Abstract
Most metazoan messenger RNAs encoding histones are cleaved, but not polyadenylated at their 3′ ends. Processing in mammalian cell extracts requires the U7 small nuclear ribonucleoprotein (U7 snRNP) and an unidentified heat-labile factor (HLF). We describe the identification of a heat-sensitive protein complex whose integrity is required for histone pre-mRNA cleavage. It includes all five subunits of the cleavage and polyadenylation specificity factor (CPSF), two subunits of the cleavage stimulation factor (CstF), and symplekin. Reconstitution experiments reveal that symplekin, previously shown to be necessary for cytoplasmic poly(A) tail elongation and translational activation of mRNAs during Xenopus oocyte maturation, is the essential heat-labile component. Thus, a common molecular machinery contributes to the nuclear maturation of mRNAs both lacking and possessing poly(A), as well as to cytoplasmic poly(A) tail elongation.
Keywords: Symplekin, polyadenylation, 3′-end processing, U7 snRNP, histone mRNA, Cajal body
During the S phase of the cell cycle, histone mRNA levels are up-regulated to meet the need for histones to package newly synthesized DNA. Increased transcription of the histone genes, increased efficiency of 3′-end formation, and stabilization of mature histone messenger RNAs all contribute (for review, see Marzluff and Duronio 2002). In metazoa, most histone transcripts are not polyadenylated. Rather, their 3′ ends are produced by a simple endonucleolytic cleavage event that is not followed by polymerization of adenylate residues onto the resulting 3′-hydroxyl group (Birchmeier et al. 1984; Krieg and Melton 1984; Gick et al. 1986).
The signal for 3′-end processing of histone pre-mRNAs consists of a conserved stem-loop positioned upstream of the cleavage site and a purine-rich histone downstream element (HDE) (for review, see Marzluff and Duronio 2002). The HDE is recognized through base pairing to the 5′ end of the U7 small nuclear RNA (Schaufele et al. 1986; Bond et al. 1991), which is incorporated into a rib-nucleoprotein (RNP) of the Sm class (U7 snRNP). The stem-loop is bound by stem-loop-binding protein (SLBP), which helps recruit the U7 snRNP to the pre-mRNA (Dominski et al. 1999). A zinc finger protein (ZFP100) facilitates this recruitment (Dominski et al. 2002; Pillai et al. 2003) by bridging between SLBP and Lsm11, a unique component of the U7-specific Sm core, in which the spliceosomal SmD1 and SmD2 proteins are replaced by Lsm10 and Lsm11 (Pillai et al. 2001, 2003). The site of endonucleolytic attack, which usually occurs after a CA dinucleotide in the histone pre-mRNA sequence, is located 10-11 nucleotides (nt) upstream of the intermolecular U7/HDE RNA duplex (Scharl and Steitz 1994, 1996). In mammalian nuclear extracts, SLBP is not required for in vitro processing of pre-mRNA substrates with strong complementarity to the 5′ end of U7 snRNA (Streit et al. 1993; Dominski et al. 1999).
In addition to the U7 snRNP (Strub et al. 1984; Mowry and Steitz 1987), an unidentified heat-labile factor (HLF) (Gick et al. 1987; Lüscher and Schümperli 1987) is absolutely essential for histone pre-mRNA processing in extracts of mammalian cells. The HLF was described almost 20 years ago (Gick et al. 1987) and is distinct from the U7 snRNP in that it is resistant to micrococcal nuclease and is not depleted by anti-Sm antibodies. It is inactivated by treatment of nuclear extract for 15 min at 50°C, while the U7 snRNP remains stable and active under these conditions (Gick et al. 1987). The HLF is present in small amounts in extracts prepared from G1-arrested cells, but is highly enriched in extracts from exponentially growing cells (Lüscher and Schümperli 1987).
We describe purification of the HLF from HeLa cell nuclear extract and show that it is a large complex that shares subunits with the well-characterized CPSF and CstF complexes (Takagaki et al. 1989) involved in the cleavage and polyadenylation of the majority of messenger RNAs. Most unexpected was the identification of symplekin as the heat-sensitive component, a protein that has been implicated in nuclear polyadenylation (Takagaki and Manley 2000; Xing et al. 2004) and assigned an essential role in the elongation of poly(A) tails in the cytoplasm (Barnard et al. 2004).
Results
Fractionation of the HLF reveals polyadenylation components and symplekin
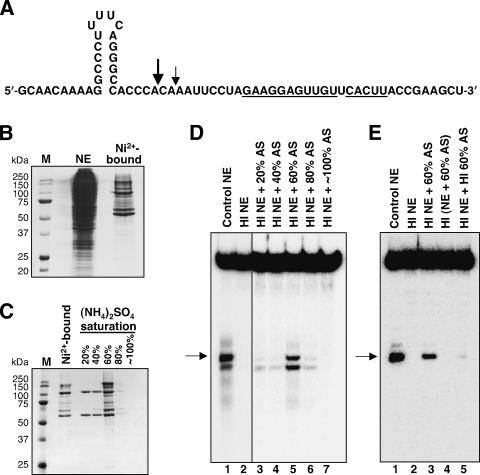
To identify factors required for histone mRNA 3′-end processing, we used an RNA substrate derived from the mouse histone H4-12 pre-mRNA, whose HDE has excellent complementarity to the 5′ end of U7 snRNA (Fig. 1A) and thus undergoes SLBP-independent cleavage in vitro (Streit et al. 1993). In preliminary experiments, we had used a Ni2+-affinity resin to purify the transcription factor Yin Yang 1 (YY1) (Shi et al. 1991) because it had been shown to activate transcription of histone genes (Eliassen et al. 1998; Last et al. 1999) and has zinc fingers homologous to those of ZFP100 (Dominski et al. 2002). Since YY1 contains 11 consecutive histidines, we incubated crude HeLa cell nuclear extract with the resin under stringent conditions: 400 mM salt and 20 mM imidazole (Fig. 1B). Although subsequent experiments indicated no direct role for YY1 in histone pre-mRNA processing, we discovered that the material eluted from the Ni2+-affinity resin could complement the processing activity of a heat-inactivated extract (data not shown).
Figure 1.
