Abstract
Salmonella enterica serovar Enteritidis is a major cause of food-borne diseases associated with consumption of shell eggs. Clinical isolates of S. enterica serovar Enteritidis exhibit a wide spectrum of virulence in mice. A highly virulent isolate (SE2472) was previously shown to be more resistant in vitro than other clinical isolates to acidified sodium nitrite (ASN), a generator of reactive nitrogen and oxygen intermediates (RNI/ROI). SE2472 is also more resistant to S-nitrosoglutathione (GSNO) and hydrogen peroxide (H2O2) than an ASN-susceptible isolate of S. enterica serovar Enteritidis (SE8743). To investigate the molecular basis for the RNI/ROI resistance of S. enterica serovar Enteritidis, we transformed a genomic DNA library of SE2472 into SE8743. A plasmid clone conferred upon SE8743 enhanced resistance to ASN, GSNO, and H2O2. The DNA insert in the clone encoded ArcA, a global regulator. An arcA mutant of SE2472 was constructed and was found to be more susceptible to GSNO and hydrogen peroxide but not more susceptible to ASN than wild-type SE2472. The susceptibility of the arcA mutant to GSNO and H2O2 was complemented by a plasmid harboring arcA. The coding sequence of the arcA gene in SE2472 and the coding sequence of the arcA gene in SE8743 were identical, suggesting that the difference in resistance to RNI/ROI maybe due to the activity of genes regulated by ArcA. No significant difference in virulence between the wild type and the arcA mutant of SE2472 was observed in mice. These observations show that arcA is essential for resistance of S. enterica serovar Enteritidis to nitrosative and oxidative stress. However, additional genetic loci may contribute to the resistance to RNI/ROI and unusually high virulence for mice of SE2472.
The facultatively intracellular pathogen Salmonella is a leading cause of food-borne diseases in the United States. Between 1993 and 1997, Salmonella caused more outbreaks and cases of food poisoning than all other bacteria combined (6), and Salmonella enterica serovar Enteritidis was responsible for more outbreaks, cases, and deaths than any other Salmonella serovar during this period. Most of the cases were associated with consumption of contaminated eggs laid by infected chickens (6, 33, 38). Chickens are probably infected on poultry farms by field mice, which serve as a reservoir for S. enterica serovar Enteritidis (10, 23, 25). Therefore, infection of mice is believed to be a link in the natural transmission cycle of S. enterica serovar Enteritidis.
S. enterica serovar Enteritidis causes systemic infections in mice. In orally infected mice, bacteria can be recovered from many organs, including the liver, spleen, intestine, ovaries, and blood. Death can occur rapidly (<12 h after infection), although the exact cause of death is not entirely clear. We have previously reported that clinical isolates of S. enterica serovar Enteritidis exhibit a wide range of 50% lethal doses (LD50) after oral challenge in BALB/c mice (29). In contrast to S. enterica serovar Typhimurium isolates, which usually have an oral LD50 of approximately 105 organisms (S. Lu and L. W. Riley, unpublished results), clinical S. enterica serovar Enteritidis isolates may have oral LD50 of less than 100 organisms. S. enterica serovar Enteritidis isolates are quite heterogeneous with respect to virulence-associated phenotypes determined in vitro, and these in vitro phenotypes correlate poorly with virulence in mice. However, one clinical isolate (SE2472) with an oral LD50 of 16 organisms was found to be significantly more resistant than other isolates to acidified sodium nitrite (ASN), which generates a variety of reactive nitrogen intermediate (RNI) and reactive oxygen intermediate (ROI) products (29). Isolate SE2472 was also found to be more resistant to other RNI/ROI generators, such as S-nitrosoglutathione (GSNO) and hydrogen peroxide (H2O2), than ASN-susceptible isolate SE8743.
Microbes have evolved a variety of strategies to defend themselves against toxic reactive oxygen and nitrogen products in aerobic and anaerobic environments. For pathogens, differences in the responses to such stresses in vivo may determine the clinical outcome for an infected mammalian host. Generation of RNI and ROI is essential for the defense against Salmonella in mice (12, 17, 37). De Groote et al. have observed oxygen-dependent and -independent antimicrobial activities of different redox forms of NO against isogenic wild-type and mutant Salmonella strains in vitro (11). In Escherichia coli and Salmonella, at least four global regulators are responsible for much of the response to molecular oxygen. These regulators include the OxyR, SoxRS, Fnr, and ArcAB systems (2, 28, 31). OxyR and SoxRS are activated by reactive oxygen species, such as hydrogen peroxide and superoxide, respectively (7, 22). Disruption of the transcriptional regulators oxyR and soxS affects the oxidative stress responses in vitro but does not appear to affect the virulence of S. enterica serovar Typhimurium in mice (19, 39). However, disruption of genes encoding bacterial products directly involved in detoxification or electron transport and energy transduction (zwf and sodC) has been shown to increase the susceptibility of S. enterica serovar Typhimurium to RNI and to killing by mice (18, 30). Insertion mutations in the metL gene rendered S. enterica serovar Typhimurium hypersusceptible to the nitric oxide (NO) donor compound GSNO, and such a strain became attenuated in mice (13). These observations indicate the importance of RNI in Salmonella infection control in vivo.
We investigated the molecular basis of the RNI/ROI resistance of S. enterica serovar Enteritidis isolate SE2472. In this report, we provide evidence that ArcA, the response regulator of the ArcAB global regulatory system, is essential for nitrosative and oxidative stress resistance in S. enterica serovar Enteritidis SE2472.
MATERIALS AND METHODS
Reagents.
Growth media for bacteria were purchased from Difco Laboratories (Detroit, Mich.). Anaerobic peptone-yeast extract medium was obtained from Anaerobe Systems (Morgan Hill, Calif.). Chemicals and antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise indicated. GSNO was obtained from Alexis Co. (San Diego, Calif.). Restriction and modifying enzymes for manipulating DNA were obtained from New England Biolabs (Beverly, Mass.). Custom oligonucleotides were purchased from Operon (Alameda, Calif.) or Sigma Genosys (The Woodlands, Tex.).
