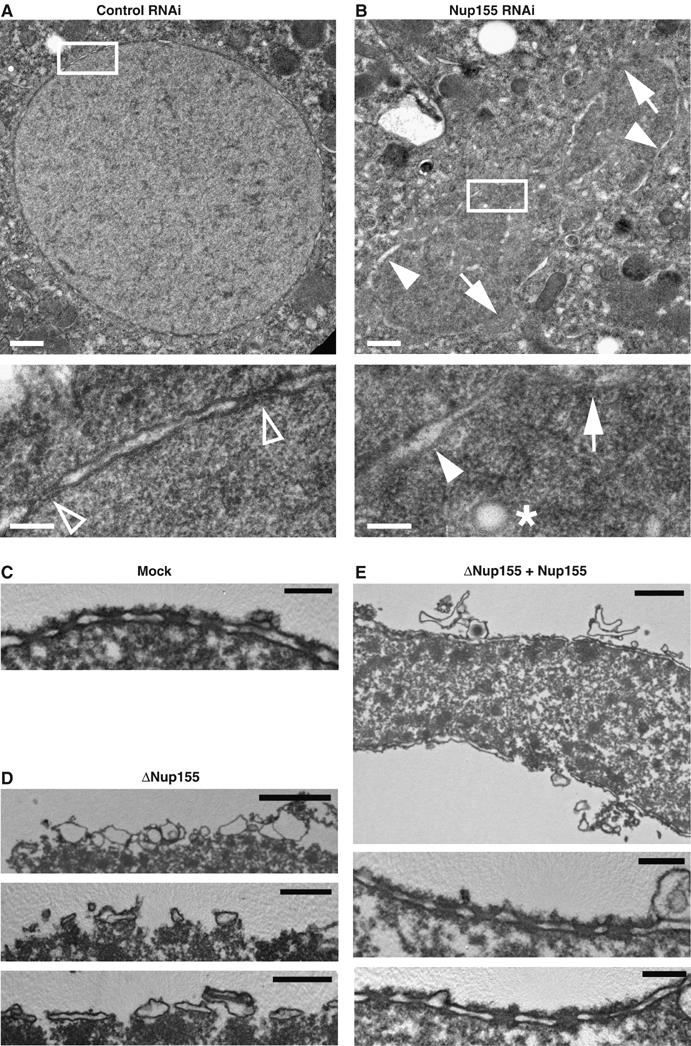
TEM analysis of NE defects in vivo and in vitro. (A, B) Control (A) or Nup155-depleted (B) C. elegans embryos were fixed by high-pressure freezing and processed for TEM. White boxes in upper images indicate the area shown at higher magnification below. In the higher magnifications, chromatin is localized to the lower right half of the micrographs. In panel A, empty triangles point to NPCs, which were not visible in Nup155 RNAi samples. In panel B, arrows point to areas where the chromatin surface is not covered by membranes while filled triangles indicate chromatin surface that is covered by membranes without NPCs. An asterisk indicates a membrane vesicle or tubule present inside the chromatin space. Scale bars, 0.5 μm (upper images) and 0.1 μm (lower images). (C–E) In vitro-assembled nuclei as in Figure 4A were processed for TEM. (C) Mock depleted. Scale bar, 0.2 μm. (D) Nup155 depleted. Scale bars, 2 μm (upper image) and 0.5 μm (middle and lower images). (E) Nup155 depleted followed by addition of recombinant Nup155. Scale bars, 1 μm (upper image) and 0.2 μm (middle and lower images).
Ultrastructural defects in the absence of Nup155
To analyse these phenotypes in detail, we analysed C. elegans embryos and in vitro-reconstituted nuclei by transmission electron microscopy (TEM). Focusing on young embryos (two- to eight-cell stages), we found that control samples contained well-preserved, spherical nuclei with an even distribution of nuclear pores (Figure 6A). In contrast, and as expected from our light microscopy observations, the chromatin structure in embryos depleted of Nup155 was highly irregular, and in several cases, we observed chromatin trapped in the cleavage furrow between two cells (Figure 6B). Importantly, a continuous NE never enclosed the chromatin in these embryos and NPCs were not observed. Instead, some of the chromatin surface was covered with membranes whereas other areas were membrane-free. The chromatin in Nup155 RNAi samples contained spherical membrane structures not found in control embryos that probably represent either docked membrane vesicles or tubules (Figure 6B).
