Abstract
The fission yeast cell-polarity regulator tea1p is targeted to cell tips by association with growing microtubule ends. Tea1p is subsequently anchored at the cell cortex at cell tips via an unknown mechanism that requires both the tea1p carboxy-terminus and the membrane protein mod5p. Here, we show that a tea1p-related protein, tea3p, binds independently to both mod5p and tea1p, and that tea1p and mod5p can also interact directly, independent of tea3p. Despite their related structures, different regions of tea1p and tea3p are required for their respective interactions with an essential central region of mod5p. We demonstrate that tea3p is required for proper cortical localization of tea1p, specifically at nongrowing cell tips, and that tea1p and mod5p are independently required for tea3p localization. Further, we find that tea3p fused to GFP or mCherry is cotransported with tea1p by microtubules to cell tips, but this occurs only in the absence of mod5p. These results suggest that independent protein–protein interactions among tea1p, tea3p and mod5p collectively contribute to tea1p anchoring at cell tips via a multistep and multimode mechanism.
Keywords: cell polarity, kelch repeat, mCherry, S. pombe, tdTomato
Introduction
Communication between microtubules and the actin cytoskeleton is a common feature of eukaryotic cell polarity (Drubin and Nelson, 1996; Yarm et al, 2001; Chang and Peter, 2003; Small and Kaverina, 2003). In many cases, proteins that are transported on the plus ends of microtubules, such as EB1 and CLIP-170, forge interactions with proteins localized at sites on the plasma membrane, facilitating this communication (Schuyler and Pellman, 2001; Gundersen, 2002; Carvalho et al, 2003; Akhmanova and Hoogenraad, 2005; Watanabe et al, 2005).
In the fission yeast Schizosaccharomyces pombe, tea1p is a key mediator between microtubules and the actin cytoskeleton (Hayles and Nurse, 2001; Chang and Peter, 2003). After a variety of stresses, tea1Δ cells become bent or branched (Verde et al, 1995; Mata and Nurse, 1997; Niccoli et al, 2003; Sawin and Snaith, 2004), in part because microtubules are unable to specify positional information to the cell cortex (Sawin and Nurse, 1998; Feierbach et al, 2004; Sawin and Snaith, 2004). In addition, during steady-state growth, tea1Δ mutants exhibit a mostly monopolar growth pattern, unlike wild-type cells, which exhibit monopolar growth early in the cell cycle and bipolar growth later in the cell cycle (Mitchison and Nurse, 1985; Mata and Nurse, 1997; Glynn et al, 2001). tea1Δ mutants are also defective in the cortical localization of other polarity factors, including the actin-binding protein bud6/aip3 (Glynn et al, 2001; Jin and Amberg, 2001), the formin for3p (Feierbach and Chang, 2001; Feierbach et al, 2004), the SH3-domain protein tea4p/wsh3p (Martin et al, 2005; Tatebe et al, 2005) and the tea1p-related protein tea3p (Arellano et al, 2002).
Tea1p is a 1147-amino-acid protein. Its N-terminus contains six kelch repeats, which are found in many proteins, including actin-binding proteins, and are likely involved in protein–protein interactions (Adams et al, 2000; Prag and Adams, 2003; Li et al, 2004). The C-terminal regions of tea1p are predicted to be largely alpha-helical coiled coil. Tea1p also displays limited functional and sequence similarity to ezrin, a member of the ERM (ezrin–radixin–moesin) family, which has been shown to link the plasma membrane with the underlying actin cytoskeleton in animal cells (Vega and Solomon, 1997; Bretscher et al, 2002).
In vivo, tea1p displays a dynamic pattern of localization, reflecting its function in cell polarity. Tea1p is transported to cell tips on the growing plus ends of microtubules (Behrens and Nurse, 2002; Snaith and Sawin, 2003; Feierbach et al, 2004), dependent on the kip2-like kinesin tea2p and the CLIP-170 homologue tip1p (Browning et al, 2000, 2003; Brunner and Nurse, 2000; Busch et al, 2004). Once at tips, tea1p is unloaded from microtubules and retained at the cell cortex. This retention is defective in mutants lacking either the membrane protein mod5p or the C-terminal 200 amino acids of tea1p (tea1Δ200; Behrens and Nurse, 2002; Martin and Chang, 2003; Snaith and Sawin, 2003; also see below). Cortically associated tea1p then contributes to the organization of actin filaments at cell tips via interactions with proteins such as tea4p/wsh3p and bud6p/aip3p, thereby ensuring proper bipolar growth (Glynn et al, 2001; Verde, 2001; Martin et al, 2005; Snaith and Sawin, 2005; Tatebe et al, 2005).
Currently, there are three major outstanding issues in relation to how tea1p functions in microtubule-mediated cell polarity in fission yeast. At the beginning of the tea1p ‘pathway' is the question of how tea1p is associated with and transported on microtubule plus ends, and at the end of the pathway is the question of how tea1p interacts with the actin cytoskeleton. Here, we address a question at the center of the tea1p pathway, namely, how does tea1p become anchored at the cortex at cell tips?
An important factor in tea1p anchoring is the protein mod5p, which was first identified in a screen for mutations in S. pombe that affect cell shape (Snaith and Sawin, 2003). In mod5Δ cells, tea1p is transported normally to cell tips but fails to accumulate there (Snaith and Sawin, 2003). Mod5p, which is unusually rich in serine, threonine and proline residues and thus may be an intrinsically unstructured protein (Dyson and Wright, 2005), is localized to membranes at cell tips, via a C-terminal prenylation sequence. Interestingly, in tea1Δ cells, mod5p is no longer restricted to cell tips but rather spreads out around the entire cell cortex. This has led to the suggestion of a positive feedback loop linking mod5p and tea1p localization, which might ensure high-fidelity polarized growth in the context of dynamic tea1p delivery to cell tips (Snaith and Sawin, 2003). However, the molecular mechanisms underlying tea1p localization at the cell cortex remain largely unclear. For example, although there is genetic evidence for an interaction between mod5p and tea1p, a clear biochemical interaction has not yet been shown. In particular, live-cell analyses of GFP-tagged mod5p and tea1p have demonstrated that the two proteins do not completely colocalize at cell tips, and that tea1p at the cortex is highly dynamic (Snaith and Sawin, 2003). Thus, even if tea1p and mod5p do interact physically, whether directly or indirectly, other proteins may also be involved in regulating their localization.
In this work, we have sought to further understand the mechanisms regulating tea1p localization at cell tips by identifying protein–protein interactions involving mod5p. From a two-hybrid screen, we identified the tea1p-related protein tea3p as a mod5-interacting protein. In these experiments, we also found that mod5p interacts with tea1p and that tea1p interacts with tea3p. In vivo, each of the pairwise interactions occurring between any two of these three proteins can take place in the absence of the third protein, and in a series of localization-dependency studies, we found that each of these proteins contributes to the proper localization and function of the other two, but in distinct ways. These results suggest that a complex network of interactions among tea1p, mod5p and tea3p is involved in regulating the localization of these three proteins, ultimately leading to correctly anchored tea1p at cell tips.