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are shown in Table 1. S. enterica serovar Enteritidis isolates SE2472 and SE8743 were kindly provided by Duc Vugia and Sharon Abbott of the Department of Health Services, State of California, and have been described previously (29). E. coli DH5α (Gibco/BRL, Gaithersburg, Md.) was used as the host for all recombinant DNA manipulations. Plasmid vector pRB3-273C (3) was used to construct a genomic library from isolate SE2472. S. enterica serovar Typhimurium LB5000 is a restriction-deficient LT2 derivative (5). Bacteriophage P22 was used for generalized transduction. Plasmids pKD4 and pKD46 used for mutagenesis of S. enterica serovar Enteritidis were generously provided by Barry Wanner (Purdue University, West Lafayette, Ind.). Plasmid pKD4 contains the kanamycin resistance gene sequence, and pKD46 contains the Red recombinase gene. All bacterial strains were grown aerobically with shaking at 225 rpm unless otherwise stated. Anaerobic cultures were grown by using the Hungate method and peptone-yeast extract medium obtained from Anaerobe Systems.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44λ−thi-1 gyrA96 relA1 | Gibco/BRL |
| S. enterica serovar Enteritidis strains | ||
| SE2472 | Clinical isolate, mouse virulent and ASN resistant | 29 |
| SE2472arcA− | arcA::kan derivative of SE2472 | This study |
| SE8743 | Clinical isolate, ASN susceptible | 29 |
| S. enterica serovar Typhimurium LB5000 | Restriction-deficient strain | 5 |
| Plasmids | ||
| pKD4 | Apr Kanr oriRγ | 9 |
| pKD46 | Apr, containing the Red recombinase of λ phage | 9 |
| pRB3-273C | Apr, low- to medium-copy-number plasmid for Salmonella | 3 |
| pRB3arcA | Derivative of pRB3-273C containing arcA | This study |
Construction of S. enterica serovar Enteritidis genomic DNA library and screening of transformants for plasmids conferring ASN resistance.
Salmonella genomic DNA was purified as described previously (32). Plasmid DNA was purified by the alkaline lysis method (36). DNA fragments used for cloning were purified by using Geneclean II obtained from Bio101 (Vista, Calif.) after gel electrophoresis. A genomic DNA library of SE2472 was constructed by partially digesting the SE2472 genomic DNA with Sau3AI and cloning the 0.7- to 2.5-kb DNA fragments into BamHI-digested plasmid pRB3-273C. The library was introduced into E. coli DH5α by electroporation. Approximately 5 × 103 recombinant DH5α colonies were obtained, which were estimated to represent approximately 85% of the S. enterica serovar Enteritidis genome. Plasmid DNA was purified from the DH5α host and transformed into S. enterica serovar Typhimurium LB5000. Plasmid DNA was purified from LB5000 and transformed into SE8743 (15). The resulting library was plated onto Luria-Bertani (LB) agar with 100 μg of ampicillin per ml, and colonies were scraped from the plate and exposed to 15 mM sodium nitrite at pH 5.0 (ASN). After exposure to ASN, the Salmonella culture was plated onto LB agar supplemented with ampicillin and incubated overnight at 37°C. All colonies that survived the first round of ASN stress were scraped from the plates for another round of selection. After three rounds of screening, plasmid inserts were amplified from the surviving bacteria by PCR and sequenced by using an ABI310 sequencer and a Big-dye sequencing kit (Applied Biosystems, Foster City, Calif.).
Sequence analysis of arcA loci of SE2472 and SE8743.
To compare the arcA locus of SE2472 and the arcA locus of SE8743, we amplified the genomic DNA of each isolate with primers PK2F3 (5′-AGGTAGCAAACATGCAGACC) and PK2R2 (5′-CAGGAGGGAAAGCGAGGC). This pair of primers was designed to amplify a DNA sequence from 11 bp upstream to 100 bp downstream of arcA. The PCR products were then sequenced with the ABI310 sequencer and a Big-dye sequencing kit (Applied Biosystems).
Mutagenesis of S. enterica serovar Enteritidis.
Mutagenesis of the arcA gene of S. enterica serovar Enteritidis SE2472 was carried out by using the gene disruption method described by Datsenko and Wanner (9), except that 10 mM arabinose was used to induce expression of the Red recombinase instead of 1 mM arabinose as described by Datsenko and Wanner. Oligonucleotide primers 5′-TCTTATCGTTGAAGACGAGTTGGTAACACGCAACACGTTGAAAA GTATTTTCGAAGCGGAGTGTAGGCTGGAGCTGCTTC and 5′-CTTATCGTTGAAGACGAGTTGGTAACACGCAACACGTTGAAAAGTATTTTCG AAGCGGAGTGTAGGCTGGAGCTGCTTC were used to amplify the kanamycin resistance gene, and pKD4 was used as the template. These oligonucleotide primers contain a sequence of the kanamycin resistance gene and nucleotides 17 to 76 and 667 to 726 of the arcA gene, respectively. The arcA sequence from nucleotide 77 to nucleotide 666 was replaced with the kanamycin resistance gene by homologous recombination between the genomic DNA and the PCR product. Individual colonies obtained after electroporation were cultured in 1 ml of superbroth (1) overnight. The cultures were pelleted and resuspended in 100 μl of water. The resuspended bacteria were then boiled for 10 min and centrifuged for 10 min, and the supernatant was used to characterize the targeted region by performing PCR with flanking primers PK2F3 and PK2R2 and internal primers K1 and K2 (9). To confirm the interruption of the arcA locus, the junctions of the recombination sites were sequenced. Once homologous recombination was confirmed, the arcA mutation was transduced into a fresh culture of SE2472 by using phage P22, and individual phage-free transductants were selected for further analysis.
Assays of bacterial survival after exposure to nitrosative, oxidative and other stresses.
Freshly transformed bacteria were cultured in 2 ml of LB medium at 37°C overnight with shaking. Antibiotics were added as appropriate. Portions (20 μl) of the overnight cultures were added to 2-ml portions of LB medium containing one of the following chemicals: sodium nitrite, hydrogen peroxide, sodium chloride, or sodium dodecyl sulfate (SDS). Survival in the presence of GSNO was assayed in M9 minimal medium with 1:1,000 dilutions of overnight cultures. Survival in the presence of sodium nitrite and GSNO was assayed at pH 5.0, while other stress assays were performed at pH 7.0. In all assays the cultures were grown aerobically with shaking at 225 rpm. After exposure to RNI, ROI, or other stresses, aliquots of cultures were diluted and plated in triplicate. Bacterial colonies were enumerated by determining the number of CFU after overnight incubation in order to determine the bacterial concentrations.