HLF activity is retained on Ni2+-affinity resin. (A) The histone H4-12 pre-mRNA substrate used in the in vitro processing assays. The sequence that is complementary to the 5′-end of U7 snRNA is underlined. Large and small arrows indicate the alternative (major and minor) positions of endonucleolytic scission (Streit et al. 1993). (B) Coomassie brilliant blue-stained SDS-PAGE of the crude nuclear extract (NE) starting material (1/250th) and of the material eluted from Ni2+-NTA agarose (1/50th). Lane M shows molecular weight marker proteins. (C) Saturated (NH4)2SO4 was added progressively to the material eluted from the Ni2+ resin to achieve the indicated percent saturation values. Precipitated proteins were resuspended and visualized by Coomassie staining after separation on 10% SDS-PAGE. (D) Complementation of in vitro processing. HeLa nuclear extract (NE) was incubated at 50°C for 15 min to generate heat-inactivated extract (HI), which was used for processing of a uniformly labeled H4-12 pre-mRNA substrate either without supplementation (lane 2) or after addition of the indicated ammonium sulfate (AS) precipitates (lanes 3-7). Lane 1 shows processing in the extract before heat inactivation. The arrow indicates the upstream fragment (30 nt) produced by cleavage at the major site indicated in A; additional bands above and below are degradation products of the downstream fragment (36 nt). (E) The 60% ammonium sulfate fraction containing HLF activity (60% AS) is inactivated by heat treatment (15 min at 50°C). In vitro processing using a 5′-end labeled H4-12 pre-mRNA substrate. (Lane 1) Non-heat-treated extract. (Lane 2) Heat-inactivated extract. (Lane 3) Heat-inactivated extract complemented with the non-heat-treated 60% AS fraction. (Lane 4) A nontreated extract and the 60% AS fraction were mixed and incubated together at 50°C. (Lane 5) The extract and the 60% AS fraction were heat treated separately, then combined prior to addition of the processing substrate. The arrow indicates the upstream product resulting from cleavage at the major site; the faint band above results from processing at the minor position shown in A.
To further purify the HLF, the Ni2+-resin eluate was fractionated by ammonium sulfate precipitation (Fig. 1C). While the resulting precipitates alone possessed no ability to process the H4-12 pre-mRNA substrate (data not shown), complementation of a heat-inactivated nuclear extract (Fig. 1D) revealed the highest HLF activity in the 60% saturated ammonium sulfate fraction (Fig. 1D, lane 5). This activity was completely destroyed by heat treatment after mixing with the nuclear extract (Fig. 1E, lane 4) and almost completely lost by exposure to 50°C separate from the extract (Fig. 1E, lane 5). Attempts to further purify the HLF activity on anion- or cation-exchange resins resulted in poor separation from the bulk of the proteins and gradual but significant loss of activity (data not shown).
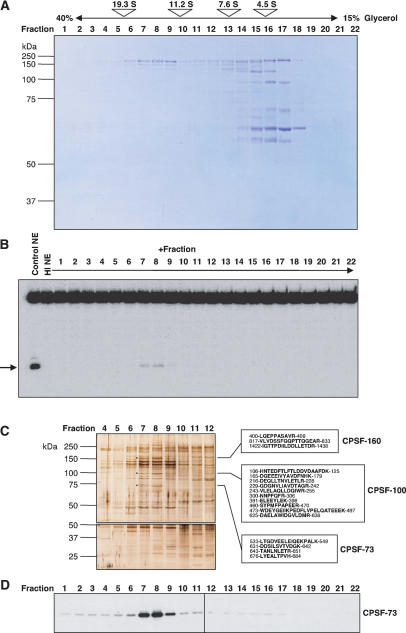
We therefore used glycerol gradient centrifugation to fractionate the 60% ammonium sulfate precipitate containing HLF activity. While the majority of proteins sedimented near the top of the gradient (slower than 7.6S) (Fig. 2A), fraction 8 (14.7S) had the greatest ability to restore processing to a heat-inactivated HeLa nuclear extract, followed closely by fraction 7; trace HLF activity was found in fraction 9 (Fig. 2B). Silver staining of these glycerol gradient fractions separated by SDS-PAGE revealed only a few bands whose profile closely mirrored HLF activity (Fig. 2C, bands indicated by dots). These comigrating proteins were selected for analysis by tandem mass spectrometry. After tryptic digestion, LC MS/MS identified the polypeptides migrating at ∼150, 100, and 75 kDa as subunits of the cleavage and polyadenylation specificity factor: CPSF-160 (three peptide matches), CPSF-100 (10 peptide matches), and CPSF-73 (four matching peptides), respectively (Fig. 2C). Western blotting revealed a precise match between HLF activity and the distribution of CPSF-73 across all fractions of the gradient (Fig. 2D).
Figure 2.
HLF activity sediments at 14.7S. (A) Protein profile of the 60% ammonium sulfate precipitate after 15%-40% glycerol gradient fractionation. Proteins were separated by 10% SDS-PAGE and stained with Coomassie. Above is shown the distribution of size markers in a parallel gradient: porcine thyroglobulin (19.3S, 669 kDa), bovine catalase (11.2S, 232 kDa), bovine lactate dehydrogenase (7.6S, 140 kDa), and bovine serum albumin (4.5S, 66 kDa). It should be noted that purified CPSF has a sedimentation coefficient of 11.5S (Bienroth et al. 1991; Murthy and Manley 1992), even though the predicted combined molecular weight of its subunits (excluding hFip1) is 357 kDa. (B) Complementation of in vitro processing by glycerol gradient fractions. Glycerol gradient fractions shown in A were mixed with heat-inactivated extract and incubated with a 5′-end labeled H4-12 substrate. The arrow indicates the major upstream cleavage product. We calculate (accounting for the dilution in the gradient) that fractions 7, 8, and 9 (14.7S fraction) combined contain slightly more HLF activity than the starting 60% ammonium sulfate precipitate. This indicates that there is no significant HLF stimulatory activity sedimenting with the bulk of the protein at ∼4.5S in our gradients. (C) SDS-PAGE (7% gel, top; 13% gel, bottom) of the fractions in the vicinity of HLF activity in the glycerol gradient shown in A was followed by silver staining. The boxes show tryptic peptide sequences identified by LC MS/MS analysis for the bands marked with black dots. Numbers indicate the amino acid positions within the sequences of the identified proteins. (D) Western blot analysis of all fractions from the glycerol gradient of A using anti-CPSF-73 antibody.