Results
Pairwise interactions between tea1p, tea3p and mod5p
To better understand the mechanisms underlying tea1p localization. we performed a two-hybrid screen using essentially full-length mod5p as bait (see Supplementary data). Out of 11 plasmids isolated, 10 contained fragments of the tea3 gene, previously implicated in cell polarity (Arellano et al, 2002). Interestingly, tea3p is structurally related to tea1p. Both proteins contain the protein–protein interaction kelch domain in their N-termini (Adams et al, 2000; Prag and Adams, 2003; Li et al, 2004) and long regions of predicted coiled coil in their C-termini. We confirmed the interaction between mod5p and tea3p in GST pulldown experiments and co-immunoprecipitation assays, using extracts from fission yeast simultaneously expressing GST-mod5p and tea3p-GFP (Supplementary Figure 1A and Figure 1A).
Figure 1.
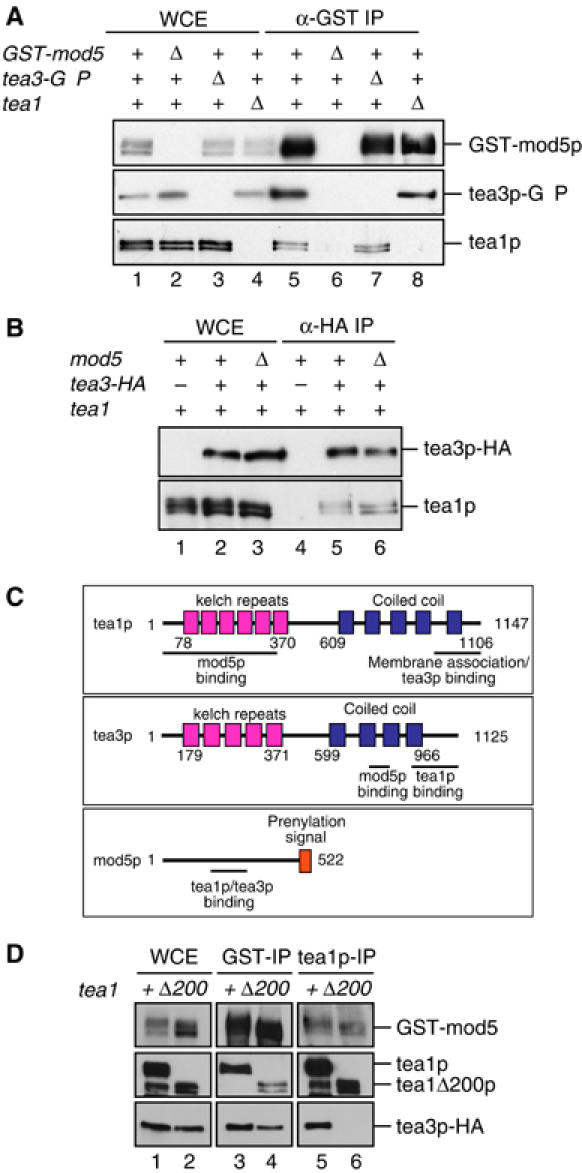
Tea1p, mod5p and tea3p form independent complexes in vivo. (A) GST-mod5p was immunoprecipitated from soluble protein extracts of wild-type cells expressing GST-mod5p and tea3p-GFP (lanes 1 and 5), mod5Δ cells expressing tea3p-GFP (lanes 2 and 6), tea3Δ cells expressing GST-mod5p (lanes 3 and 7) and tea1Δ cells expressing GST-mod5p and tea3p-GFP (lanes 4 and 8). The resulting immunocomplexes were analyzed for GST-mod5p, tea3p-GFP and tea1p. Whole-cell extract (WCE) fractions are shown in lanes 1–4 and immunoprecipitates in lanes 5–8. Immunoprecipitates were loaded 30 × relative to WCE sample. (B) Tea3p-HA was immunoprecipitated from soluble protein extracts of wild-type cells (lanes 1 and 4), wild-type cells expressing tea3p-HA (lanes 2 and 5) and mod5Δ cells expressing tea3p-HA (lanes 3 and 6). The resulting immunocomplexes were analyzed for tea3p-HA and tea1p. WCE fractions are shown in lanes 1–3 and immunoprecipitates in lanes 4–6. Immunoprecipitates were loaded 20 × relative to WCE. (C) Schematic diagram summarizing interactions between tea1p, tea3p and mod5p. (D) GST-mod5p (lanes 3 and 4) or tea1p (lanes 5 and 6) was immunoprecipitated from soluble protein extracts of either wild-type (lanes 1, 3 and 5) or tea1Δ200 (lanes 2, 4 and 6) cells expressing GST-mod5p and tea3p-HA. The resulting immunocomplexes were analyzed for GST-mod5p, tea1p (or tea1Δ200p) and tea3p-HA. WCE fractions are shown in lanes 1 and 2 and immunoprecipitates in lanes 3–6. Immunoprecipitates were loaded 30 × relative to WCE.
Arellano et al (2002) reported that tea3p bound to tea1p in the yeast two-hybrid system, but they did not verify this biochemically. We found that immunoprecipitation of tea3p-HA from fission yeast cell extracts co-precipitated tea1p (Figure 1B). In addition, we found that tea1p was co-precipitated in immunoprecipitates from cells expressing GST-mod5p (Figure 1A). Thus, tea1p, tea3p and mod5p all associate with each other in vivo.
Based on these results, we wanted to examine whether tea3p might act as a molecular ‘bridge' between tea1p and mod5p. We therefore used deletion strains to test whether each of the pairwise protein–protein interactions observed in immunoprecipitation experiments could occur in the absence of the third protein. As expected from two-hybrid results, tea1p interacted with tea3p in the absence of mod5p, and mod5p interacted with tea3p in the absence of tea1p (Figure 1A and B). Interestingly, however, we also found that tea1p interacted with mod5p in the absence of tea3p (Figure 1A). Thus, each of the pairwise interactions among tea1p, tea3p and mod5p is independent of the third protein.
Through a combination of immunoprecipitation experiments, two-hybrid analysis and in vitro binding studies, we mapped the regions of tea1p, tea3p and mod5p involved in binding to each other (Supplementary Figures 1B–F, 2A and B). The results are summarized in Figure 1C. Four important points emerged from these experiments. First, all of the observed interactions are likely to be direct. Second, a central region of mod5p (amino acids 156–205) is required for binding to both tea1p and tea3p. Third, even though tea1p and tea3p are structurally related, binding of tea1p to mod5p is mediated by the N-terminus of tea1p (amino acids 1–352), while binding of tea3p to mod5p is mediated by a central coiled-coil region of tea3p (amino acids 739–785). Fourth, binding of tea1p to tea3p is mediated by the C-termini of both proteins (amino acids 948–1147 of tea1p and amino acids 901–1125 of tea3p). These last two points are particularly salient because Behrens and Nurse (2002) have shown that deletion of the tea1p C-terminus (tea1Δ200) prevents anchoring of tea1p to the cell cortex. In this context, our mapping data suggest that the anchoring defect of tea1Δ200 mutants correlates not with a failure to bind mod5p but rather with a failure to bind tea3p and/or other proteins (see Discussion).