Mouse oral infection studies.
Mice were infected with wild-type SE2472 and its derivatives as previously described (29). The bacteria were diluted in phosphate-buffered saline and used for infection. Groups of eight 6- to 8-week-old BALB/c and C3H/HeN mice (Charles River Laboratories, Kingston, N.Y.) were used for intragastric infection experiments. Each mouse received approximately 3 × 105 to 5 × 105 organisms in 0.25 ml of phosphate-buffered saline through a feeding needle. The mice were monitored for 2 weeks after infection, and mortality during this period was recorded.
RESULTS
Construction of the library and screening clones for ASN resistance.
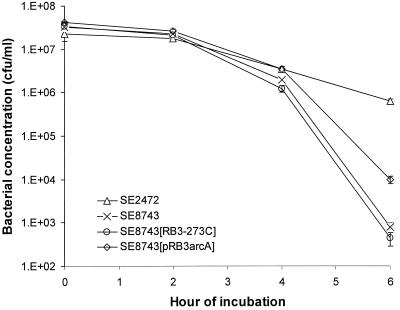
We showed previously that the clinical isolate S. enterica serovar Enteritidis SE2472 is more resistant to killing than other clinical S. enterica serovar Enteritidis isolates after exposure for 6 h to medium containing 20 mM sodium nitrite at pH 5 (29). To identify genes associated with resistance to ASN, we constructed a genomic DNA library of SE2472 using plasmid pRB3-273C. We obtained approximately 5 × 103 independent clones with an average DNA insert size of 1.5 kb, representing approximately 85% of the genome. The library was transformed into SE8743, an S. enterica serovar Enteritidis isolate susceptible to ASN (29). The SE8743 transformants were subjected to selection in ASN medium, and resistant clones were recovered and characterized. We found one plasmid clone, pK2, that consistently conferred a survival advantage upon transformed SE8743 exposed to ASN. SE8743 transformed with pK2 exhibited more than 10-fold-greater survival after 6 h of exposure to 15 mM sodium nitrite at pH 5.0 than untransformed or vector pRB3-273C-transformed SE8743. However, SE8743 containing pK2 still exhibited a lower level of survival than wild-type resistant isolate SE2472 (Fig. 1). The growth kinetics of SE8743 transformed with pK2 and the growth kinetics of SE8743 transformed with the vector pRB3-273C were identical in LB broth (data not shown).
FIG. 1.
ArcA increases resistance of S. enterica serovar Enteritidis to ASN. Cultures of SE2472, SE8743, and SE8743 transformed with vector pRB3-273C or plasmid pRB3arcA were grown in the presence of 15 mM sodium nitrite in pH 5.0 LB medium, and bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to ASN. At least three experiments were performed, and the results of a representative experiment performed in triplicate are shown. The error bars indicate standard deviations.
Characterization of plasmid pK2.
Plasmid pK2 contained an approximately 1.7-kb insert. The insert was sequenced and was found to contain a complete 714-bp open reading frame that was highly homologous to genes encoding the global regulator ArcA of E. coli and S. enterica serovar Typhimurium. In addition to the arcA coding sequence, the insert also contained 423 bases upstream and 543 bases downstream of the arcA sequence. The S. enterica serovar Enteritidis arcA gene exhibited 91 and 99% nucleotide sequence homology to the E. coli and S. enterica serovar Typhimurium arcA genes, respectively. The amino acid sequence of the S. enterica serovar Enteritidis ArcA protein differed from that of S. enterica serovar Typhimurium ArcA at position 127 (alanine → valine) and from that of E. coli ArcA at positions 127 (alanine → valine) and 237 (glutamine → glutamic acid). We renamed plasmid pK2 pRB3arcA to reflect the nature of the insert.
pRB3arcA provides GSNO and H2O2 resistance to SE8743.
Since plasmid pRB3arcA provided ASN resistance to SE8743, we determined whether this plasmid also mediated resistance to other RNI/ROI products. Survival of SE8743 transformed with pRB3arcA was assayed in the presence of 4 mM GSNO at pH 5.0 and in the presence of 5 mM hydrogen peroxide at neutral pH. Compared to untransformed SE8743 and SE8743 transformed with vector pRB3-273C alone, SE8743 transformed with pRB3arcA exhibited enhanced survival under both conditions (Fig. 2). After 7 h of exposure to 4 mM GSNO at pH 5.0, the concentration of viable SE8743 transformed with pRB3arcA decreased less than 1 log10. In contrast, the concentration of SE8743 transformed with pRB3-273C decreased from more than 106 CFU/ml to approximately 102 CFU/ml over the same period, and untransformed SE8743 was completely killed. This indicated that pRB3arcA conferred at least a 3-log10-fold survival advantage on SE8743 in the presence of the stress induced by 4 mM GSNO (Fig. 2A). Interestingly, pRB3arcA-transformed SE8743 exhibited even better survival than resistant isolate SE2472, further demonstrating that overexpression of ArcA effectively increased the resistance of S. enterica serovar Enteritidis to GSNO (Fig. 2A). Exposure to 5 mM hydrogen peroxide for 2 h at pH 7.0 resulted in about a 1-log10 reduction in the concentration of viable SE8743 organisms transformed with pRB3arcA, and the bacterial count returned to the original level at the end of the assay (6 h). Exposure of untransformed SE8743 or SE8743 transformed with pRB3-273C to the same conditions resulted in sharp decreases, about 3 log10 after 2 h and more than 4 log10 after 6 h. Therefore, by the end of the assay SE8743 transformed with pRB3arcA had a survival advantage of 4-log10-fold compared to bacteria transformed with vector pRB3-273C (Fig. 2B). In contrast to the effect of plasmid pRB3arcA on GSNO resistance, pRB3arcA did not increase the level of resistance of transformed SE8743 to hydrogen peroxide to the level of resistance of SE2472 (Fig. 2B). This result suggests that overexpression of ArcA might be more effective against RNI than against ROI.