The surprising finding that CPSF-73 (Jenny et al. 1996), CPSF-100 (Jenny et al. 1994), and CPSF-160 (Murthy and Manley 1995) comigrate with HLF activity at 14.7S, similar to the previously reported ∼14S for the HLF (Gick et al. 1987) versus the 11.5S value determined independently by two groups for the purified CPSF complex (Bienroth et al. 1991; Murthy and Manley 1992), prompted us to test for the presence of other components of the cleavage and polyadenylation machinery. Western blot analysis of the same glycerol gradient fractions (Fig. 3A) revealed that the other two CPSF subunits, CPSF-30 (Barabino et al. 1997) and hFip1 (Kaufmann et al. 2004), also peak in fraction 8, as do two subunits of the cleavage stimulation factor (Takagaki et al. 1990), CstF-77 (Takagaki and Manley 1994), and CstF-64 (Takagaki et al. 1992). The 50-kDa third subunit of CstF (Takagaki and Manley 1992), as well as the subunits of the heterodimeric cleavage factor Im (CF Im 72, 68, 59, and CF Im 25) (Rüegsegger et al. 1998) and two of the identified subunits of cleavage factor IIA (hClp1 and hPcf11) (de Vries et al. 2000) did not comigrate with HLF activity but were detected in lighter fractions (Fig. 3A).
Figure 3.
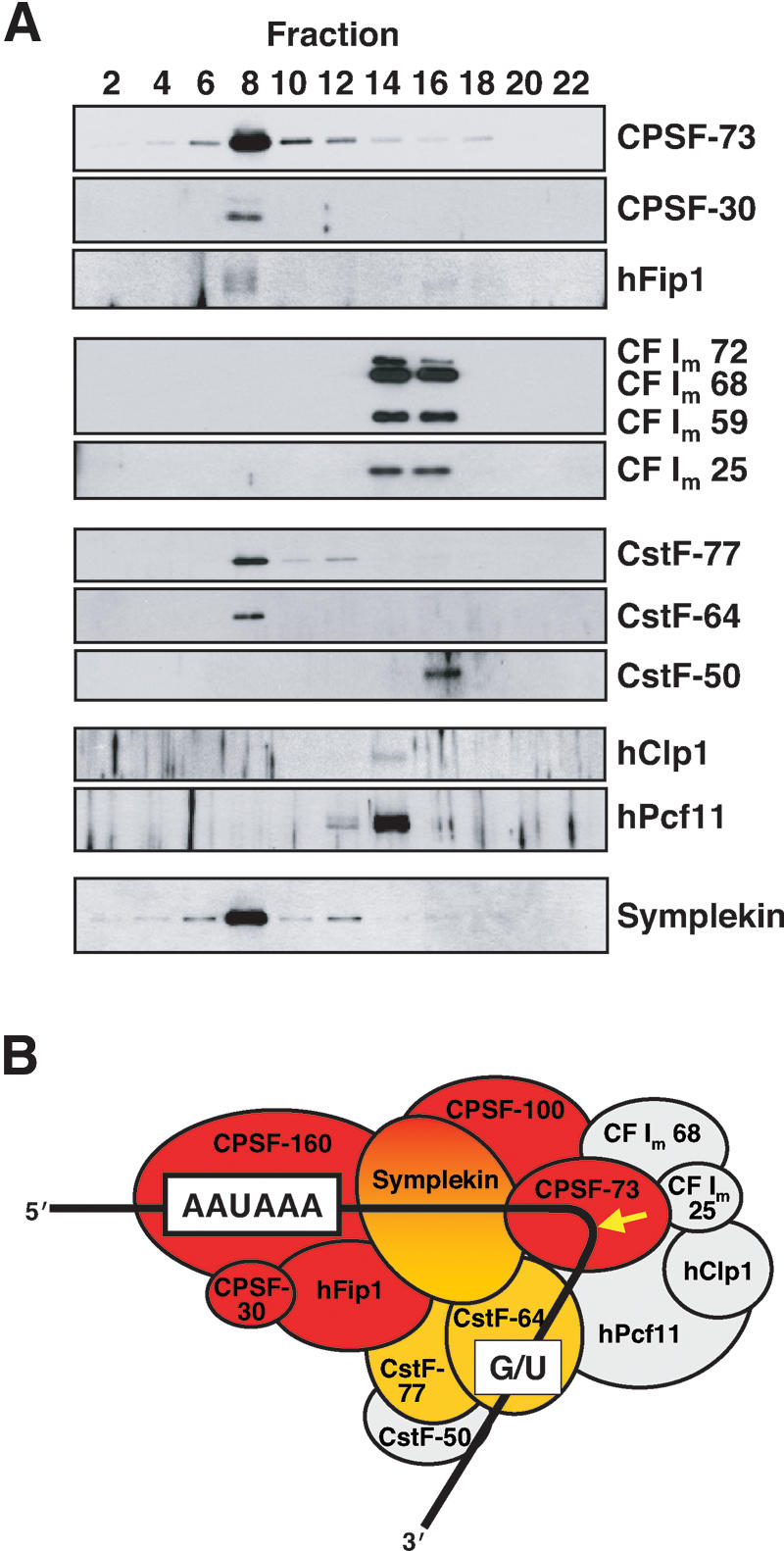
CPSF, CstF subunits, and symplekin cofractionate with HLF activity. (A) Western blots of fractions from a glycerol gradient separating the 60% ammonium sulfate precipitate, as in Figure 2. Only even-numbered fractions were analyzed for the presence of the proteins listed on the right. HLF activity peaked in fraction 8 (data not shown). (B) Diagram depicting proteins assigned to the cleavage and polyadenylation machinery, excluding poly(A) polymerase (Ryan et al. 2004). It has been previously observed that nuclear PAP does not copurify with either CPSF or CstF (Takagaki et al. 1989, and references therein). Consensus sequences and the site of cleavage (arrow) prior to polyadenylation are shown. HLF proteins concentrated in fraction 8, as identified by either mass spectrometry (Fig. 2C) or Western blotting (A) are colored red (CPSF) and orange (CstF). Proteins peaking elsewhere in the gradient are colored gray.