Xenopus nuclear assembly reactions were also processed for TEM. In control reactions, we observed nuclei with a continuous double bilayer containing NPCs (Figure 6C). In the absence of Nup155, no continuous membrane bilayer was present (Figure 6D). Instead, membrane vesicles that interacted with the chromatin surface but remained at this initial docking step without further fusion were present. In some areas, the docked vesicles flattened and formed unconnected patches (Figure 6D). Importantly, no NPCs were found at the periphery of the chromatin templates in Nup155-depleted samples, confirming our confocal microscopy observations that neither NPCs nor closed nuclear membranes formed in the absence of Nup155. Both processes were restored upon readdition of recombinant Nup155. Membranes fused to continuous bilayers with regularly spaced NPCs (Figure 6E). We conclude that the functional role of Nup155 in NE and NPC formation is conserved from C. elegans to vertebrates.
Nup155 dynamics
Having demonstrated an absolute requirement for Nup155 in NE formation, we investigated the behaviour of Nup155 during NE assembly. Initially, we generated a C. elegans strain expressing an N-terminally GFP-tagged Nup155 fusion protein. As expected, the pronuclei showed accumulation of GFP-Nup155 at the nuclear rim (Figure 7A, −4:40). The pronuclear envelopes broke down when the embryos entered prometaphase as visualized by the influx of cytosolic macromolecules (data not shown) and GFP-Nup155 began to dissociate from the pronuclei (Figure 7A, −2:20). GFP-Nup155 was not detectable on metaphase chromosomes (Figure 7A, 0:00). Recruitment of GFP-Nup155 to chromatin was observed 60–80 s after anaphase onset, initiating at the apical surfaces of chromatin facing the endoplasmic reticulum during anaphase (n=5 embryos) (Figure 7A, 1:20, see also Figure 5A). At this stage, spindle microtubules still cover the more central regions of the chromatin. The GFP-Nup155-covered area rapidly expanded to cover the entire periphery of the chromatin, followed by nuclear growth (Figure 7A, 2:20–6:40). The behaviour of GFP-Nup155 was clearly different from YFP-Nup107, which remained associated with chromatin throughout mitosis (Figure 2C). Rather, GFP-Nup155 behaved similarly to two other nucleoporins, GFP-Nup35 and GFP-Nup45/Nup58 (Supplementary Figure S3).
Figure 7.
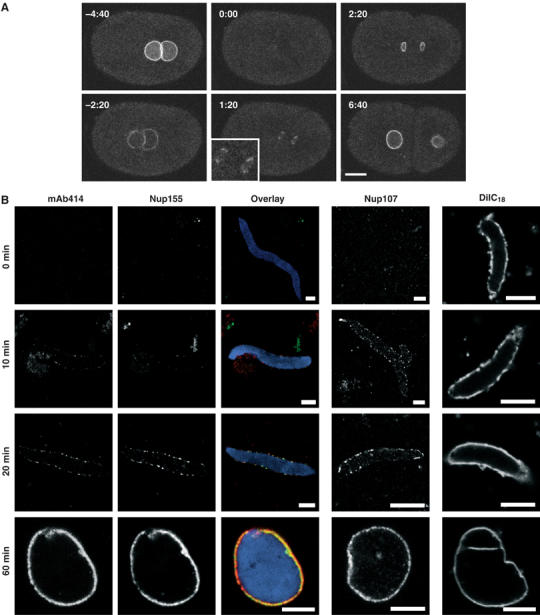
Dynamics of Nup155. (A) Still images from time-lapse microscopy recording of C. elegans embryo expressing GFP-Nup155. Images show progression from pronuclear meeting in the one-cell embryo to the two-cell stage. The inset shows a magnification of initial GFP-Nup155 recruitment to the two reforming nuclei during anaphase (1:20). See Supplementary video 8. Scale bar, 10 μm. (B) NE assembly reactions were fixed in 4% paraformaldehyde at the time points indicated. The three columns on the left, which show immunodetection of Nup155 (green), mAb414 (red) and DAPI (blue), are identical samples and merged in the overlaid images. Parallel reactions for Nup107 immunodetection or that were stained with the membrane dye DiIC18 before fixation are on the right. Scale bars, 5 μm.