FIG. 2.
ArcA increases the resistance of S. enterica serovar Enteritidis to GSNO and hydrogen peroxide. Cultures of SE2472, SE8743, and SE8743 transformed with vector pRB3-273C or plasmid pRB3arcA were grown in the presence of 4 mM GSNO in pH 5.0 M9 minimal medium (A) or in the presence of 5 mM hydrogen peroxide in pH 7.0 LB medium (B). Bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to GSNO (A) or H2O2 (B). At least three experiments were performed, and the results of a representative experiment performed in triplicate are shown. The error bars indicate standard deviations.
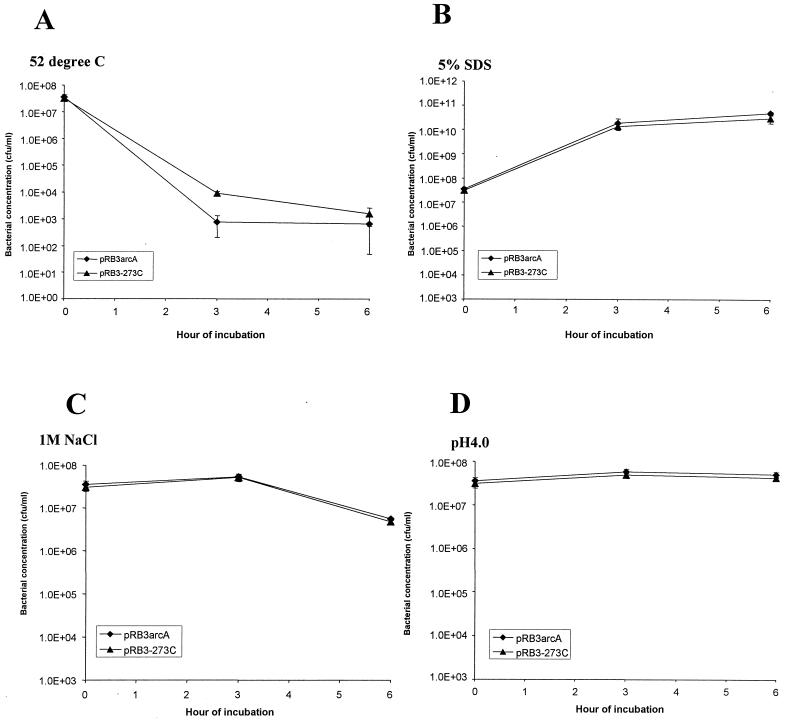
pRB3arcA does not provide protection against general stress.
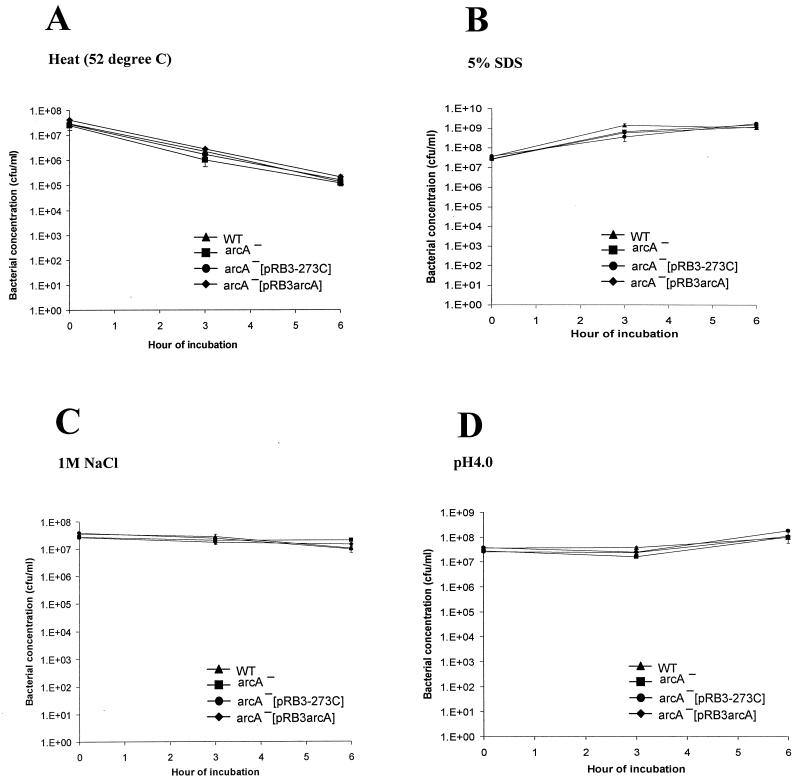
We determined whether the protective effect of pRB3arcA was specific for RNI/ROI stresses or provided protection against stresses in general. Standard assays for general stress conditions used by other investigators were used in this study (16, 35). SE8743 transformed with pRB3arcA was exposed to heat (52°C), salt (1 M NaCl), acid (pH 4.0), or detergent (5% SDS), and the survival of this organism was compared to the survival of SE8743 transformed with the pRB3-273C vector. No differences in survival and growth were observed between S. enterica serovar Enteritidis transformed with pRB3arcA and S. enterica serovar Enteritidis transformed with pRB3-273C under any of these conditions (Fig. 3). Of all the stress conditions tested, incubation at 52°C was the most detrimental to the bacteria. Approximately 3 to 4 log10 of killing occurred in the first 3 h, and there was no further change between 3 and 6 h. S. enterica serovar Enteritidis transformed with pRB3arcA was slightly more susceptible to thermal stress than the strain containing the vector alone. Salt or acid stress had little or no killing effect on S. enterica serovar Enteritidis, although these stresses inhibited the growth of the bacteria. These results demonstrate that plasmid pRB3arcA does not provide a survival and growth advantage to S. enterica serovar Enteritidis under the nonoxidative stress conditions which we evaluated. This finding supports the hypothesis that the protection of SE8743 associated with pRB3arcA is specifically directed against RNI/ROI stresses.
FIG. 3.