The profile of cleavage factors present in fraction 8 (colored proteins in Fig. 3B) was remarkably similar to that of a large complex immunoprecipitated by Takagaki and Manley (2000) from nuclear extracts using anti-CstF-64 antibodies. In the coimmunoprecipitate, they also found symplekin, a protein migrating at 150 kDa that was recently implicated in the cytoplasmic polyadenylation of mRNAs occurring upon maturation of Xenopus oocytes (Barnard et al. 2004). We asked whether symplekin might be a component of the 14.7S HLF complex by Western blotting (Fig. 3A, bottom). It was indeed detected, peaking in fraction 8.
Heat compromises the integrity and processing activity of the complex
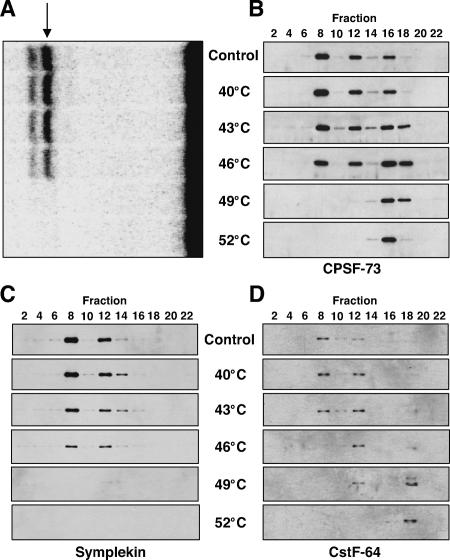
We reasoned that heat lability of a multimolecular complex could be due either to its dissociation into subcomplexes or to irreversible denaturation of one or more particular components. To examine the effect of heat on the 14.7S HLF complex, we performed glycerol gradient fractionation of a crude HeLa nuclear extract after exposure to different temperatures for 15 min. As reported by Gick et al. (1987), incubation of our extract at temperatures above 46°C abolished processing of the SLBP-independent H4-12 pre-mRNA substrate (Fig. 4A). The glycerol gradient fractions from the nuclear extract treated at the different temperatures were then blotted with antibodies against CPSF-73 and CstF-64, constituents of two different multisubunit complexes (Takagaki et al. 1989) known to be involved in nuclear cleavage and polyadenylation of pre-mRNAs (Fig. 4B,D) as well as against symplekin (Fig. 4C).
Figure 4.
Heat destroys the integrity of the 14.7S HLF complex. (A) In vitro processing of histone pre-mRNA in HeLa nuclear extracts pretreated for 15 min at the indicated temperatures compared with a non-heat-treated extract (Control). The H4-12 substrate was internally 32P-labeled. The arrow indicates the upstream cleavage product. Additional bands above and below are degradation products of the downstream fragment. (B-D) The heat-treated nuclear extracts assayed in A, as well as a non-heat-treated extract (Control), were fractionated on 15%-40% glycerol gradients. Every other fraction was subjected to SDS-PAGE and immunoblotted with antibodies against CPSF-73, Symplekin, or CstF-64.
The control unheated extract showed major peaks for both CPSF-73 and CstF-64 in fraction 8, where the HLF activity concentrates (as in Fig. 2B; data not shown). Redistribution of these proteins from fraction 8 to lighter fractions in the gradients closely correlated with temperature inactivation of the processing activity (Fig. 4B,D). This argues that CPSF and CstF are components of the same rather than different active complexes that sediment at 14.7S.
In contrast, upon heat inactivation and centrifugation of the extract, the signal for symplekin (Fig. 4C) progressively disappeared, becoming completely absent at 49°C and higher. Since symplekin was not found at the very bottom of the gradients (in fraction 1; data not shown), we conclude that either it was proteolytically degraded or the epitope recognized by the monoclonal antibody used in the assay becomes altered upon denaturation. Whatever the case, the effect of heat on symplekin, manifested by its disappearance from these Western blots, is not only associated with, but precisely matches the temperature inactivation profile of the nuclear extract.
We attempted depletion with antibodies against CPSF-73 and symplekin, but in both cases were unable to deplete HLF activity. In the case of the anti-CPSF-73 antibody, similar observations were made by Dominski et al. (2005); they were able to immunoprecipitate the protein only after boiling the extract in the presence of SDS and subsequent dilution of the detergent. In the case of the anti-symplekin antibody, Barnard et al. (2004) were able to immunodeplete from Xenopus oocyte extract the cytoplasmic polyadenylation activity. We used much higher amounts of the same antibody and still did not achieve immunodepletion of the HLF, which may indicate that the two complexes are quite different, and in the HLF complex, the epitope recognized by the monoclonal anti-symplekin antibody is not accessible.
Symplekin reconstitutes processing in heat-treated extract
To address which component is responsible for the heat lability of the HLF complex in the processing of histone pre-mRNAs, we performed complementation assays. Each of the cleavage and polyadenylation factor subunits identified in fraction 8 of the glycerol gradient of the 60% saturated ammonium sulfate precipitate was synthesized from its cloned cDNA by in vitro transcription/translation (TNT) in rabbit reticulocyte lysate. Symplekin is shown as an example in Figure 5; it is detected only when the TNT was programmed with symplekin cDNA. All eight TNTs were then tested both singly and in multiple combinations for their ability to reconstitute cleavage activity in heat-inactivated extract.
Figure 5.
In vitro-translated symplekin restores processing activity in heat-inactivated extract. (Left) Western blots analyzing the presence of symplekin (top) or CPSF-73 (bottom) in equal volumes of non-heat-treated nuclear extract (lane 1), heat-inactivated extract (lane 2), symplekin cDNA-programmed TNT reaction (lane 3), or an unprogrammed TNT reaction (lane 4). Note that the symplekin band does not disappear immediately after heat treatment, suggesting that its heat-induced inactivation precedes its degradation during the lengthy centrifugation in the glycerol gradients. The right panel shows in vitro processing of an internally 32P-labeled H4-12 pre-mRNA substrate in control non-heat-inactivated nuclear extract (lane 1), heat-inactivated (HI) extract (lane 2), HI extract supplemented with a transcription/translation reaction (TNT) programmed with symplekin cDNA (lane 3), or HI extract supplemented with an unprogrammed TNT reaction (lane 4).