We next tested whether tea1p, tea3p and mod5p all coexist in a single protein complex in vivo. This was not entirely straightforward to determine, because physical analysis of tea1p shows it to be present in complexes covering a wide range of molecular sizes (Feierbach et al, 2004). Moreover, because each of the three proteins concerned can interact independently with the other two, immunoprecipitation of any one protein will co-precipitate both of the other two, even if a three-way complex does not exist. We therefore examined whether a three-way complex could be identified when one of the pairwise interactions was disrupted. Interestingly, we found that tea3p could not be co-immunoprecipitated with tea1Δ200p, even though both proteins could bind to mod5p in the same cell extract (Figure 1D). This suggests that a three-way complex of tea1p, tea3p and mod5p may be present only transiently in vivo, if at all. However, we also found that the absence of a three-way complex in vivo is unlikely to be a result of tea1p and tea3p competing for potentially overlapping binding sites on mod5p, as a three-way complex could be demonstrated artificially, in a yeast ‘bridging two-hybrid' assay (Supplementary Figure 2C).
The central region of mod5p is required for function
We next wanted to determine to what extent the protein–protein interactions identified might mediate the localization and/or function of mod5p, tea3p and tea1p, in order to understand how these interactions contribute to the proper cortical anchoring of tea1p at cell tips.
The only recognizable amino-acid sequence motif in mod5p is a C-terminal prenylation signal that is essential for mod5p function and localization (Snaith and Sawin, 2003). To identify other functionally significant regions, we fused GFP to a series of 50-amino-acid internal deletions spanning the mod5p open reading frame (i.e., those used in mapping studies; Supplementary Figure 2A) and expressed the internal-deletion mutant proteins individually in mod5Δ cells. In a quantitative polarity maintenance assay, nearly all of the deletions behaved like wild-type mod5p; only cells expressing either mod5Δ156–205p or mod5Δ206–255p, which fail to bind tea1p or tea3p (Supplementary Figure 2A and B), were unable to maintain polarity (Supplementary Figure 3A).
We also studied the localization of tea1p and the GFP-mod5p internal-deletion proteins themselves in each internal-deletion mutant strain. In nearly all cases, both tea1p and the mutant versions of mod5p were localized as in wild-type cells (Figure 2A, B, E and F; additional data not shown). However, in cells expressing either mutant mod5Δ156–205p or mod5Δ206–255p, tea1p localization resembled that of mod5Δ cells, with tea1p present on microtubule plus ends but failing to accumulate at the cell cortex (Figure 2C and D and Supplementary Figure 4E). These same two mutants were also defective in mod5 localization (Figure 2G and H), with the mutant GFP-mod5p spread out around the plasma membrane in the same manner as full-length wild-type mod5p in tea1Δ cells (Snaith and Sawin, 2003). Tea1p localization was not altered from wild type when any of the internal-deletion proteins was expressed in mod5+ rather than mod5Δ backgrounds, even though the localization of GFP-tagged mod5Δ156–205p and mod5Δ206–255p remained aberrant in these experiments (Figure 2I–L; additional data not shown). These results suggest that, in addition to the C-terminal prenylation site, the central region of mod5p, which interacts with both tea1p and tea3p, contains the major determinants for both mod5p localization and function.
Figure 2.
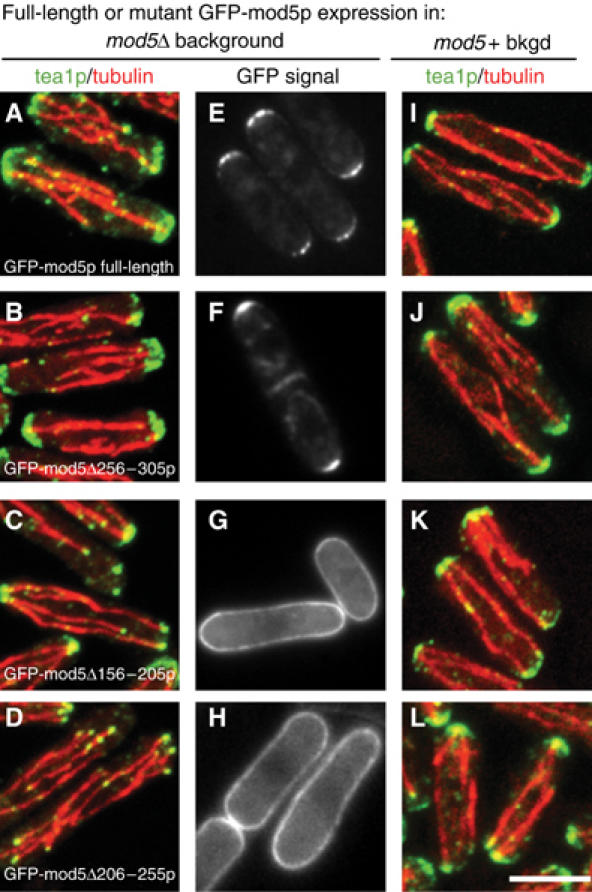
Mod5p amino acids 156–255 are essential for the localization of tea1p and mod5p. The localization of tea1p (green) and microtubules (red) in (A–D) mod5Δ cells and (I–L) wild-type cells, expressing different mutant versions of GFP-mod5p. (E–H) The localization of GFP-mod5p (and mutant versions) in mod5Δ cells. (A, E, I) Wild-type GFP-mod5p; (B, F, J) GFP-mod5Δ256–305p; (C, G, K) GFP-mod5Δ156–205p; (D, H, L) GFP-mod5Δ206–255p. The scale bar represents 5 μm.
We previously showed that restriction of mod5p to cell tips is dependent on tea1p and also partially on tea3p (Supplementary Figure 3B and C; Snaith and Sawin, 2003). In light of our results showing that mod5p can bind to tea1Δ200p, we examined the localization of GFP-mod5p in tea1Δ200 cells. GFP-mod5p was spread out around the membrane (Supplementary Figure 3D), suggesting that restriction of mod5p to cell tips requires not only the tea1p–mod5p interaction but also the stable binding of tea1p at the cell cortex.
Tea1p and mod5p are independently required for different aspects of tea3p localization
We next sought to investigate the roles played by tea1p and mod5p in the localization of tea3p. In wild-type cells, the majority of tea3p-GFP was confined to the cell tip region, with a few cytoplasmic dots also present, confirming previous results (Figure 3A; Arellano et al, 2002). In contrast, in tea1Δ cells, tea3p-GFP was still partially enriched at cell tips but also displayed a punctate staining evenly spread around the plasma membrane (Figure 3B; Arellano et al, 2002), reminiscent of GFP-mod5p localization in tea1Δ mutants (Snaith and Sawin, 2003). In addition, we observed an increase in the cytoplasmic tea3p-GFP signal (Figure 3B). In tea1Δ200 cells, the same mislocalization of tea3p-GFP was seen (Figure 3C), consistent with the carboxy-terminal region of tea1p being required for binding to tea3p (Supplementary Figure 1F).
Figure 3.
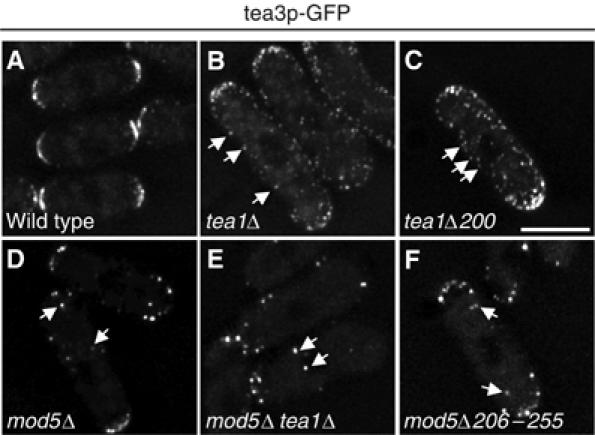
Localization dependencies of tea3p-GFP. Localization of tea3p-GFP in (A) wild-type, (B) tea1Δ, (C) tea1Δ200, (D) mod5Δ, (E) mod5Δ tea1Δ and (F) mod5Δ206–255 cells. The scale bar represents 5 μm.