ArcA does not protect S. enterica serovar Enteritidis from heat, detergent, salt, or low pH. Cultures of SE8743 transformed with vector pRB3-273C or plasmid pRB3arcA were subjected to 52°C (A), 5% SDS (B), 1 M NaCl (C), or pH 4.0 in LB medium (D). Bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to the stress conditions. At least two experiments were performed, and the results of a representative experiment performed in triplicate are shown. The error bars indicate standard deviations.
Comparison of arcA sequences in isolates SE2472 and SE8743.
Since isolate SE2472 exhibited greater resistance against RNI/ROI stress than SE8743 and since this resistance could be transferred to susceptible isolate SE8743 by plasmid pRB3arcA, we compared the sequence of the arcA locus of SE2472 and the sequence of the arcA locus of SE8743. The coding sequences of the arcA locus were the same in the two isolates.
Construction of an arcA mutant.
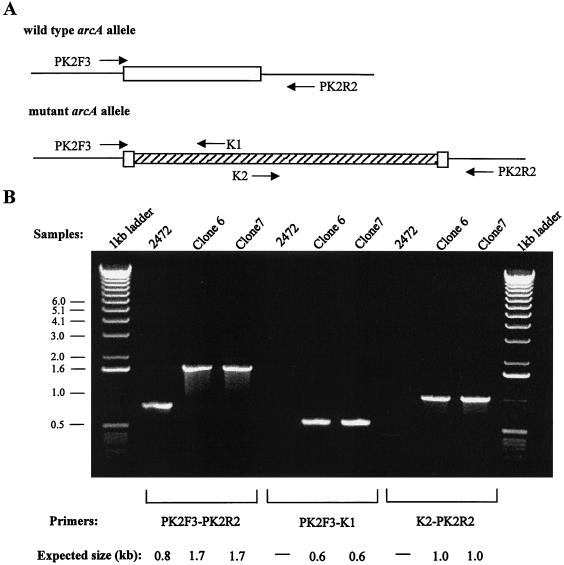
We demonstrated that overexpression of arcA on a multicopy plasmid protects S. enterica serovar Enteritidis against RNI and ROI stresses. It was therefore of interest to determine whether arcA is essential for bacterial defense against these stresses at the physiological level of expression. An arcA mutant of S. enterica serovar Enteritidis was generated by using the Red recombinase system described by Datsenko and Wanner (9). The DNA sequence encoding ArcA amino acids 28 to 223 (of a total of 238 amino acids) was successfully replaced with a kanamycin resistance cassette. Four candidate mutant colonies were screened by PCR performed with both flanking primers and internal kanamycin resistance cassette primers. The primers and expected size of a PCR product obtained following the gene replacement are shown in Fig. 4A. All four colonies displayed the expected pattern after PCR, and two of the patterns are shown in Fig. 4B. To confirm that the expected homologous recombination event occurred, we sequenced the junction regions of the kanamycin resistance and arcA genes. The sequences observed at both the upstream and downstream junction regions were exactly as predicted.
FIG. 4.
Generation of an arcA mutation in S. enterica serovar Enteritidis SE2472. (A) Structures of the wild-type and mutant arcA alleles. The coding region of arcA is represented by an open box, and Kanr is represented by a cross-hatched box. The positions and designations of the primers used to characterize the mutants are shown next to the alleles. (B) Characterization of two arcA mutant clones by PCR. The genomic DNA of wild-type strain SE2472 and mutant clones 6 and 7 were PCR amplified by using the primers shown in panel A. Numbers on the left are molecular sizes in kilobases.
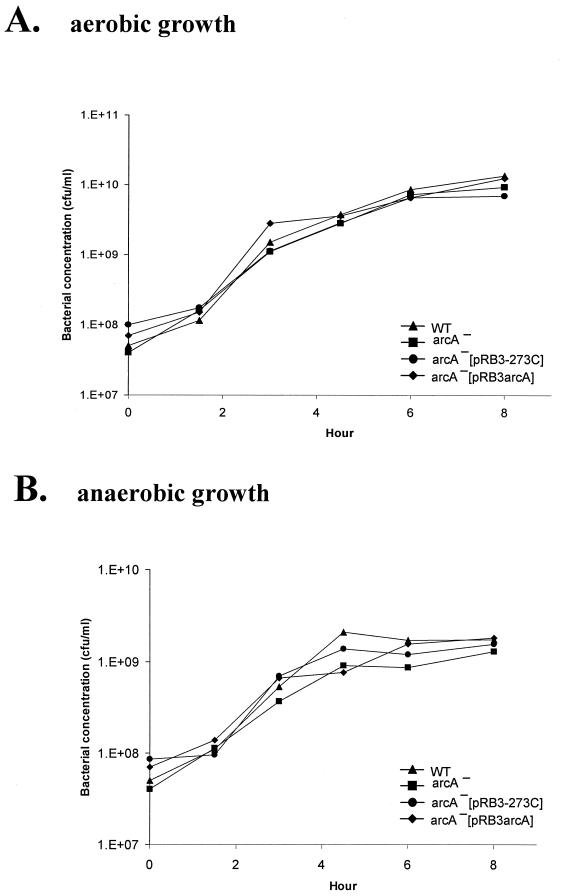
Morphology and growth characteristics of the arcA mutant.
The S. enterica serovar Enteritidis arcA mutant exhibited the same smooth colony morphology that wild-type bacteria exhibited. However, the colonies were smaller than those of the wild type. Since the growth kinetics of an arcA mutant of Salmonella have not been reported previously, we compared the growth curve of the arcA mutant to that of wild-type strain SE2472. The wild type and the arcA mutant of SE2472 showed similar growth kinetics when they were grown either aerobically or anaerobically (Fig. 5). When the organisms were grown in minimal or LB medium, the optical density of the log-phase culture of the arcA mutant was less than the optical density of the wild-type culture. However, the bacterial concentrations were similar when the organisms were enumerated by plating (data not shown). This observation showed that an arcA mutant of SE2472 grows normally in culture and provided the basis for the analysis of resistance of the arcA mutant to RNI and ROI.
FIG. 5.