Symplekin was the only protein that restored cleavage of the histone H4-12 pre-mRNA substrate (Fig. 5; data not shown for the proteins that did not complement). Its addition alone to the heat-treated nuclear extract was sufficient. Moreover, addition of in vitro synthesized symplekin to heat-inactivated extract caused the redistribution of CPSF-73 to heavier fractions in the glycerol gradient, coincident with the rescued processing activity (data not shown). Together, these results demonstrate that symplekin is the temperature-sensitive component of the HLF necessary for 3′-end maturation of histone messages. It is absolutely required for this processing event, but most likely acts only in concert with the other polypeptides, such as CPSF-73, with which it stably associates.
Discussion
We have discovered that symplekin is the heat-labile component of a multimolecular complex required for histone mRNA 3′-end formation. The HLF assembly also contains many, but not all factors previously assigned to the nuclear cleavage and polyadenylation machinery that fashions the polyadenylated ends of most mRNAs. The eight proteins have a combined predicted molecular weight of 694 kDa, consistent with a single complex sedimenting at 14.7S; this suggests that we have identified the majority, if not all constituents of the HLF complex (Fig. 6). The functional integrity of the complex appears to depend on symplekin, since heat-induced disassembly and loss of activity are coupled and addition of in vitro-synthesized symplekin rescues histone pre-mRNA processing in a heat-inactivated extract.
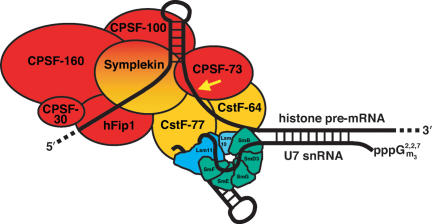
Figure 6.
Model for histone pre-mRNA processing. Components of the HLF identified in this study are colored red and orange. The U7 snRNA is depicted base pairing with the histone pre-mRNA downstream element. We propose that the U7 snRNP orients the histone pre-mRNA for cleavage (arrow) by CPSF-73 through contacts between its Sm proteins and the CstF subunits in HLF (see text). Proteins of the Sm ring that are shared with spliceosomal snRNPs are shown in green; the U7-specific Lsm10 and Lsm11 proteins (Pillai et al. 2001, 2003) are shown in shades of blue.
Symplekin was initially identified as a component of tight junctions in polar epithelial cells and Sertoli cells and is also present in the nuclei of all types of cultured mammalian cells and tissues examined (Keon et al. 1996). Takagaki and Manley (2000) first suggested its involvement (perhaps in a scaffolding role) in mRNA 3′-end formation by showing interactions with CstF-77 and CstF-64, as well as with CPSF-160, CPSF-100, and CPSF-73 (Takagaki and Manley 2000; Hofmann et al. 2002). Importantly, symplekin has been shown to be required for poly(A) tail elongation of mRNAs containing a cytoplasmic polyadenylation element (CPE), a regulated process that occurs in the cytoplasm during oocyte maturation (Barnard et al. 2004), as well as during cell cycle progression (Groisman et al. 2002) and synaptic stimulation of neurons (Wu et al. 1998; Huang et al. 2002). The data of Barnard et al. (2004) clearly demonstrate that cytoplasmic symplekin associates with CPSF, as well as with CPEB, an RNA-binding protein that recognizes the CPE.
What are the roles of the various polyadenylation factors we have identified as HLF components in histone pre-mRNA processing? The major functions of CPSF and CstF, respectively, in recognizing the upstream AAUAAA and downstream G/U-rich region that flank the cleavage site in polyadenylation substrates (Murthy and Manley 1995; Takagaki and Manley 1997), seem irrelevant, since these features are absent in histone pre-mRNAs. The crude nuclear extracts from which we have isolated the HLF are, in fact, markedly reduced in their activity to cleave polyadenylation substrates (data not shown). Instead, the CPSF and CstF components may serve to forge a link between the U7 snRNP and the endonuclease that cleaves downstream of the CA dinucleotide in the pre-mRNA substrate, producing the 3′-OH terminus (Scharl and Steitz 1994) of the mature histone mRNA.
CPSF-73 seems most likely to be the endonuclease that creates the 3′ ends of histone mRNAs since it is a member of the metallo-β-lactamase family of Zn2+-dependent hydrolytic enzymes (Callebaut et al. 2002). This assignment is strongly supported by the studies of Dominski et al. (2005), who have recently characterized a U7 snRNP-dependent cross-link between the cleavage site of a histone pre-mRNA substrate and CPSF-73. In the case of polyadenylated mRNAs, Ryan et al. (2004) have reported both protein-RNA cross-linking data and mutational analyses that implicate CPSF-73 as the catalytic subunit that executes the cleavage step prior to polyadenylation. Moreover, the same specificity—creation of a 3′-OH downstream of CA—is seen for cleavage/polyadenylation substrates (Sheets et al. 1987) as for histone pre-mRNAs. Our identification of all the subunits of CPSF as components of the HLF assembly provides additional support for the conclusion that CPSF-73 is the 3′-endonuclease in histone pre-mRNA processing.
It is interesting that CstF-50 is missing from the HLF complex. CstF-50 exhibits extensive homology to mammalian G-protein β-subunits (Takagaki and Manley 1992), including seven WD40 domains, which X-ray analysis shows form toroidal structures (Wall et al. 1995; Lambright et al. 1996; Sondek et al. 1996). This architecture is reminiscent of the heteroheptameric ring of the Sm protein core (Kambach et al. 1999; Raker et al. 1999). We hypothesize that the U7 snRNP may replace CstF-50 in the assembled processing complex, interacting with the other CstF subunits via its Sm protein ring (Fig. 6). In this scenario, symplekin would act as a scaffold protein, similar to its proposed role in the cytoplasmic polyadenylation complex (Barnard et al. 2004). By making documented contacts with three of the five subunits of CPSF (Hofmann et al. 2002) and the remaining two subunits of CstF (Takagaki and Manley 2000), symplekin would bring the endonuclease (CPSF-73) and the U7 snRNP together into a single assembly. Since the higher-order complex containing HLF and the U7 snRNP would be extremely large, it is possible that it represents the elusive molecular measuring device (Scharl and Steitz 1994, 1996) that spans from the duplex formed between U7 and the HDE to the site of cleavage 11 nt upstream in the histone pre-mRNA.