Because mod5p is required for proper tea1p localization, and tea1p is required for tea3p localization, we suspected that mod5p would also be necessary for the correct localization of tea3p. Interestingly, however, the manner of tea3p-GFP mislocalization in mod5Δ mutants was different from that in tea1Δ mutants. In mod5Δ cells, the tea3p-GFP signal became highly punctate, with a few bright dots of tea3p-GFP visible near cell tips and also throughout the cell, suggesting that mod5p is required for a stable membrane localization of tea3p (Figure 3D). Consistent with this, we found that tea3p-GFP localization in mod5Δ tea1Δ double mutants resembled tea3p-GFP in mod5Δ single mutants, although tea3p-GFP localization to cell tips was more compromised than in single mutants (Figure 3E). These results support the notion that tea1p can promote the localization of tea3p to cell tips, while mod5p acts to stably integrate tea3p at the cell cortex, to the extent that the mislocalization of mod5p (i.e., in tea1Δ mutants) will recruit tea3p to ectopic cortical sites. We also found that in cells expressing mod5Δ156–205p or mod5Δ206–255p, tea3p-GFP was delocalized in a pattern similar to that present in mod5Δ cells (Figure 3F; data not shown). In conclusion, the overall tip localization of tea3p is dependent both on binding to mod5p and on the carboxy-terminus of tea1p.
Microtubule-based transport of tea3p to cell tips requires both tea1p and the absence of mod5p
Because tea1p binds to tea3p and facilitates its localization to cell tips, and tea1p is transported to cell tips by association with microtubule plus ends (Behrens and Nurse, 2002; Snaith and Sawin, 2003; Feierbach et al, 2004), we investigated whether tea3p is similarly transported to cell tips along microtubules. In wild-type cells, tea3p-GFP displayed little or no apparent movement toward cell tips (Supplementary Movie 1). Previously, however, we demonstrated that levels of tea1p-GFP transport to cell tips are elevated in mod5Δ cells relative to wild-type cells, and we argued that this might be due to the fact that an inability of tea1p to anchor at the cortex could result in a higher free cytoplasmic pool of tea1p able to associate with microtubules (Snaith and Sawin, 2003). We therefore followed tea3p-GFP in mod5Δ cells.
Here, we observed significant motion of cytoplasmic tea3p-GFP particles. Most of the fainter particles of tea3p-GFP showed rapid movements with no clear directionality. However, many of the bright particles of tea3p-GFP exhibited linear movements, both toward and away from cell tips (Figure 4A and Supplementary Movie 2). To test whether this movement was dependent on the actin or microtubule cytoskeletons, we treated cells with latrunculin B or methyl 2-benzimidazolecarbamate (carbendazim; MBC). Treatment of cells with latrunculin B for up to an hour had no significant effect on the movement of tea3p-GFP (data not shown). By contrast, within 5 min of MBC treatment, the movement of bright tea3p-GFP particles was reduced, and after 20 min it was almost abolished (data not shown), suggesting that tea3-GFP was moving on microtubules.
Figure 4.
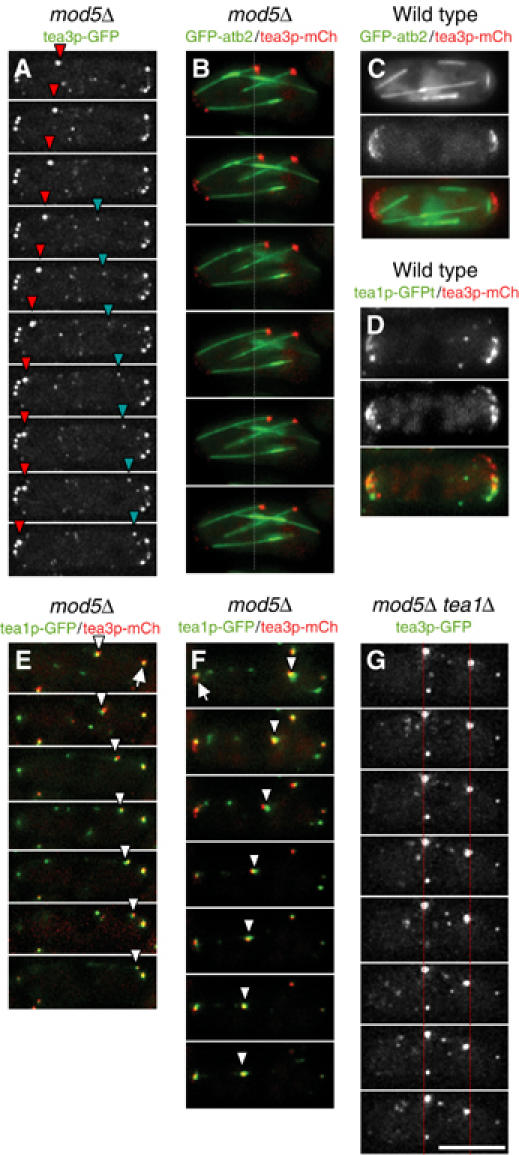
Microtubule-dependent movement of tea3p in mod5Δ. (A) Time-lapse movie frames of tea3p-GFP in mod5Δ cells at 15 s intervals. Red and turquoise arrowheads mark traveling particles of tea3p-GFP. (B) Time-lapse movie frames of GFP-atb2p (green) and tea3p-mCh (red) in mod5Δ cells at 15 s intervals. Gray dashed line indicates the starting position of the traveling tea3p-mCh particle. (C) Localization of GFP-atb2p (green) with tea3p-mCh (red) in wild-type cells. (D) Localization of tea1p-GFP (green) with tea3p-mCh (red) in wild-type cells. (E, F) Time-lapse movie frames of tea1p-GFP (green) and tea3-mCh (red) in mod5Δ cells at 15 intervals. Traveling particles of colocalized tea1p-GFP and tea3p-mCh are indicated by white arrowheads and static particles are indicated by white arrows. (G) Time-lapse movie frames of tea3p-GFP in mod5 tea1Δ cells at 15 s intervals. Red dotted lines indicate reduced movement of tea3p-GFP. The scale bar represents 5 μm.
To confirm this, we developed a series of tagging plasmids derived from novel variants of the red fluorescent protein dsRed (Shaner et al, 2004; see Supplementary data) and used these to construct strains carrying tea3p tagged with the variant mCherry (tea3p-mCh), in order to image tea3p in conjunction with GFP-tubulin (GFP-atb2p; Adachi et al, 1986). In wild-type cells, tea3p-mCh accumulated at cell tips, consistent with the localization of tea3p-GFP, although in further experiments we found that fusion of tea3p with mCherry may in fact compromise some more subtle aspects of tea3p behavior as compared to fusion with GFP (data not shown). When we imaged tea3p-mCh together with GFP-tubulin in mod5Δ mutants, tea3p-mCh particles were readily detected on the ends of growing microtubules (Figure 4B). In wild-type cells, however, tea3p-mCh particles failed to colocalize with microtubule ends (Figure 4C).