ArcA mutant of S. enterica serovar Enteritidis exhibited normal growth under both aerobic and anaerobic conditions. Wild-type strain SE2472 (WT), the SE2472 arcA mutant (arcA−), and vector pRB3-273C- and plasmid pRB3arcA-transformed arcA mutant SE2472 (arcA−[pRB3-273C] and arcA−[pRB3arcA], respectively) were grown in peptone-yeast extract medium either aerobically (A) or anaerobically (B). The concentrations of bacteria were determined by sampling and plating after incubation.
Resistance of arcA mutant of S. enterica serovar Enteritidis to RNI and ROI.
Since S. enterica serovar Enteritidis transformed with an arcA plasmid exhibited enhanced resistance to RNI and ROI, we sought to determine if endogenous arcA is essential for the bacterial defense against nitrosative and oxidative stresses. The arcA mutant was exposed to GSNO, hydrogen peroxide, or acidified nitrite, and its survival was compared to that of the wild-type bacteria. All assays were performed under aerobic conditions with vigorous shaking since ArcA is known to be the global regulator induced under anaerobic conditions and anaerobic conditions may affect bacterial resistance to other stresses.
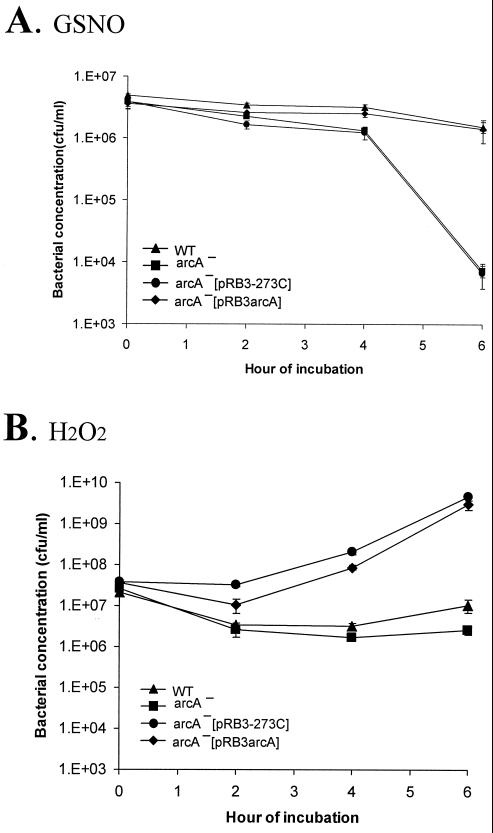
After 4 h of exposure to 4 mM GSNO at pH 5.0 in M9 medium, the arcA mutant decreased over 4 logs, while the wild-type strain decreased minimally (Fig. 6A). To demonstrate that the observed phenotype was specifically attributable to the arcA mutation, we transformed pRB3arcA into the arcA mutant and tested the transformants in the same assay. While the pRB3-273C vector-transformed arcA mutant exhibited the same level of survival as the untransformed mutant, the pRB3arcA plasmid fully compensated for the deficit, indicating that the increased susceptibility to GSNO was indeed due to the arcA mutation (Fig. 6A).
FIG. 6.
ArcA mutant of S. enterica serovar Enteritidis is more susceptible to GSNO and hydrogen peroxide. Cultures of SE2472 (WT), the SE2472 arcA mutant (arcA−), and vector pRB3-273C- and plasmid pRB3arcA-transformed arcA mutant SE2472 (arcA−[pRB3-273C] and arcA−[pRB3arcA], respectively) were grown in the presence of 4 mM GSNO in pH 5.0 M9 minimal medium (A) or in the presence of 5 mM hydorgen peroxide in pH 7.0 LB medium (B). Bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to GSNO (A) or H2O2 (B). At least three experiments were performed, and the results of a representative experiment performed in triplicate are shown. The error bars indicate standard deviations.
We also tested whether the arcA mutant was more susceptible to ROI. The arcA mutant was exposed to 5 mM hydrogen peroxide at pH 7.0. The concentration of mutant bacteria decreased about 1 log10 after the initial 2 h of exposure, remained constant after 4 h, and eventually returned to the level in the starting culture. In contrast, the concentration of wild-type bacteria slightly decreased initially, and then growth of the bacteria resumed. By 6 h after exposure, there was a more-than-2-log10 difference in the concentrations of wild-type and arcA mutant bacteria. The survival and growth disadvantage of the arcA mutant bacteria in the presence of hydrogen peroxide were fully compensated for by transformation with pRB3arcA (Fig. 6B).
The resistance of the arcA mutant to ASN was also analyzed. However, no difference between the wild type and the arcA mutant could be consistently demonstrated in the ASN assay with sodium nitrite concentrations of 10 to 35 mM at pH 5.0.
arcA mutant of S. enterica serovar Enteritidis is not deficient in resistance against heat, salt, detergent, and acid.
To determine the specificity of the increased susceptibility of the SE2473 arcA mutant to RNI/ROI, we determined the susceptibility of the S. enterica serovar Enteritidis arcA mutant to heat (52°C), salt (1 M NaCl), detergent (5% SDS), or acid (pH 4.0). No difference in survival or growth was detected between the arcA mutant and wild-type strain SE2472 under any of these conditions (Fig. 7).
FIG. 7.
ArcA mutation does not make S. enterica serovar Enteritidis susceptible to heat, detergent, salt, or low pH. Cultures of SE2472 (WT), the SE2472 arcA mutant (arcA−), and vector pRB3-273C- and plasmid pRB3arcA-transformed arcA mutant SE2472 (arcA−[pRB3-273C] and arcA−[pRB3arcA], respectively) were subjected to 52°C (A), 5% SDS (B), 1 M NaCl (C), or pH 4.0 in LB medium (D). Bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to the stress conditions. At least two experiments were performed, and the results of a representative experiment performed in triplicate are shown. The error bars indicate standard deviations.
arcA mutant of S. enterica serovar Enteritidis is not significantly attenuated in virulence during oral infection of mice.
To evaluate the effect of arcA on the virulence of S. enterica serovar Enteritidis in vivo, we orally infected mice with the SE2472 arcA mutant and compared the ability of this organism to cause death in mice with that of wild-type strain SE2472. Both BALB/c and C3H/HeN mice were used. The arcA mutant of SE2472 was not attenuated in orally infected BALB/c mice (data not shown). The arcA mutant bacteria were slightly attenuated in orally infected C3H/HeN mice compared to wild-type strain SE2472 and an pRB3arcA-complemented arcA mutant. However, the difference was not statistically significant (data not shown).