Finally, we investigated the recruitment of Nup155 relative to other nucleoporins and membranes by time-course experiments in Xenopus egg extracts. Nup155, Nup107, mAb414 antigens, DiIC18 stained membranes and Pom121 (data not shown) were followed through NPC and NE formation (Figure 7B). Chromatin was decondensed for 10 min in the absence of membranes and the first time point (t=0) was taken and immediately fixed when membranes, energy mix and glycogen were added to start the assembly reaction. Nup107, together with Pom121, appeared early relative to mAb414 antigens on the chromatin surface after 5–10 min (Figure 7B, and data not shown; see also Antonin et al, 2005). Nup155 and mAb414 antigens were first visible at the nuclear periphery between 10 and 20 min. Their initial appearance at the chromatin template is different from that of Nup107 and Pom121, as they are only seen in peripheral foci and do not coat the chromatin surface as Nup107 and Pom121 do (Figure 7B, see also Figure 4C and D; Walther et al, 2003; Antonin et al, 2005). Monitoring the nuclear membranes by DiIC18 staining, we found that membranes immediately dock to chromatin when added to the assembly reaction. The fusion of vesicles to form a closed NE occurred after approximately 20 min (Figure 7B, right column) concomitant with Nup155 and mAb414 antigens accumulating at the nuclear periphery.
Discussion
We demonstrate that Nup155 is required for postmitotic NE and NPC formation in both nematodes and vertebrates. Depletion of Nup155 either by RNAi from C. elegans embryos or biochemically from Xenopus egg extracts effectively prevented the accumulation of all the nucleoporins tested at the nuclear periphery during nuclear assembly, thereby abolishing NPC assembly. Prior work has identified important early steps in NPC assembly that involve the Nup107–160 complex and Pom121 (Harel et al, 2003; Walther et al, 2003; Antonin et al, 2005). Nup155 is recruited subsequently to Nup107 and Pom121, together with other soluble nucleoporins, and its recruitment is critical for NPC assembly to proceed. In the absence of Nup155, nuclear membranes were still able to dock to the chromatin surface. However, we observed severe defects in membrane fusion resulting in a lack of nuclear enclosure both in vivo and in vitro.
Depletion of Nup155 causes defects in nuclear morphology, segregation and viability
In agreement with our previous observations (Galy et al, 2003), depletion of Nup155 from C. elegans embryos caused 100% embryonic lethality and severe nuclear morphology defects. The pronuclei were separated from the centrosomes in Nup155-depleted embryos, similar to the phenotypes observed upon depletion of components of the Ran GTPase cycle (Askjaer et al, 2002). Rather than being closely aligned in the centre of the zygote, the pronuclei were positioned randomly, underlining the importance of the NE in the nuclear anchoring of centrosomes. Moreover, severe DNA segregation defects were observed, which are a likely cause of the embryonic lethality induced by Nup155 RNAi. Defects in DNA segregation have also been observed when nuclear transport receptors or nuclear lamin was depleted (Liu et al, 2000; Askjaer et al, 2002; Geles et al, 2002). DNA segregation depends on coordinated events of DNA replication and condensation, sister chromatid cohesion and incorporation into the mitotic spindle via kinetochore fibres. Since pronuclear formation was abnormal in the absence of Nup155 (see below), it is likely that one or more of these processes were perturbed, arguing that the defects in DNA segregation upon Nup155 depletion were probably indirect. We note that the yeast proteins Nup157p and Nup170p, which are both homologues of metazoan Nup155, have been linked to chromosome transmission fidelity (Kerscher et al, 2001). However, the fact that yeast contains two Nup155 homologues and that yeast NPCs do not disassemble during mitosis prevents straightforward analysis of their potential role in mitotic NPC and NE assembly activity. Neither NUP157 nor NUP170 is an essential gene but simultaneous deletion of both is lethal (Aitchison et al, 1995; Kenna et al, 1996). The observation that deletion of either NUP157 or NUP170 is also lethal in combination with distinct sets of other nucleoporin mutations suggests that the two Nup155 homologues may have both overlapping and distinct functions (Aitchison et al, 1995; Kenna et al, 1996). Reduction of Nup170p expression in the absence of the integral membrane nucleoporin Pom152p caused formation of distorted NEs with fewer NPCs (Aitchison et al, 1995), implicating NUP170 in NPC biogenesis. Moreover, the diameter for passive diffusion through NPCs is increased in nup170Δ yeast, possibly because several nucleoporins are less stably associated with the NPC in the absence of Nup170 (Shulga and Goldfarb, 2003).