We next used tea3p-mCh to test whether tea3p was cotransported with tea1p to cell tips. In wild-type cells, some cortical particles of tea3p-mCh colocalized with tea1p-GFP, while others remained distinct from tea1p-GFP (Figure 4D), suggesting that there may be distinct populations of each protein, with an overlapping subset. In mod5Δ cells, particles of tea3p-mCh colocalized with tea1p-GFP as they translocated both toward and away from the cell tips (Figure 4E and F). (As described in Supplementary data, relatively long exposure times were required during time-lapse acquisition, resulting in occasional misalignment of the moving tea3p-mCh and tea1p-GFP signals.) Thus, tea3p and tea1p can move together.
Finally, we tested whether the movement of tea3p-GFP seen in mod5Δ cells was dependent on tea1p. In mod5Δ tea1Δ double mutant cells, the linear movement of bright particles of tea3p-GFP was almost completely abolished (Figure 4G and Supplementary Movie 3). Collectively, these results suggest that tea3p does not significantly associate with tea1p or microtubules in the cytoplasm in wild-type cells. However, in mod5Δ cells, where tea3p is no longer sequestered at cells tips, tea3p can associate with tea1p in the cytoplasm and as a result is cotransported with tea1p along microtubules to cell tips.
Mod5p and tea3p have distinct but overlapping functions in cell polarity and tea1p anchoring
Both mod5Δ and tea3Δ mutants have been shown to have growth-polarity defects, but thus far each mutant has been examined in a distinct type of polarity assay (Arellano et al, 2002; Snaith and Sawin, 2003). To determine whether mod5p and tea3p act in a single common pathway, we tested several mutants side by side in both assays.
In return-to-growth experiments involving cells previously grown to stationary phase, wild-type cells re-establish polarity axes at pre-existing cell ends, and tea1Δ mutants form branched cells (Browning et al, 2000; Snaith and Sawin, 2003). By contrast, mod5Δ mutants re-establish polarity normally when microtubules are intact but form branches when microtubules are disrupted with MBC, which impairs delivery of tea1p to cell tips (Snaith and Sawin, 2003). We found that tea3Δ cells, like mod5Δ cells, formed branches only in the presence of MBC (Figure 5A). If tea3p and mod5p both acted in a strictly linear genetic pathway to regulate cell polarity, we might expect that the phenotype of the mod5Δ tea3Δ double mutant would resemble the single mutants. However, a significant fraction of mod5Δ tea3Δ mutants (20%) formed branched cells even without microtubule disruption, much more than occurs in either single mutant (Figure 5A). This suggests that although mod5p and tea3p both interact and contribute to growth polarity in fission yeast, each protein may make a distinct contribution to cell polarity and tea1p function.
Figure 5.
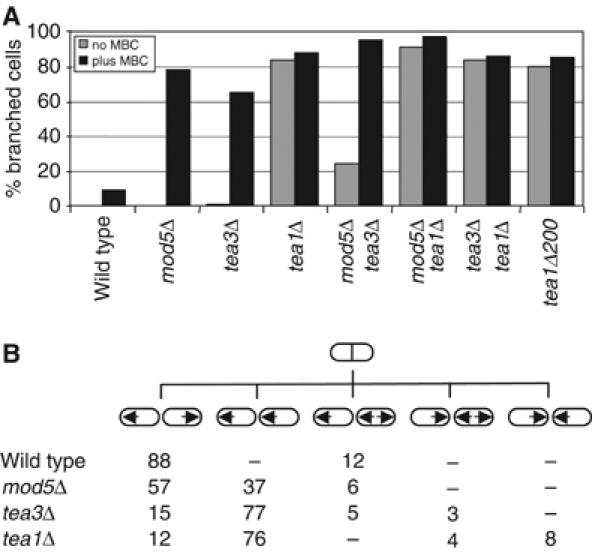
Mod5p and tea3p have distinct but overlapping functions. (A) Wild-type, mod5Δ, tea3Δ, tea1Δ, mod5Δ tea3Δ, mod5Δ tea1Δ, tea3Δ tea1Δ and tea1Δ200 cells were depolarized by growth to stationary phase and returned to fresh medium in the absence (gray bars) or presence (black bars) of 50 μg/ml MBC for 3 h at 32°C. The percentage of branched cells in each sample was counted, n=200. (B) Percentage of daughter cell pairs displaying illustrated initial growth patterns after septation in wild-type (n=133), mod5Δ (n=228), tea3Δ (n=165) and tea1Δ cells (n=194). Arrows indicate direction of growth.
In these experiments, we also found that tea1Δ200 cells formed branches both in the presence and absence of MBC (Figure 5A) and thus resemble tea1Δ rather than mod5Δ or tea3Δ mutants with regard to polarity defects. Because tea1Δ200p can still bind to mod5p (Supplementary Figure 1E), this suggests that the essential function of the C-terminus of tea1p in anchoring tea1p may be required subsequent to the tea1p–mod5p interaction. In addition, because tea1Δ200p fails to bind tea3p but the tea1Δ200 phenotype is more extreme than the tea3Δ phenotype, this indicates that the tea1p carboxy-terminus is likely to interact functionally not only with tea3p but also with other proteins as well (see Discussion).
The second assay involved time-lapse observations of growth patterns in the different mutants. Whereas tea3Δ cells mainly exhibit monopolar growth, in a pattern similar to tea1Δ mutants (Glynn et al, 2001; Arellano et al, 2002), we found that the majority of mod5Δ cells display wild-type growth patterns, with less than 50% of cells showing the aberrant growth patterns characteristic of tea1Δ and tea3Δ mutants (Figure 5B). This result strongly supports a view of overlapping but distinct functions for mod5p and tea3p. Moreover, since the monopolar growth defects in mod5Δ mutants are much less severe than in tea1Δ mutants, these data also suggest that at least some ‘bipolar growth' functions of tea1p can bypass the need for mod5p at cell tips (see Discussion).
While we have shown that mod5p is required for anchoring of tea1p at cell tips (Snaith and Sawin, 2003), Arellano et al (2002) reported that tea3p does not play a role in tea1p localization. Our studies with these proteins prompted us to re-examine the localization of tea1p in tea3Δ mutants. We found that although tea1p is present at both tips of tea3Δ cells, tea3Δ cells have only 76% of the tea1p levels seen at cell tips in wild-type cells, while tea1p localization in mod5Δ tea3Δ double mutants resembles tea1p localization in mod5Δ cells (Supplementary Figure 4). Because this reduction of tip-localized tea1p in tea3Δ mutants could reflect a role for tea3p in anchoring tea1p at cell tips, we examined tea1p levels at cell tips in wild-type cells and tea3Δ mutants after microtubule disruption by MBC, where new tea1p delivery to cell tips is prevented. Under these conditions, tea1p is rapidly lost from both cell tips in mod5Δ mutants (HA Snaith, unpublished data). Whereas wild-type cells gradually lost tea1p symmetrically from both cell tips (Figure 6A and D), tea3Δ cells rapidly lost tea1p from one tip, such that after 5 min of MBC treatment, nearly all tea3Δ cells displayed monopolar tea1p localization (Figure 6B and E). To determine whether this asymmetric loss of tea1p was simply due to the monopolar growth pattern of tea3Δ cells, we assayed tea1p localization in the pak1/shk1/orb2 mutant orb2-34, which also exhibits strongly monopolar growth (Verde et al, 1995; Kim et al, 2003). In contrast to tea3Δ cells, orb2-34 cells lost tea1p symmetrically after treatment with MBC (Figure 6C and F). We also examined actin and tea1p localization in MBC-treated tea3Δ cells to determine whether tea1p was lost from the growing tip, the nongrowing tip or randomly from either tip (Figure 6I–N). In nearly all tea3Δ cells, tea1p was lost from the nongrowing, actin-poor tip; the small number of cases in which loss of tea1p was symmetrical (7%, n=100) corresponded to those few tea3Δ cells that had initiated bipolar growth. These results indicate that tea3p is required for the proper cortical anchoring of tea1p, specifically at nongrowing cell tips.