Our findings rationalize previous observations that symplekin and other polyadenylation factors are enriched in or intimately associated with Cajal bodies (Schul et al. 1996, 1999; Gall et al. 1999; Hofmann et al. 2002). Cajal bodies (formerly called coiled bodies) are highly conserved nuclear organelles that are particularly prominent in rapidly cycling cells. The U7 snRNP has been shown to preferentially reside in Cajal bodies both in Xenopus oocytes (where they are also named spheres or C snurposomes) (Wu and Gall 1993) and in mammalian cells (Frey and Matera 1995). Snurposomes also contain SLBP (Abbott et al. 1999). The tight association of Cajal bodies with histone gene loci was noted long ago for the lampbrush chromosomes of newt (Gall et al. 1981) and of Xenopus (Callan et al. 1991). In mammalian cells, Cajal bodies preferentially localize at both histone loci and gene arrays encoding U1 and U2 snRNAs (Frey and Matera 1995). Our characterization of the proteins present in the HLF complex (see Fig. 6) suggests that Cajal bodies contain the complete set of processing components for histone mRNA 3′-end maturation, neatly packaged for delivery to chromosomal sites of histone gene transcription.
The HLF required for 3′-end processing of histone pre-mRNAs has been reported to be cell-cycle regulated with its level low in G1-arrested cells and high during exponential growth (Lüscher and Schümperli 1987). So far, the only component of the HLF complex we have characterized that has been reported to fluctuate during the cell cycle is CstF-64 (Martincic et al. 1998). It or any other untested component could contribute to the cell cycle-dependent variation in HLF activity. On the other hand, Marzluff and Duronio (2002) have argued that the cell cycle regulation of SLBP could account for the differing levels of histone 3′-end processing during the cell cycle; conflicting data have been reported regarding the heat sensitivity of SLBP (Melin et al. 1992; Dominski et al. 1999).
Our discovery that symplekin and certain other polyadenylation factors participate in histone mRNA 3′-end maturation raises the question of how many cellular roles these versatile proteins play. They appear to exchange molecular partners depending on their cellular location—nucleoplasm, cytoplasm, or Cajal body-associated. For instance, in the nucleoplasm, the poly(A) polymerase (PAP), which does not copurify with CPSF or CstF (Takagaki et al. 1989), is engaged; in the cytoplasm, GLD-2 is the polymerase recruited for extension of poly(A) tails (Barnard et al. 2004); when Cajal body-associated, no nucleotide addition occurs after cleavage. It will be interesting to analyze the detailed intermolecular contacts in these various 3′-end forming machines and to learn whether some of the interactions are mutually exclusive; for example, does symplekin or the U7 snRNP displace CstF-50 from the other two CstF subunits in the histone system? Are all of the HLF components pictured in Figure 6 essential for activity? Have all necessary subunits been identified? The finding that shared factors contribute to the nuclear processing of mRNAs with and without 3′ poly(A) and to the regulated elongation of poly(A) tails in the cytoplasm points to a common evolutionary origin for the 3′-end formation apparatus in all eukaryotic cells.
Materials and methods
Preparation of HeLa nuclear extract
A procedure for the preparation of Drosophila Kc cell nuclear extract (Price et al. 1987) was followed, with modifications. HeLa cells were washed with 5-10 pellet volumes of ice-cold PBS. The collected cells were resuspended in 5 pellet volumes of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.1% saturated PMSF) and incubated on ice for 10 min. After lysing the the cells with 10 strokes in a Dounce homogenizer (Kontes, type B tight-fitting pestle), the nuclei were pelleted by centrifugation for 10 min at 2500 rpm in a Sorvall SS34 rotor at 4°C. The nuclear pellet received 1.6 vol of buffer A, and the nuclei were resuspended by 20 strokes in the Dounce homogenizer. To the suspension (transferred into a centrifuge tube) was added 0.1 vol of 4 M (NH4)2SO4, and the mixture was incubated with gentle mixing at 4°C for 30 min. After centrifugation at 18,000 rpm in a Sorvall SS34 rotor for 60 min at 4°C, the clear supernatant was transferred to a new tube, and 0.3 g of solid (NH4)2SO4 was added per milliliter of extract. The suspension was incubated for 20 min at 4°C with end-to-end rotation. After centrifugation at 12,000 rpm in a Sorvall SS34 rotor for 10 min at 4°C, the pellet was resuspended in 1.5-2 starting nuclear volumes of buffer A and dialyzed against buffer D (20% glycerol, 20 mM HEPES at pH 7.9, 90 mM KCl, 10 mM (NH4)2SO4, 0.2 mM EDTA, 1 mM DTT, 0.1% saturated PMSF). The crude nuclear extract was stored at -80°C.
Purification of the HLF
HeLa nuclear extract (2.5 mL) was dialyzed against 500 mL dialysis buffer (20% glycerol, 50 mM HEPES at pH 7.9, 400 mM KCl, 20 mM imidazole, 0.1% saturated PMSF) for 1 h at 4°C. The extract was diluted to 5 mL with dialysis buffer and then added to a centrifuge tube containing 1 mL Ni2+-nitrilotriacetic acid (Ni2+-NTA) metal-affinity resin (QIAGEN) that had been equilibrated with dialysis buffer. After incubation for 1 h at 4°C with end-to-end rotation, the resin was collected by centrifugation and washed three times with 10 mL of dialysis buffer. Bound material was eluted with 2 mL of elution buffer (20% glycerol, 50 mM HEPES at pH 7.9, 400 mM KCl, 500 mM imidazole, 0.1% saturated PMSF).