Nup155 dynamics
Our observation that GFP-Nup155 in C. elegans embryos apparently dissociated completely from chromatin during mitosis reflects the initial observations made of Nup155 in mammalian cells (Radu et al, 1993). In worms, GFP-Nup155 was recruited to the chromatin approximately 1 min after anaphase onset. Because of the very rapid divisions in early C. elegans embryos, this timing cannot be directly compared to that of nucleoporin recruitment in other organisms. However, we have observed very similar behaviour for two other nucleoporins that are predicted to be soluble in mitosis, GFP-Nup35 and GFP-Nup45/Nup58. In vertebrate cells, the products of the alternatively spliced Nup45/Nup58 gene form an NPC subcomplex together with Nup54 and Nup62 (Vasu and Forbes, 2001). This subcomplex is recruited to the reforming NE later than Pom121 but earlier than gp210 and Tpr (Bodoor et al, 1999; Haraguchi et al, 2000). Monitoring nuclear reconstitution in vitro, we were able to confirm the relative order of recruitment of several nucleoporins (Bodoor et al, 1999; Haraguchi et al, 2000; Walther et al, 2003; Antonin et al, 2005) and to place Nup155 isochronically with Nup62 and other mAb414 antigens in the context of NPC assembly. Thus, we observed a similar relative timing of recruitment of Nup155 in vivo and in vitro.
Nup155 is required for NPC and NE formation
NPCs were not detectable by electron microscopy or immunofluorescence analysis in Xenopus nuclei assembled in the absence of Nup155, and all nucleoporins tested did not measurably localize to the NE. In contrast, the early recruitment of both Nup107 and Pom121 on the chromatin surface still occurred, but both failed to undergo the later accumulation at the chromatin periphery seen in nondepleted extracts. This difference in response to Nup155 depletion correlates with the order of recruitment of different nucleoporins. Thus, the initial targeting of Nup107 and Pom121 to chromatin in the in vitro system appears independent of Nup155 but the downstream recruitment steps involving additional NPC components and organization did not take place. RNAi-mediated depletion of Nup155 in C. elegans embryos also prevented NPC assembly. In particular, we noted a strong defect in recruitment of Nup35 and mAb414 nucleoporins. Nup35 was recently shown to interact with Nup93, Nup155, Nup205 and lamin B and at least the Nup35–Nup93 interaction was shown to be direct (Hawryluk-Gara et al, 2005).
Analysing the fate of nuclear membranes in the absence of Nup155 both in vitro and in vivo revealed a block in nuclear membrane fusion. Although docking of membranes to the chromatin surface seemed to occur normally, subsequent fusion steps did not advance to completion, neither did we observe insertion of NPCs into the membranes. Importantly, the inhibition of NPC and NE assembly was efficiently reversed by addition of purified, recombinant Nup155. Such dramatic inhibition of NE assembly has previously been observed for only one other nucleoporin, the integral membrane protein Pom121 (Antonin et al, 2005). In contrast to Pom121, we found no evidence that Nup155 is an integral membrane protein, consistent with work by Hawryluk-Gara et al (2005).
Nup155 acts as quality control sensor linking NPC and NE assembly
Two different models for NPC assembly that are relevant for the work presented here have been proposed. According to the ‘NPC insertion' model, NPCs are assembled into pre-existing double membranes (Macaulay and Forbes, 1996; Goldberg et al, 1997), whereas the ‘pre-NPC' model suggests that postmitotic NPC assembly initiates on chromatin independent of membrane fusion (Sheehan et al, 1988; Walther et al, 2003). The fact that the number of NPCs increases at S-phase argues that NPCs can indeed be assembled subsequent to NE formation (Maul et al, 1972). However, accumulating evidence supports the ‘pre-NPC' model as the postmitotic NPC biogenesis pathway in metazoans. NPC assembly in the NE requires binding of the Nup107–160 complex to chromatin prior to membrane closure (Walther et al, 2003). In the absence of the Nup107–160 complex, recruitment of nucleoporins is blocked and NPC assembly does not take place although membrane fusion to form a closed NE is not inhibited (Harel et al, 2003; Walther et al, 2003). Similarly, Pom121 is recruited very early to chromatin before NE closure. Immunodepletion of Pom121 prevents both NPC and NE assembly; however, the block to NE formation can be released by codepletion of the Nup107–160 complex. This indicates that in the presence of the Nup107–160 complex, the absence of Pom121 can be sensed and leads to a block in NPC and NE assembly (Antonin et al, 2005). The phenotype of Nup155 depletion is very similar to that seen on removal of Pom121, suggesting that the soluble Nup155 may also participate in the proposed NE assembly checkpoint. Unfortunately, for technical reasons related to the stringency of the conditions needed for Nup155 depletion, we have been unable to test the effect of codepleting Nup155 and the Nup107–160 complex.