Figure 6.
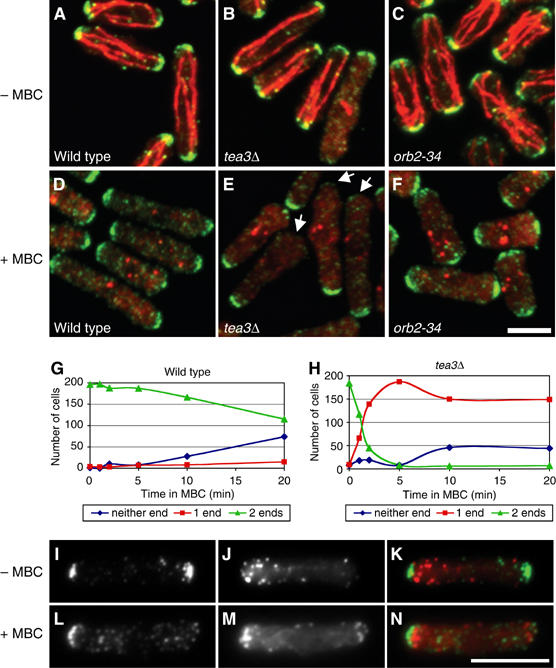
Tea1p is preferentially lost from the nongrowing tip in tea3Δ cells. (A–H) Treatment of (A, D) wild-type, (B, E) tea3Δ and (C, F) orb2-34 cells with MBC for (A–C) 0 min or (D–F) 5 min. Cells are stained for tea1p (green) and microtubules (red). Time course of tea1p loss from cell tips in (G) wild-type and (H) tea3Δ cells, showing number of cells with detectable levels of tea1p at one cell tip, two tips or neither tip, for each strain. n=200 for each strain. (I–N) Treatment of tea3Δ cells with MBC for (I–K) 0 min or (L–N) 5 min. Cells are stained with anti-tea1p antibodies (I, L) and Alexa-labeled phalloidin (J, M). Merged imaged are shown with tea1p in green and phalloidin in red (K, N). The scale bars represent 5 μm.
Discussion
Multistep and multimode tea1p anchoring
We have demonstrated multiple interactions among tea1p, tea3p and mod5p and examined their roles in cell polarity and in regulating protein localization at cell tips, especially that of tea1p. Because interactions between any two of these three proteins can occur in vivo in the absence of the third protein, while mutation and/or deletion of any one protein characteristically alters the localization of each of the other two, it might appear difficult to place these interactions at specific points in a pathway regulating tea1p anchoring. However, by taking into account our additional findings—for example, the asymmetric cell-tip loss of tea1p in tea3Δ mutants, the cotransport of tea3p with tea1p only in the absence of mod5p and the behavior of tea1Δ200p—we have formulated a multistep and multimode model for the efficient anchoring of tea1p at cell tips (Figure 7). The model is ‘multistep' because it postulates that efficient anchoring of tea1p at the cortex occurs in several stages, and ‘multimode' because we propose that the mechanisms of tea1p anchoring are different in the presence or absence of mod5p, and at growing versus nongrowing cell tips. According to the model, tea1p is initially in a transport complex on microtubule plus ends, in association with other plus-end binding proteins (Figure 7A; also see below; Busch et al, 2004; Feierbach et al, 2004). Upon reaching cell tips, tea1p interacts, via its N-terminus, with a central region of mod5p (Figure 7B). This interaction is important for the anchoring of tea1p at the cortex (Snaith and Sawin, 2003) but is by itself insufficient for the correct steady-state localization of tea1p and mod5p. Tea1p then further interacts, via its C-terminus, with cortically localized tea3p. This may occur at both cell tips but is functionally important primarily at nongrowing tips (Figure 7C). Our data further indicate that tea3p is itself anchored at cell tips by a strong interaction with a central region of mod5p (Figure 7D). We also suggest that at growing cell tips, there may be a parallel, tea3p-independent pathway of tea1p retention, involving both a tea1p–mod5p interaction and an interaction of tea1p with other cortical proteins (Figure 7E). Finally, we propose that in addition to the mod5p-dependent pathway of cortical tea1p anchoring, a complementary mechanism exists whereby tea1p and tea3p functionally interact both in the cytoplasm and at the cortex to ‘bypass' mod5p (Figure 7F). Tea1p anchored at cell tips (Figure 7G) would then be capable of making further interactions with its ‘downstream' effectors, perhaps also further strengthening its retention as a result.
Figure 7.
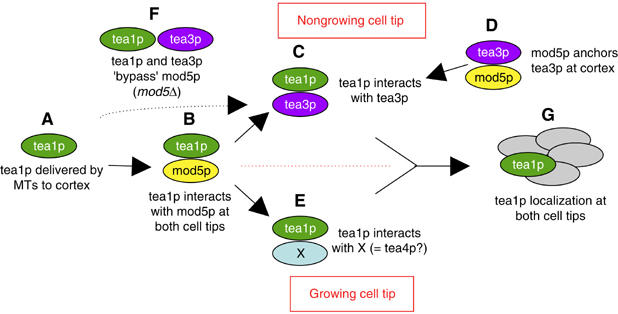
Multistep cortical retention of tea1p. A model of protein–protein interactions regulating tea1p localization at cell tips. (A) Tea1p arrives at the cell cortex on microtubule plus ends. (B) The tea1p N-terminus interacts with mod5p at both cell tips. (C) At nongrowing cell tips, the tea1p C-terminus interacts with tea3p, which is itself anchored at the cortex (D) via interaction with mod5p. (E) At growing cell tips, tea1p interacts with unknown factors (X) that provide a function similar to that of tea3p at nongrowing tips. (F) A partially functional interaction between tea1p and tea3p can occur without mod5p, but tea1p and tea3p will be poorly anchored. (G) Together, these interactions lead to proper tea1p anchoring at both cell tips. See text for further details.
Structure of the model
Because of the diverse nature of the results contributing to our model, we discuss more specific aspects of the model below.
A cortical interaction between tea3p and mod5p. Several pieces of evidence support the idea that tea3p and mod5p form a stable cortical complex. First, tea3p and mod5p have a robust biochemical interaction both in the presence and absence of tea1p; indeed, tea3p was the only strong interactor with mod5p identified in our two-hybrid screen. Second, we note that the delocalized tea3p seen in tea1Δ cells is nevertheless still associated with the cell cortex, in a pattern closely resembling that of mod5p (Figure 3; Arellano et al, 2002). As this cortical localization is lost when both tea1p and mod5p are deleted, it seems likely that the aberrant cortical localization of tea3p in tea1Δ mutants is due to its stable interaction with mod5p.