DISCUSSION
In a previous report, we described an analysis of a collection of clinical isolates of S. enterica serovar Enteritidis that exhibited a wide range of LD50 for oral infection of BALB/c mice (29). The results of in vitro virulence assays showed little correlation with these oral LD50. However, isolate SE2472, which had the lowest LD50 (16 organisms), was observed to be most resistant to ASN. In the present study, SE2472 was found to be more resistant than a less virulent isolate (SE8743) to GSNO and hydrogen peroxide in liquid culture assays. We initiated a study to search for the molecular mechanisms of RNI/ROI resistance of SE2472. In this study, the global regulator ArcA was found to be associated with the oxidative and nitrosative stress response. The evidence that arcA is essential for resistance of S. enterica serovar Enteritidis to RNI and ROI is as follows: (i) an RNI/ROI-susceptible clinical isolate of S. enterica serovar Enteritidis (SE8743) became more resistant when it was transformed with a multicopy plasmid containing arcA, (ii) disruption of arcA by a kanamycin resistance cassette in an RNI/ROI-resistant isolate of S. enterica serovar Enteritidis (SE2472) made the isolate susceptible to GSNO and H2O2, and (iii) transformation of an arcA mutant of SE2472 with the arcA gene carried on a multicopy plasmid restored the isolate’s resistance to both GSNO and H2O2.
We observed, however, that, while transformation of RNI-susceptible isolate SE8743 with a plasmid expressing arcA made it resistant to ASN, disruption of arcA in relatively RNI-resistant isolate SE2472 did not significantly affect its susceptibility to ASN, suggesting that arcA is not required for resistance of SE2472 to ASN. Instead, the arcA mutant of SE2472 became susceptible to GSNO, a donor of RNI products. Other workers have shown that GSNO mediates oxygen-independent cytostasis of S. enterica serovar Typhimurium and that another NO redox form, NO·, has no antimicrobial activity against this organism (11). Thus, Salmonella appears to exhibit different patterns of susceptibility to different redox forms of NO. The disruption of arcA in SE2472 may have affected a bacterial product that mediates resistance to the bacteriostatic redox form of NO generated by GSNO (presumably nitrosonium [NO+] [11]), whereas the susceptibility of SE8743 to ASN may be mediated by different NO redox forms that are overcome by the multicopy effect of arcA. It is also possible that the difference in the genetic backgrounds of SE8743 and SE2472 is responsible for the difference in susceptibility to ASN of SE8743 transformed with multicopy arcA and SE2472 disrupted in a singe copy of gene arcA.
ArcA is one component of the E. coli and Salmonella global control systems that respond to molecular oxygen, including the OxyR, SoxRS, Fnr, and ArcAB systems (2, 28, 31). Together with Fnr, ArcAB regulates anaerobic growth of E. coli. ArcA is the transcriptional regulator, and ArcB is a transmembrane protein that contains receiver and transmitter domains. Signals recognized by ArcB, thought to be membrane potential or metabolites generated during anaerobic growth, may activate the system (4, 20, 21, 26–28). Recently, Georgellis et al. reported that the redox signals for the ArcAB system are quinones which are membrane-associated electron carriers (21). Anaerobic induction of ArcAB appears to be dependent on Fnr, which also regulates many of the ArcAB-regulated genes involved in cellular adaptation to anaerobic growth (31). Together, ArcAB and Fnr repress genes involved in aerobic metabolism, but ArcAB may also induce positive regulation of the cytochrome d oxidase operon (8, 31). The function of ArcA in aerobic growth has not been reported. Here we provide evidence for the first time that ArcA is important for RNI/ROI resistance under aerobic conditions.
In addition to ArcAB and Fnr, OxyR and SoxRS are the other major global control systems that regulate genes in response to oxidative stress. Both OxyR and SoxRS systems are also involved in resistance to RNI (14, 24, 34). SoxRS appears to be important for RNI resistance in E. coli but not in S. enterica serovar Typhimurium (19). Both OxyR and SoxR can be directly modified by S nitrosylation (14, 24). This report provides evidence for the first time that arcA of the ArcAB regulon is important for Salmonella resistance to nitrosative stress. In an anaerobic environment, nitrogen oxides can serve as electron acceptors. It would be interesting to determine whether expression or activity of the ArcAB system is induced by RNI and whether the sensor ArcB is modified by nitrosylation, as OxyR and SoxR are.
Although we found that arcA is necessary for resistance of S. enterica serovar Enteritidis to both nitrosative stress and oxidative stress in vitro, we did not detect significant attenuation of an arcA mutant S. enterica serovar Enteritidis strain in mice. It is possible that an arcA mutation is compensated for by other factors in vivo. As mentioned above, many of the ArcA-regulated genes are coregulated by the Fnr modulon. Since the RNI/ROI-susceptible arcA mutant strain of S. enterica serovar Enteritidis was able to kill mice in most of the experiments, we concluded that nitrosative and oxidative stresses in vivo are distinct from the stresses examined in vitro in this study or that other factors induced in vivo compensate for the susceptibility to stresses observed in vitro. The observation that an arcA mutant is not clearly attenuated in vivo is not entirely surprising. Two other global transcriptional regulators, OxyR and SoxS, have been shown to be nonessential for virulence of S. enterica serovar Typhimurium in vivo, even though both are essential for resistance to RNI or ROI in vitro (19, 39).
Since all S. enterica serovar Enteritidis isolates carry arcA and the sequence of this gene is the same in SE2472 (RNI/ROI resistant) and SE8743 (RNI/ROI susceptible), the relative RNI/ROI resistance of SE2472 compared to that of other clinical isolates cannot be attributed to this gene alone. Introduction of arcA on a multicopy plasmid was not sufficient for isolate SE8743 to become as resistant to ASN and hydrogen peroxide as SE2472, further indicating that other unidentified loci play a role. Nevertheless, our analyses showed that arcA is necessary for SE2472 to resist some redox forms of RNI, as well as ROI. In the future, it would be of interest to identify ArcA-regulated genes involved in RNI/ROI resistance. The arcA mutant which we constructed provides an opportunity to study such genes. It is becoming clear that the study of the role of global regulators in stress response and Salmonella pathogenesis requires detailed analyses of the interactions of all of these systems in vivo and of the overlapping regulation of the genes that they control.
