Abstract
Reactive nitrogen and oxygen species (O2•−, H2O2, NO• and ONOO−) have been strongly implicated in the pathophysiology of neurodegenerative and mitochondrial diseases. In the present study, we examined the effects of nitrosative and/or nitrative stress generated by DETA-NO {(Z)-1-[2-aminoethyl-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate}, SIN-1 (3-morpholinosydnonimine hydrochloride) and SNP (sodium nitroprusside) on U87MG glioblastoma cybrids carrying wt (wild-type) and mutant [A3243G (Ala3243→Gly)] mtDNA (mitochondrial genome) from a patient suffering from MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes). The mutant cybrids had reduced activity of cytochrome c oxidase, significantly lower ATP level and decreased mitochondrial membrane potential. However, endogenous levels of reactive oxygen species were very similar in all cybrids regardless of whether they carried the mtDNA defects or not. Furthermore, the cybrids were insensitive to the nitrosative and/or nitrative stress produced by either DETA-NO or SIN-1 alone. Cytotoxicity, however, was observed in response to SNP treatment and a combination of SIN-1 and glucose-deprivation. The mutant cybrids were significantly more sensitive to these insults compared with the wt controls. Ultrastructural examination of dying cells revealed several characteristic features of autophagic cell death. We concluded that nitrosative and/or nitrative stress alone were insufficient to trigger cytotoxicity in these cells, but cell death was observed with a combination of metabolic and nitrative stress. The vulnerability of the cybrids to these types of injury correlated with the cellular energy status, which were compromised by the MELAS mutation.
Keywords: caspase-independent, electron transport chain, MELAS, mitochondrial DNA mutation, nitric oxide donor, reactive nitrogen and oxygen species
Abbreviations: AIF, apoptosis inducing factor; CFDA, 5-carboxyfluorescein diacetate; CMH2DCFDA, 5-6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate; COX, cytochrome c oxidase; DETA-NO, (Z)-1-[2-aminoethyl-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate; DiOC6, 3,3′-dihexyloxacarbocyanine; ETC, electron transport chain; GD, glucose-deprivation; hsp, heat-shock protein; MELAS, mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes; MEM, minimum essential medium; mtDNA, mitochondrial genome; mtNOS, mitochondrial nitric oxide synthase; PI, propidium iodide; PMOXR, plasma membrane oxidoreductase; RNOS, reactive nitrogen and oxygen species; ROS, reactive oxygen species; SIN-1, 3-morpholinosydnonimine hydrochloride; SNP, sodium nitroprusside; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling; wt, wild-type
INTRODUCTION
Single base mutations and/or rearrangements in the mtDNA (mitochondrial genome) cause mitochondrial dysfunction and contribute to the pathogenesis of several neuromuscular and neurological disorders. MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) is a maternally inherited multisystem disorder, recognized as a clinical syndrome more than 20 years ago [1]. Patients with the MELAS syndrome exhibit a wide spectrum of clinical symptoms, including stroke-like episodes, headache, vomiting, seizures, lactic acidosis, dementia, muscle weakness, deafness, growth retardation and diabetes mellitus. Pathological findings in MELAS patients most frequently include ragged-red fibres in the skeletal muscle [2], cerebellar cortical degeneration, calcification in the basal ganglia and vascular changes known as ‘mitochondrial angiopathy’ [1]. The most common cause of the syndrome is the A to G transition at nucleotide 3243 [3] and T to C transition at nucleotide 3271 [4] that affects the mitochondrial tRNALeu gene. This, in turn, impairs mitochondrial protein synthesis and reduces the activities of respiratory chain complexes containing mitochondrially encoded subunits [5]. Indeed, decreased production of ATP has been observed in cultured myoblasts from MELAS patients [6]. The exact mechanisms by which these mutations result in cellular injury or why they preferentially affect tissues with high-energy requirement is still not clear.
Several neurodegenerative and mitochondrial diseases have been associated with excessive production of RNOS (reactive nitrogen and oxygen species), of which ROS (reactive oxygen species) are produced mainly in the mitochondria as by-products of oxidative phosphorylation [7,8]. Because of the toxic nature of these species, mitochondria have evolved strong anti-oxidant defence systems; however, their detoxifying capacity is not always sufficient to offset the high rates of oxidative stress. The formation of superoxide anions (O2•−) occurs via the transfer of a free electron from the ETC (electron transport chain) to molecular oxygen. Complexes I and III produce most of the O2•−, which is detoxified by MnSOD (where SOD stands for superoxide dismutase) or cytosolic Cu/ZnSOD to produce H2O2. Mitochondria also strongly depend on GSH to combat the oxidative stress; for example, H2O2 is converted into H2O by glutathione peroxidase and GSH. Some laboratories have provided evidence that mitochondria possess their own nitric oxide synthase [mtNOS (mitochondrial nitric oxide synthase)] and may serve as an endogenous source of nitric oxide (NO•) [9,10]. Under pathological conditions, NO• might react with O2•− to produce peroxynitrite (ONOO−) resulting in the oxidation, nitrosation and nitration of proteins leading to mitochondrial dysfunction [11]. This, however, is still controversial since other laboratories have not been able to confirm the presence of mtNOS [12,13].