In our time-course experiments, Nup155 was recruited to chromatin significantly later than Nup107 or Pom121. Moreover, depletion of Nup155 did not prevent chromatin association of Nup107 or Pom121. Many nucleoporins are recruited at a similar time as Nup155, presumably reflecting NPC assembly. Moreover, although membranes dock to chromatin very early in nuclear assembly, fusion to form a closed NE happens concomitantly with Nup155 accumulation. In yeast, a direct interaction between Nup157p and the yeast homologue of vertebrate Nup160, Nup120p, was recently demonstrated (Lutzmann et al, 2005), suggesting that metazoan Nup155 might also bind the Nup107–160 complex. However, despite several attempts, we have not been able to identify any Nup155 interaction partners by co-immunoprecipitation from Xenopus extracts, leaving this possibility open.
In conclusion, this study strengthens the evidence for the recently proposed checkpoint that links the interconnected processes of NE and NPC assembly in the Xenopus egg extract system (Antonin et al, 2005) and extends this concept by demonstrating that Nup155, a soluble nucleoporin that is recruited late in NPC assembly, must be present in order for NE membrane assembly to proceed to completion. Given that assembly of an NE lacking NPCs would be a lethal event, the existence of such a checkpoint has considerable logic. Aside from Nup155, none of the other soluble nucleoporins that have been tested by depletion from Xenopus extracts have led to a similar phenotype (Finlay and Forbes, 1990, 1991; Powers et al, 1995; Grandi et al, 1997; Walther et al, 2001, 2002). Our data may either suggest that rather late steps in NPC assembly are monitored prior to NE membrane formation or that Nup155, the Nup107–160 complex and Pom121 have a direct role in controlling NE membrane and NPC assembly. The concordant results obtained from C. elegans and the Xenopus in vitro system suggest that the NPC–NE assembly checkpoint is conserved. Based on our results, we propose the existence of either a protein or a signalling event that brings together the chromatin-associated Nup107–160 complex, membrane-bound Pom121 and soluble Nup155. An important challenge is now to determine the nature of this control mechanism and to understand how integration of NPC and NE assembly is achieved.
Materials and methods
C. elegans strains and RNAi
Strain XA3541 expressing GFP-β-tubulin and YFP-lamin was generated by crossing WH204 gfp∷tbb-2 (Strome et al, 2001) with XA3502 yfp∷lmn-1 (Galy et al, 2003). XA3506 expressing YFP-CeNup107 was generated by transforming DP38 unc-119(ed3) (Maduro and Pilgrim, 1995) with plasmids pUP9 (Galy et al, 2003) and pPAG3. Full-length genomic Nup107/npp-5 sequence was PCR amplified and used to replace his-11 downstream of GFP in pJH4.52 (Strome et al, 2001) giving rise to plasmid pPAG1. The GFP sequence of pPAG1 was then replaced with YFP from pPAG6 (Galy et al, 2003) to construct pPAG3. Plasmids pID npp-8, pID npp-19 and pID npp-4 were generated by Gateway cloning (Invitrogen) using plasmid pID-3.01B (Pellettieri et al, 2003) and PCR-amplified full-length genomic sequences. Transformation of DP38 with these plasmids yielded strains XA3546 expressing GFP-CeNup155, XA3547 expressing GFP-CeNup35 and XA3548 expressing GFP-CeNup45/CeNup58. Primer sequences are available upon request. Other strains used in this study were C. elegans Bristol strain N2, XA3501 gfp∷tbb-2; gfp∷his-11 (Askjaer et al, 2002) and XA3507 gfp∷lem-2 (Galy et al, 2003). dsRNA-mediated interference targeting CeNup155 was performed at 20°C for 26–32 h (Galy et al, 2003).