The C-terminus of tea1p is important subsequent to the tea1p–mod5p interaction. Tea1Δ200p is found at microtubule plus ends at both cell tips but is not properly anchored at the cortex (Behrens and Nurse, 2002). We had initially expected this to be due to a failure of tea1Δ200p to bind to membrane-associated mod5p, but instead we found that tea1Δ200p can interact with mod5p in vivo. This suggests that the ‘failure point' of tea1Δ200p in a tea1p-anchoring pathway occurs after the tea1p–mod5p interaction, and that interactions of the tea1p C-terminus with other proteins are additionally required for tea1p anchoring. Because tea1Δ200p fails to interact with tea3p, we have placed a tea1p–tea3p interaction subsequent to the tea1p–mod5p interaction at nongrowing cell tips, which accords with the demonstrated role for tea3p in the retention of tea1p at these sites. Furthermore, since deletion of tea3p does not phenocopy tea1Δ200, we propose that there may also be a ‘parallel mode' of tea3p-independent tea1p retention at the growing cell tip (see below).
Parallel tea1p-anchoring systems at growing and nongrowing tips. Our results suggest that there are both common and distinct elements in the mechanisms regulating tea1p localization at growing versus nongrowing cell tips. Whereas mod5p and the tea1p C-terminus are each required for tea1p anchoring at both cell tips, tea3p is required primarily for tea1p anchoring at nongrowing tips, consistent with its preferential localization to nongrowing tips (Arellano et al, 2002). Thus, at growing cell tips, other tea1p-binding proteins may perform functions analogous to those performed by tea3p at nongrowing tips. Further support for the suggestion that a parallel pathway at growing cell tips involves the tea1p–mod5p interaction comes from the observation that mod5Δ tea3Δ double mutants form ectopic branches at a much higher frequency than tea3Δ single mutants (Figure 3A). This indicates that mod5p makes additional contributions to polarity fidelity outside a strictly ‘tea3p-specific' pathway. Recently, a novel SH3-domain protein, tea4p/wsh3p, has been identified that may perform the ‘tea3p-analogous' function at growing cell tips; interestingly, tea4p, like tea3p, binds to the C-terminus of tea1p (Martin et al, 2005; Tatebe et al, 2005).
The tea1p–tea3p interaction can ‘bypass' mod5p. This suggestion is based not only on the strong tea1p–tea3p interaction seen in mod5Δ cells but also on mutant cell-polarity phenotypes. Both tea1Δ and tea3Δ mutants have more severe monopolar growth defects than mod5Δ (Figure 5B), indicating that tea1p and tea3p still retain some function in organizing growth polarity in the absence of mod5p, even if they are not stably anchored at cell tips during steady-state growth. Further support comes from the observation that mod5Δ tea3Δ double mutants form ectopic branches at a much higher frequency than mod5Δ single mutants (Figure 5A). Moreover, antipodal growth persists in mod5Δ mutants as long as there is constant delivery of tea1p to cell tips by microtubules (Snaith and Sawin, 2003). Most importantly, our proposal for a ‘mod5p bypass' is directly supported by the observation that tea3p is cotransported with tea1p to cell tips when mod5+ is deleted (Figure 4).
Limits of the model. An important issue not explicitly addressed in the model is how the hypothesized positive feedback between tea1p and mod5p localization may operate (Snaith and Sawin, 2003). We have shown that two mod5 internal-deletion mutants fail to be restricted to cell tips even when tea1p is properly localized at the cortex, but we cannot yet distinguish whether this is directly due to their failure to bind to tea1p, or to tea3p, or both. This will benefit from further studies.
Concluding remarks
In summary, a complex set of pairwise protein–protein interactions among tea1p, tea3p and mod5p is essential for the correct cortical anchoring of tea1p after its delivery to cell tips by microtubules. In many eukaryotic systems, proteins associated with microtubules are subsequently targeted to the cell cortex, and it is likely that this is achieved by multiple mechanisms (Schuyler and Pellman, 2001; Gundersen, 2002; Carvalho et al, 2003; Akhmanova and Hoogenraad, 2005; Watanabe et al, 2005). In relation to this, we note that a key point of our ‘mod5p bypass' model is that the two proposed mechanisms of achieving a tea1p–tea3p interaction at cell tips are not only complementary but also mutually exclusive. Moreover, the exclusive aspect of their relationship emerges from the properties of the system as a whole. In wild-type cells, tea1p interacts with tea3p primarily at the cell cortex, because tea3p is sequestered there by mod5p. However, in, and only in, mod5Δ cells, tea3p is no longer similarly sequestered, and tea1p can therefore interact with tea3p in the cytoplasm to form stable complexes that then travel to the cortex. Importantly, even if the interaction of tea1p and tea3p in mod5Δ cells is less productive than in wild-type cells (mod5Δ cells still exhibit some defects in polarized growth), both scenarios result in an interaction of tea1p and tea3p at the cell cortex, which would then allow tea1p to interact with effector binding partners. In the context of ‘systems cell biology', this may reflect a kind of homeostatic design principle by which desired outputs can be achieved, even with some degree of fine-tuning, in the face of changing amounts of a critical system component, in this case mod5p.
Materials and methods
General methods
S. pombe methods were as described (Moreno et al, 1991). Molecular biology methods were essentially as described (Sambrook and Russell, 2001). Epitope-tagged mod5+ and tea3+ strains were constructed by PCR-based gene targeting as described (Bähler et al, 1998). Tea3p-GFP, tea3Δ and tea1Δ200 strains were gifts from P Nurse, The Rockefeller University, NY. The two-hybrid library was a kind gift from T Nakamura, Osaka City University, Japan, and pRSETB-mCherry and pRSETB-tdTomato were gifts of R Tsien, UCSD, CA. Complete lists of plasmids and strains used in this work appear as Supplementary data.
Antibodies
Anti-GST antibodies were obtained from Amersham (Little Chalfont, UK); anti-HA 12CA5 antibodies were a gift from I Stancheva, University of Edinburgh, UK, and anti-TAT1 hybridoma supernatant was a gift from K Gull, University of Oxford, UK (Woods et al, 1989). Affinity-purified tea1p antibodies were used for Western blotting, and serum was used for immunofluorescence (Snaith and Sawin, 2003). Anti-tea1p antibodies were purified against a 6-His fusion of tea1p amino acids 554–1147 (tea1p-C) expressed in bacteria (Mata and Nurse, 1997). Tea1p-C was isolated as inclusion bodies, solubilized in SDS and coupled to Affigel 15 as described (Sawin et al, 1992). Alexafluor-conjugated phalloidin and secondary antibodies were obtained from Molecular Probes (Eugene, OR).
Immunoprecipitation studies
For immunoprecipitation experiments from yeast cell extracts, GST-mod5p was expressed from the nmt41 promoter to steady-state levels. Tea1p, tea3p-GFP and tea3p-HA were all expressed from their endogenous promoters. A 400–500 mg portion of frozen cell pellets was ground with a mortar and pestle under liquid nitrogen and extracts were prepared in 20 mM NaHEPES pH 7.5, 50 mM K-acetate, 200 mM NaCl, 1 mM EDTA, 2 mM MgCl2 and 0.2% Triton X-100. A 5–8 mg portion of total protein was used in each immunoprecipitation in a final volume of 500 μl. Samples were incubated with 5 μg of anti-HA 12CA5, anti-GST or anti-tea1p antibodies, and 25 μl of protein A or protein G sepharose beads as appropriate, for 2–3 h at 5°C. All samples were separated on 8% SDS–PAGE and subjected to Western blotting.