Saturated (NH4)2SO4 solution was added to the eluate to sequentially achieve the indicated saturation percentages. After each addition, the sample was thoroughly and carefully mixed, incubated on ice for 15 min and then centrifuged at 13,400 × g for 10 min at 4°C. The pellet after each spin was dissolved in 0.5 mL of 20% glycerol, 50 mM HEPES (pH 7.9), 1 mM DTT, 0.1% saturated PMSF, stored at -80°C, and used for complementation of in vitro processing reactions. Glycerol gradients (15%-40%, 10 mL) were prepared in gradient buffer (50 mM HEPES at pH 7.9, 50 mM (NH4)2SO4, 1 mM DTT, 0.1% saturated PMSF). Either 250 μL of the dissolved 60% ammonium sulfate precipitate or 62.5 μL crude nuclear extract were diluted to 1 mL with gradient buffer prior to loading on the gradient. Centrifugation was for 35.5 h at 36,000 rpm in a SW 41 Ti rotor. Fractions (0.5 mL) were collected from the bottom of the gradients, stored at -80°C, and used for complementation of processing reactions and for SDS-PAGE and Western blotting.
Protein identification
In-gel trypsin digestion of Coomassie-stained gel pieces containing individual bands that comigrated with activity and LC MS/MS analysis of the resulting peptides were performed by the staff of W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Following the in-gel trypsin digestion, the samples were separated on a 100-μm I.D. Atlantis C18 column (Waters). The MS/MS spectra were searched using the Mascot distiller (to locate and combine MS/MS spectra) and the Mascot database search algorithm.
RNA substrates and in vitro processing
The H4-12 histone pre-mRNA substrate was synthesized by T7 transcription. For internally labeled RNA, transcription was in the presence of GpppG cap analog and [α-32P]UTP; 5′-end labeled RNA was generated using T4 polynucleotide kinase and [γ-32P]ATP from dephosphorylated H4-12 pre-mRNA transcripts. Labeled substrates were purified on denaturing polyacrylamide gels prior to addition to in vitro processing reactions (final volume 20 μL) containing 20% HeLa nuclear extract, 10 mM EDTA, 4 μg total yeast RNA, and 1-5 fmol 32P-labeled substrate. Heat inactivation of the extracts was for 15 min at 50°C (unless indicated otherwise) prior to assembly of the reaction. For complementation assays, the processing reactions were supplemented with 5 μL of purified HLF fractions or 3 μL in vitro transcription/translation (TNT) reaction. In vitro processing was for 2 h at 30°C. After Proteinase K digestion and precipitation with ethanol, RNAs were subjected to electrophoresis in 15% polyacrylamide gels in the presence of 8 M urea.
Antibodies and Western blotting
Monoclonal anti-symplekin antibody (raised against a peptide encompassing amino acids 914-1080) was from BD Biosciences. Rabbit polyclonal affinity-purified antibodies against CPSF-73, CstF-77, and CstF-50 were generous gifts of David Bentley (University of Colorado, Aurora). Rabbit sera against CPSF-30, hFip1, hClp1, CF Im 68, CF Im 25, and hPcf11 were generous gifts of Walter Keller (University of Basel, Basel, Switzerland). Monoclonal anti-CstF-64 antibody was from Clinton MacDonald (Texas Tech University, Lubbock). All Western blots were performed using standard procedures, except in the case of the anti-CstF-64 blot, where Tween 20 was omitted from all buffers.
In vitro translation
Plasmids containing verified full-length cDNA sequences downstream of an SP6 promoter for human CPSF-160, CPSF-73, CPSF-30, hFip1, symplekin, and CstF-64, as well as for mouse CPSF-100 and CstF-77, were obtained from Open Biosystems. In vitro protein synthesis was performed using the TNT SP6 Quick coupled transcription/translation rabbit reticulocyte system (Promega). The efficient translation of each protein was confirmed by labeling with [35S]methionine in a parallel reaction.
Acknowledgments
We are extremely grateful to D. Bentley, W. Keller, and C. MacDonald for antibodies and to R. Samstein for assistance in preparing nuclear extract. We thank W. Marzluff for sharing unpublished results; K. Tycowski, N. Conrad, S. Mili, and S. Vasudevan for critical reading of the manuscript; and A. Miccinello for secretarial help. This work was supported by National Institutes of Health grant GM26154 to J.A.S., who is an investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1371105.
References
- Abbott J., Marzluff, W.F., and Gall, J.G. 1999. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol. Biol. Cell 10: 487-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S.M., Hübner, W., Jenny, A., Minvielle-Sebastia, L., and Keller, W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes & Dev. 11: 1703-1716. [DOI] [PubMed] [Google Scholar]
- Barnard D.C., Ryan, K., Manley, J.L., and Richter, J.D. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641-651. [DOI] [PubMed] [Google Scholar]
- Bienroth S., Wahle, E., Suter-Crazzolara, C., and Keller, W. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266: 19768-19776. [PubMed] [Google Scholar]
- Birchmeier C., Schümperli, D., Sconzo, G., and Birnstiel, M.L. 1984. 3′ editing of mRNAs: Sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc. Natl. Acad. Sci. 81: 1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U.M., Yario, T.A., and Steitz, J.A. 1991. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 5: 1709-1722. [DOI] [PubMed] [Google Scholar]
- Callan H.G., Gall, J.G., and Murphy, C. 1991. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma 101: 245-251. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Moshous, D., Mornon, J.P., and de Villartay, J.P. 2002. Metallo-β-lactamase fold within nucleic acids processing enzymes: The β-CASP family. Nucleic Acids Res. 30: 3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H., Rüegsegger, U., Hübner, W., Friedlein, A., Langen, H., and Keller, W. 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19: 5895-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Zheng, L.X., Sanchez, R., and Marzluff, W.F. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19: 3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Erkmann, J.A., Yang, X., Sanchez, R., and Marzluff, W.F. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 16: 58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang, X., and Marzluff, W.F. 2005. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell (in press). [DOI] [PubMed]
- Eliassen K.A., Baldwin, A., Sikorski, E.M., and Hurt, M.M. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18: 7106-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R. and Matera, A.G. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. 92: 5915-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G., Stephenson, E.C., Erba, H.P., Diaz, M.O., and Barsacchi-Pilone, G. 1981. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma 84: 159-171. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Bellini, M., Wu, Z., and Murphy, C. 1999. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10: 4385-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer, A., Keller, W., and Birnstiel, M.L. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5: 1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer, A., Vasserot, A., and Birnstiel, M.L. 1987. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. 84: 8937-8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I., Jung, M.Y., Sarkissian, M., Cao, Q., and Richter, J.D. 2002. Translational control of the embryonic cell cycle. Cell 109: 473-483. [DOI] [PubMed] [Google Scholar]