Live embryo imaging and immunofluorescence
Live recordings and immunofluorescence of C. elegans embryos were performed as described (Galy et al, 2003). Rabbit polyclonal antibodies against CeNup35/NPP-19, CeNup153/NPP-7 and Ce-Nup96/NPP-10-C (Galy et al, 2003) and monoclonal antibody mAb414 (Covance, Berkeley, CA) were used at 1:300–1:500 dilutions. Secondary antibodies were Alexa Fluor 633-conjugated anti-rabbit goat antibodies (used at 1:2000; Molecular Probes, Eugene, OR) and Cy-3-conjugated anti-mouse goat antibodies (used at 1:500; Jackson Immunoresearch Laboratories, West Groove, PA) whereas chromatin was detected by DAPI.
Xenopus egg extract preparation
Preparation of X. laevis egg extract cytosol was performed as described (Hartl et al, 1994) except that after low-speed centrifugation the extract was centrifuged twice at 225 000 g for 40 min at 4°C to obtain the cytosol as a clear supernatant.
Total and floated membrane fractions were prepared as described (Wilson and Newport, 1988) and the membrane fraction with lowest density was collected. Sperm heads were prepared from Xenopus testis (Gurdon, 1976).
For NE assembly, 10 μl cytosol was incubated with 0.5 μl sperm heads (3000 sperm heads/μl) for 10 min at 20°C for chromatin decondensation. To start the reaction, 0.5 μl of floated membranes, 0.2 μl of energy mix (50 mM ATP, 500 mM creatine phosphate, 10 mg/ml creatine kinase) and 0.2 μl glycogen (20 mg/ml; USB Amersham) were added. The reaction was incubated for 2 h at 20°C. For immunofluorescence, reactions were fixed in 4% paraformaldehyde in PBS for 30 min on ice. Membrane labelling with DiIC18 before fixation was performed as described (Antonin et al, 2005).
After fixation, reactions were laid on top of a 30% (w/v) sucrose cushion in S250 buffer and spun onto poly-L-lysine-coated coverslips. Samples with DiIC18 stained membranes were directly mounted for confocal microscopy, whereas other samples were processed for immunofluorescence using described protocols and antibodies (Walther et al, 2003; Antonin et al, 2005).
Expression of Xenopus Nup155 and antibody generation
Full-length X. laevis Nup155 cDNA from RZPD (clone ID: IMAGp998J1010564Q3) was PCR amplified and cloned into pET28a for expression using NheI and HindIII. Primer sequences were ctagctagcatgcccagt (N-terminal) and cccaagcttgctagctcaacctctccaatt (C-terminal).
Nup155 was expressed in Escherichia coli strain BL21DE3 and purified from inclusion bodies for rabbit polyclonal antibody generation. For in vitro nuclear assembly reactions, soluble recombinant Nup155 was purified on Ni-NTA Agarose under standard conditions.
Nup155 depletion from cytosol
Preimmune and anti-Nup155 antibody sera were crosslinked to Protein A-Sepharose in the presence of 10 mM dimethylpimelimidate (Sigma). Cytosol was incubated with the antibody column in a 1:1.3 volume ratio for two rounds, each of 30 min at 4°C. Cytosol was either used immediately in NE assembly reactions or frozen with 3% (v/v) glycerol.
Transmission electron microscopy
C. elegans hermaphrodites fed bacteria expressing either control or Nup155 dsRNA were transferred to 1.5 mm by 200 μm planchettes filled with 10% dextran or yeast paste (McDonald and Muller-Reichert, 2002; Muller-Reichert et al, 2003). The nematodes were cryoimmobilized immediately using a Leica EMPact high-pressure freezer (Leica, Vienna, Austria) and stored in liquid nitrogen. Samples were freeze-substituted over 3 days at −90°C in anhydrous acetone containing 2% osmium tetroxide and 0.1% uranyl acetate and warmed gradually to room temperature (5°C per hour). After several acetone rinses, samples were infiltrated with Epon resin during 2 days and flat embedded in a thin layer of resin and polymerized at 60°C during 48 h (Müller-Reichert et al, 2003). Ultrathin sections were mounted on Formvar-coated copper grids and stained with 2% uranyl acetate in water and lead citrate.