Physiology and immunofluorescence experiments
Immunostaining with anti-tea1p serum and anti-tubulin antibodies was exactly as described (Snaith and Sawin, 2003). Images of entire cell volume were captured using a Leica TCS-SP confocal microscope (Leica Microsystems, Milton Keynes, UK). Fluorescence quantitation of anti-tea1p immunofluorescence (n=130 cells) was performed as described (Snaith and Sawin, 2003). For costaining tea3Δ cells for actin and tea1p, cells were grown in YES and fixed exactly as described (Sawin and Nurse, 1998), and then digested for 15 min at 37°C in 25 μg/ml zymolyase 20-T. Images were captured using wide-field microscopy on a Nikon TE300 system (Snaith and Sawin, 2003). Polarity re-establishment experiments were carried out as described (Snaith and Sawin, 2003). In all experiments, >200 cells were counted for each sample. Treatment with 50 μg/ml MBC was used to disrupt microtubules and 200 μM latrunculin B was used to disrupt actin filaments, during log-phase growth.
Live cell imaging
All images were collected on the Nikon TE300 microscope system. Bright-field imaging of cell growth patterns was performed as described (Glynn et al, 2001). At least 130 cell divisions (i.e., growth patterns in at least 260 daughter cells) were analyzed for each strain. For fluorescence imaging, cells were grown at 25°C in EMM and imaged at room temperature. Expression of mutant GFP-mod5p was induced for 2 days at 32°C in EMM plus 150 nM thiamine before shifting to 25°C for 4 h prior to imaging single planes. Z-series of tea3p-GFP still images were deconvolved using Deltavision Softworx (Applied Precision) and projected into a single plane. Z-series from doubly labeled tea3p-mCh tea1p-GFP or tea3p-mCh GFP-atb2p strains were projected into a single plane without further manipulation. Further details of imaging conditions are given in Supplementary data.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary data
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Acknowledgments
We thank M Masterton for invaluable help in constructing yeast strains expressing mod5p deletion proteins; K Gull, T Nakamura, P Nurse, H Ohkura I Stancheva and R Tsien for sharing strains and reagents; A Merdes and I Davis for critical reading of the manuscript; S Martin for helpful discussion of our data; and F Philp for continuing encouragement. HAS is a Caledonian Research Foundation Post-doctoral Research Fellow and KES is a Wellcome Trust Senior Research Fellow. This work was supported by the Caledonian Research Foundation and the Wellcome Trust.
References
- Adachi Y, Toda T, Niwa O, Yanagida M (1986) Differential expressions of essential and nonessential alpha-tubulin genes in Schizosaccharomyces pombe. Mol Cell Biol 6: 2168–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J, Kelso R, Cooley L (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10: 17–24 [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC (2005) Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol 17: 47–54 [DOI] [PubMed] [Google Scholar]
- Arellano M, Niccoli T, Nurse P (2002) Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr Biol 12: 751–756 [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Behrens R, Nurse P (2002) Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J Cell Biol 157: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG (2002) ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol 3: 586–599 [DOI] [PubMed] [Google Scholar]
- Browning H, Hackney DD, Nurse P (2003) Targeted movement of cell end factors in fission yeast. Nat Cell Biol 5: 812–818 [DOI] [PubMed] [Google Scholar]
- Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR (2000) Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol 151: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P (2000) CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102: 695–704 [DOI] [PubMed] [Google Scholar]
- Busch KE, Hayles J, Nurse P, Brunner D (2004) Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell 6: 831–843 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Tirnauer JS, Pellman D (2003) Surfing on microtubule ends. Trends Cell Biol 13: 229–237 [DOI] [PubMed] [Google Scholar]
- Chang F, Peter M (2003) Yeasts make their mark. Nat Cell Biol 5: 294–299 [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ (1996) Origins of cell polarity. Cell 84: 335–344 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F (2001) Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol 11: 1656–1665 [DOI] [PubMed] [Google Scholar]
- Feierbach B, Verde F, Chang F (2004) Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol 165: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Lustig RJ, Berlin A, Chang F (2001) Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr Biol 11: 836–845 [DOI] [PubMed] [Google Scholar]
- Gundersen GG (2002) Microtubule capture: IQGAP and CLIP-170 expand the repertoire. Curr Biol 12: R645–R647 [DOI] [PubMed] [Google Scholar]
- Hayles J, Nurse P (2001) A journey into space. Nat Rev Mol Cell Biol 2: 647–656 [DOI] [PubMed] [Google Scholar]
- Jin H, Amberg DC (2001) Fission yeast Aip3p (spAip3p) is required for an alternative actin-directed polarity program. Mol Biol Cell 12: 1275–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang P, Catanuto P, Verde F, Lai H, Du H, Chang F, Marcus S (2003) The kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in the fission yeast, Schizosaccharomyces pombe. J Biol Chem 278: 30074–30082 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ (2004) Crystal structure of the Kelch domain of human Keap1. J Biol Chem 279: 54750–54758 [DOI] [PubMed] [Google Scholar]
- Martin SG, Chang F (2003) Cell polarity: a new mod(e) of anchoring. Curr Biol 13: R711–R713 [DOI] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR III, Chang F (2005) Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell 8: 479–491 [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P (1997) Tea1p and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89: 939–949 [DOI] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P (1985) Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci 75: 357–376 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Niccoli T, Arellano M, Nurse P (2003) Role of Tea1p, Tea3p and Pom1p in the determination of cell ends in Schizosaccharomyces pombe. Yeast 20: 1349–1358 [DOI] [PubMed] [Google Scholar]
- Prag S, Adams JC (2003) Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sawin KE, Mitchison TJ, Wordeman LG (1992) Evidence for kinesin-related proteins in the mitotic apparatus using peptide antibodies. J Cell Sci 101 (Part 2): 303–313 [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P (1998) Regulation of cell polarity by microtubules in fission yeast. J Cell Biol 142: 457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Snaith HA (2004) Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J Cell Sci 117: 689–700 [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D (2001) Microtubule ‘plus-end-tracking proteins': the end is just the beginning. Cell 105: 421–424 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Small JV, Kaverina I (2003) Microtubules meet substrate adhesions to arrange cell polarity. Curr Opin Cell Biol 15: 40–47 [DOI] [PubMed] [Google Scholar]
- Snaith HA, Sawin KE (2003) Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423: 647–651 [DOI] [PubMed] [Google Scholar]
- Snaith HA, Sawin KE (2005) Tea for three: control of fission yeast polarity. Nat Cell Biol 7: 450–451 [DOI] [PubMed] [Google Scholar]
- Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K (2005) Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol 15: 1006–1015 [DOI] [PubMed] [Google Scholar]
- Vega LR, Solomon F (1997) Microtubule function in morphological differentiation: growth zones and growth cones. Cell 89: 825–828 [DOI] [PubMed] [Google Scholar]
- Verde F (2001) Cell polarity: a tale of two Ts. Curr Biol 11: R600–R602 [DOI] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P (1995) Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol 131: 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K (2005) Regulation of microtubules in cell migration. Trends Cell Biol 15: 76–83 [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci 93: 491–500 [DOI] [PubMed] [Google Scholar]
- Yarm F, Sagot I, Pellman D (2001) The social life of actin and microtubules: interaction versus cooperation. Curr Opin Microbiol 4: 696–702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary data
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3