- Hofmann I., Schnölzer, M., Kaufmann, I., and Franke, W.W. 2002. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13: 1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Jung, M.Y., Sarkissian, M., and Richter, J.D. 2002. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and α CaMKII mRNA polyadenylation at synapses. EMBO J. 21: 2139-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Hauri, H.P., and Keller, W. 1994. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell. Biol. 14: 8183-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia, L., Preker, P.J., and Keller, W. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514-1517. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Lührmann, R., Li, J., and Nagai, K. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375-387. [DOI] [PubMed] [Google Scholar]
- Kaufmann I., Martin, G., Friedlein, A., Langen, H., and Keller, W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23: 616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon B.H., Schäfer, S., Kuhn, C., Grund, C., and Franke, W.W. 1996. Symplekin, a novel type of tight junction plaque protein. J. Cell Biol. 134: 1003-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P.A. and Melton, D.A. 1984. Formation of the 3′ end of histone mRNA by post-transcriptional processing. Nature 308: 203-206. [DOI] [PubMed] [Google Scholar]
- Lambright D.G., Sondek, J., Bohm, A., Skiba, N.P., Hamm, H.E., and Sigler, P.B. 1996. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379: 311-319. [DOI] [PubMed] [Google Scholar]
- Last T.J., van Wijnen, A.J., Birnbaum, M.J., Stein, G.S., and Stein, J.L. 1999. Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J. Cell. Biochem. 72: 507-516. [PubMed] [Google Scholar]
- Lüscher B. and Schümperli, D. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 6: 1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K., Campbell, R., Edwalds-Gilbert, G., Souan, L., Lotze, M.T., and Milcarek, C. 1998. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl. Acad. Sci. 95: 11095-11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F. and Duronio, R.J. 2002. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14: 692-699. [DOI] [PubMed] [Google Scholar]
- Melin L., Soldati, D., Mital, R., Streit, A., and Schümperli, D. 1992. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 11: 691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K.L. and Steitz, J.A. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238: 1682-1687. [DOI] [PubMed] [Google Scholar]
- Murthy K.G. and Manley, J.L. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267: 14804-14811. [PubMed] [Google Scholar]
- ____. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes & Dev. 9: 2672-2683. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Will, C.L., Lührmann, R., Schümperli, D., and Müller, B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20: 5470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Grimmler, M., Meister, G., Will, C.L., Lührmann, R., Fischer, U., and Schümperli, D. 2003. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 17: 2321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.H., Sluder, A.E., and Greenleaf, A.L. 1987. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J. Biol. Chem. 262: 3244-3255. [PubMed] [Google Scholar]
- Raker V.A., Hartmuth, K., Kastner, B., and Lührmann, R. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 19: 6554-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger U., Blank, D., and Keller, W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1: 243-253. [DOI] [PubMed] [Google Scholar]
- Ryan K., Calvo, O., and Manley, J.L. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl E.C. and Steitz, J.A. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13: 2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Length suppression in histone messenger RNA 3′-end maturation: Processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. 93: 14659-14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin, G.M., Bannwarth, W., and Birnstiel, M.L. 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323: 777-781. [DOI] [PubMed] [Google Scholar]
- Schul W., Groenhout, B., Koberna, K., Takagaki, Y., Jenny, A., Manders, E.M., Raska, I., van Driel, R., and de Jong, L. 1996. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 15: 2883-2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W., van Der Kraan, I., Matera, A.G., van Driel, R., and de Jong, L. 1999. Nuclear domains enriched in RNA 3′-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol. Biol. Cell 10: 3815-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M.D., Stephenson, P., and Wickens, M.P. 1987. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol. Cell. Biol. 7: 1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Seto, E., Chang, L.S., and Shenk, T. 1991. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67: 377-388. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm, A., Lambright, D.G., Hamm, H.E., and Sigler, P.B. 1996. Crystal structure of a G-protein βγ dimer at 2.1Å resolution. Nature 379: 369-374. [DOI] [PubMed] [Google Scholar]
- Streit A., Koning, T.W., Soldati, D., Melin, L., and Schümperli, D. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21: 1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli, G., Busslinger, M., and Birnstiel, M.L. 1984. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 3: 2801-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y. and Manley, J.L. 1992. A human polyadenylation factor is a G protein β-subunit homologue. J. Biol. Chem. 267: 23471-23474. [PubMed] [Google Scholar]
- ____. 1994. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature 372: 471-474. [DOI] [PubMed] [Google Scholar]
- ____. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17: 3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20: 1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Ryner, L.C., and Manley, J.L. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes & Dev. 3: 1711-1724. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Manley, J.L., MacDonald, C.C., Wilusz, J., and Shenk, T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes & Dev. 4: 2112-2120. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., MacDonald, C.C., Shenk, T., and Manley, J.L. 1992. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. 89: 1403-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M.A., Coleman, D.E., Lee, E., Iñiguez-Lluhi, J.A., Posner, B.A., Gilman, A.G., and Sprang, S.R. 1995. The structure of the G protein heterotrimer Giα1β1γ2. Cell 83: 1047-1058. [DOI] [PubMed] [Google Scholar]
- Wu C.H. and Gall, J.G. 1993. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. 90: 6257-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wells, D., Tay, J., Mendis, D., Abbott, M.A., Barnitt, A., Quinlan, E., Heynen, A., Fallon, J.R., and Richter, J.D. 1998. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron 21: 1129-1139. [DOI] [PubMed] [Google Scholar]
- Xing H., Mayhew, C.N., Cullen, K.E., Park-Sarge, O.K., and Sarge, K.D. 2004. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J. Biol. Chem. 279: 10551-10555. [DOI] [PubMed] [Google Scholar]