Acknowledgments
We thank Barry Wanner of Purdue University for providing the reagents of the Red recombinase system and Duc Vugia and Sharon Abbott of the State of California Department of Health Services for providing the S. enterica serovar Enteritidis isolates.
This study was supported by grant AI43032 to L.W.R.
Editor: B. B. Finlay
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495–523. [DOI] [PubMed] [Google Scholar]
- 3.Berggren, R. E., A. Wunderlich, E. Ziegler, M. Schleicher, R. C. Duke, D. Looney, and F. C. Fang. 1995. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10:489–495. [PubMed] [Google Scholar]
- 4.Bogachev, A. V., R. A. Murtazina, and V. P. Skulachev. 1993. Cytochrome d induction in Escherichia coli growing under unfavorable conditions. FEBS Lett. 336:75–78. [DOI] [PubMed] [Google Scholar]
- 5.Bullas, L. R., and J. I. Ryu. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2000. Surveillance for foodborne-disease outbreaks—United States, 1993–1997. Morb. Mortal. Wkly. Rep. 49:1–72. [PubMed] [Google Scholar]
- 7.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605–615. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, R. H., and C. Wray. 1995. Mice as carriers of Salmonella enteritidis on persistently infected poultry units. Vet. Rec. 137:337–341. [DOI] [PubMed] [Google Scholar]
- 11.De Groote, M. A., D. Granger, Y. Xu, G. Campbell, R. Prince, and F. C. Fang. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. USA 92:6399–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Groote, M. A., T. Testerman, Y. Xu, G. Stauffer, and F. C. Fang. 1996. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science 272:414–417. [DOI] [PubMed] [Google Scholar]
- 14.Ding, H., and B. Demple. 2000. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. USA 97:5146–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, R. A., R. A. Helm, and S. R. Maloy. 1999. Increasing DNA transfer efficiency by temporary inactivation of host restriction. BioTechniques 26:892–894. [DOI] [PubMed] [Google Scholar]
- 16.Ehrt, S., M. U. Shiloh, J. Ruan, M. Choi, S. Gunzburg, C. Nathan, Q. Xie, and L. W. Riley. 1997. A novel antioxidant gene from Mycobacterium tuberculosis. J. Exp. Med. 186:1885–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, F. C. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Investig. 99:2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, F. C., A. Vazquez-Torres, and Y. Xu. 1997. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect. Immun. 65:5371–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgellis, D., O. Kwon, and E. C. Lin. 1999. Amplification of signaling activity of the arc two-component system of Escherichia coli by anaerobic metabolites. An in vitro study with different protein modules. J. Biol. Chem. 274:35950–35954. [DOI] [PubMed] [Google Scholar]
- 21.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314–2316. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guard-Petter, J., D. J. Henzler, M. M. Rahman, and R. W. Carlson. 1997. On-farm monitoring of mouse-invasive Salmonella enterica serovar Enteritidis and a model for its association with the production of contaminated eggs. Appl. Environ. Microbiol. 63:1588–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausladen, A., C. T. Privalle, T. Keng, J. DeAngelo, and J. S. Stamler. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86:719–729. [DOI] [PubMed] [Google Scholar]
- 25.Henzler, D. J., and H. M. Opitz. 1992. The role of mice in the epizootiology of Salmonella enteritidis infection on chicken layer farms. Avian Dis. 36:625–631. [PubMed] [Google Scholar]
- 26.Iuchi, S., V. Chepuri, H. A. Fu, R. B. Gennis, and E. C. Lin. 1990. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J. Bacteriol. 172:6020–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iuchi, S., Z. Matsuda, T. Fujiwara, and E. C. Lin. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4:715–727. [DOI] [PubMed] [Google Scholar]
- 28.Iuchi, S., and L. Weiner. 1996. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J. Biochem. 120:1055–1063. [DOI] [PubMed] [Google Scholar]
- 29.Lu, S., A. R. Manges, Y. Xu, F. C. Fang, and L. W. Riley. 1999. Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect. Immun. 67:5651–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg, B. E., R. E. Wolf, Jr., M. C. Dinauer, Y. Xu, and F. C. Fang. 1999. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67:436–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen, p.1526–1538. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella, vol. 1. ASM Press, Washington, D.C.
- 32.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253–263. [DOI] [PubMed] [Google Scholar]
- 33.Mishu, B., P. M. Griffin, R. V. Tauxe, D. N. Cameron, R. H. Hutcheson, and W. Schaffner. 1991. Salmonella enteritidis gastroenteritis transmitted by intact chicken eggs. Ann. Intern. Med. 115:190–194. [DOI] [PubMed] [Google Scholar]
- 34.Nunoshiba, T., and B. Demple. 1993. Potent intracellular oxidative stress exerted by the carcinogen 4-nitroquinoline-N-oxide. Cancer Res. 53:3250–3252. [PubMed] [Google Scholar]
- 35.Ruan, J., G. St. John, S. Ehrt, L. Riley, and C. Nathan. 1999. noxR3, a novel gene from Mycobacterium tuberculosis, protects Salmonella typhimurium from nitrosative and oxidative stress. Infect. Immun. 67:3276–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 37.Shiloh, M. U., J. D. MacMicking, S. Nicholson, J. E. Brause, S. Potter, M. Marino, F. Fang, M. Dinauer, and C. Nathan. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29–38. [DOI] [PubMed] [Google Scholar]
- 38.St. Louis, M. E., D. L. Morse, M. E. Potter, T. M. DeMelfi, J. J. Guzewich, R. V. Tauxe, and P. A. Blake. 1988. The emergence of grade A eggs as a major source of Salmonella enteritidis infections. New implications for the control of salmonellosis. JAMA 259:2103–2107. [PubMed] [Google Scholar]
- 39.Taylor, P. D., C. J. Inchley, and M. P. Gallagher. 1998. The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect. Immun. 66:3208–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]