In vitro nuclear assembly was performed as above but scaled up to a 60 μl reaction and processed for TEM as described (Antonin et al, 2005).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary video 4
Supplementary video 5
Supplementary video 6
Supplementary video 7
Supplementary video 8
Supplementary Video Legends
Acknowledgments
We are indebted to Vincent Galy for assistance in generating the YFP-Nup107 C. elegans strain and Peter Askjaer thanks Cayetano González for generous support. Members of the Mattaj laboratory are acknowledged for critical reading of the manuscript. Some C. elegans strains used in this work were provided by the Caenorhabditis Genetic Center, funded by the NIH National Center for Research Resources. Peter Askjaer was supported by the Deutsche Forschungsgemeinschaft, the Spanish Ministry of Education and Science (RYC-2003-001521, BFU2004-01096) and EMBL. Cerstin Franz was supported by the National German Academic Foundation and EMBL.
References
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW (1995) Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol 131: 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW (2005) The integral membrane nucleoporin Pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92 [DOI] [PubMed] [Google Scholar]
- Askjaer P, Galy V, Hannak E, Mattaj IW (2002) Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell 13: 4355–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V (2001) An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B (1999) Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci 112: 2253–2264 [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ (1990) Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell 60: 17–29 [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ (1991) A complex of nuclear pore proteins required for pore function. J Cell Biol 114: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P (2003) Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14: 5104–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TM, Wilson KL (1997) Nuclear assembly. Annu Rev Cell Dev Biol 13: 669–695 [DOI] [PubMed] [Google Scholar]
- Geles KG, Johnson JJ, Jong S, Adam SA (2002) A role for Caenorhabditis elegans importin IMA-2 in germ line and embryonic mitosis. Mol Biol Cell 13: 3138–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F, Malva C (1998) Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol 142: 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Wiese C, Allen TD, Wilson KL (1997) Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci 110: 409–420 [DOI] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E (1997) Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell 8: 2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL (2005) The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6: 21–31 [DOI] [PubMed] [Google Scholar]
- Gurdon JB (1976) Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J Embryol Exp Morphol 36: 523–540 [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci 113: 779–794 [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Hartl P, Olson E, Dang T, Forbes DJ (1994) Nuclear assembly with lambda DNA in fractionated Xenopus egg extracts: an unexpected role for glycogen in formation of a higher order chromatin intermediate. J Cell Biol 124: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW (2005) Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell 16: 2382–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna MA, Petranka JG, Reilly JL, Davis LI (1996) Yeast N1e3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol Cell Biol 16: 2025–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Hieter P, Winey M, Basrai MA (2001) Novel Role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157: 1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Gigliotti S, Fuller MT (1999) Developmental genetics of the essential Drosophila nucleoporin nup154: allelic differences due to an outward-directed promoter in the P-element 3′ end. Genetics 153: 799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell 11: 3937–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 220: 719–721 [DOI] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Stangl K, Stelter P, Toth KF, Bottcher B, Hurt E (2005) Reconstitution of Nup157 and Nup145N into the Nup84 complex. J Biol Chem 280: 18442–18451 [DOI] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ (1996) Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol 132: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D (1995) Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW (2004) Sorting out the nuclear envelope from the endoplasmic reticulum. Nat Rev Mol Cell Biol 5: 65–69 [DOI] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY-L, Borun TW (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol 55: 433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K, Muller-Reichert T (2002) Cryomethods for thin section electron microscopy. Methods Enzymol 351: 96–123 [DOI] [PubMed] [Google Scholar]
- Moore LL, Morrison M, Roth MB (1999) HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol 147: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Reichert T, Hohenberg H, O'Toole ET, McDonald K (2003) Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. J Microsc 212: 71–80 [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Reinke V, Kim SK, Seydoux G (2003) Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev Cell 5: 451–462 [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A (2005) Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell 16: 2139–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ (1995) Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol 128: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak RW (1993) Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol 121: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala HJ, Kendirgi F, Barry DM, Majerus PW, Wente SR (2004) The mRNA export factor human Gle1 interacts with the nuclear pore complex protein Nup155. Mol Cell Proteomics 3: 145–155 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ (1988) Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol 106: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Goldfarb DS (2003) Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol Cell Biol 23: 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W (2001) Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell 12: 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Vasu SK, Forbes DJ (2001) Nuclear pores and nuclear assembly. Curr Opin Cell Biol 13: 363–375 [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW (2001) The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J 20: 5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M (2002) The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol 158: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Newport J (1988) A trypsin-sensitive receptor on membrane vesicles is required for nuclear envelope formation in vitro. J Cell Biol 107: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary video 4
Supplementary video 5
Supplementary video 6
Supplementary video 7
Supplementary video 8
Supplementary Video Legends