The present study was designed to examine the effects of nitric oxide donors, namely SIN-1 (3-morpholinosydnonimine hydrochloride), DETA-NO {(Z)-1-[2-aminoethyl-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate} and SNP (sodium nitroprusside), on the viability of cybrids carrying the MELAS mtDNA mutation. The cybrid technology has been widely used to recapitulate the effects of mtDNA mutations on various cellular functions such as activities of respiratory chain complexes and cell growth. This technique involves the transfer of mitochondria from cells of affected patients to a cell line lacking mtDNA (ρ° cells). We generated human U87MGρ° glioblastoma cells [14], which were subsequently fused with enucleated fibroblasts from a MELAS patient carrying A3243G (Ala3243→Gly) mtDNA mutation in the tRNALeu gene [15]. In the present study, we have described the characterization of cybrid clones, their vulnerability to RNOS and the mechanism of cell death resulting from the cytotoxic injury.
MATERIALS AND METHODS
Generation of cybrids by fusion of enucleated fibroblasts with ρ° cells
Cybrids were generated on a background of U87MG glioblastoma (ATCC # HTB-14) devoid of their own mtDNA (ρ° cells). Parental U87MGρ+ cells were grown in Eagle's MEM (minimum essential medium; Gibco BRL, Burlington, ON, Canada) supplemented with 10% (v/v) FBS and 1× antibiotic-antimycotic solution (Gibco BRL) at 37 °C in 5% CO2. The U87MGρ° cells were obtained after 3 months of growth in the presence of 250 μg/ml ethidium bromide in complete MEM medium containing 100 μg/ml pyruvate, 50 μg/ml uridine and 4500 mg/l glucose (ρ° medium). Cells were replated once a week and the medium was changed every 2–3 days. The absence of mitochondrial DNA was determined by PCR using D-loop-specific primers (forward primer: 5′-CCCTCCCACTCCCATACTAC-3′, nucleotide position 16440–16459 and reverse primer: 5′-ACGGGCGGTGTGTACGCG-3′, nucleotide position 914–897; GenBank® accession no. D38112.1 [14]).
Primary fibroblast cultures were established from a 37 year-old female MELAS patient carrying the heteroplasmic A3243G mtDNA mutation [15]. The cells were enucleated by treatment with cytochalasin B (Sigma, St. Louis, MO, U.S.A.) and subsequently fused with ρ° cells using 50% (w/v) poly(ethylene glycol) 1500 solution (Roche Diagnostics, Laval, QC, Canada) essentially as described previously [14,16]. Cybrids containing exogenous fibroblast mitochondria were selected by culturing in MEM medium (lacking pyruvate and uridine) supplemented with 10% dialysed heat-inactivated FBS, expanded and analysed for the presence of mtDNA mutation.
DNA extraction and PCR amplification
Total cellular DNA was extracted and PCR-amplified using the following primers: 5′-ATTGACCTGCCCGTGAAGAGGCG-3′ (forward) and 5′-CACCCTTATCACAACACAAGAAC-3′ (reverse). The PCR amplification was performed at 94 °C for 1 min, 55 °C for 45 min and 72 °C for 70 s for 30 cycles. The mutation was detected after a digestion of PCR fragment with ApaI at 30 °C for 1 h [17]. The proportion of mutant mtDNA in the cybrids was quantified by densitometry using Scion Image beta 3b (Scion Corporation, Frederick, MD, U.S.A.) and expressed as percentage of mutant band volume/total band volume.
Cytochemistry of COX (cytochrome c oxidase) activity
Cybrids grown in six-well dishes for 24 h were washed in 50 mM sodium phosphate buffer (pH 7.4) and incubated with the same buffer containing 0.5 mg/ml 3′,3-diaminobenzidine and 1 mg/ml oxidized cytochrome c at 37 °C for 2–3 h. The images were captured using an Olympus IX 50 microscope.
ATP levels
Cybrids were grown in T-25 flasks, harvested in a reaction buffer containing 20 mM glycine, 50 mM MgSO4 and 4 mM EDTA (pH 7.4) and sonicated. ATP assay was carried out according to the manufacturer's instructions. Briefly, 20 μl of the sample was mixed with 2–5 μl of luciferase-luciferin solution (Thermo Labsystems Oy, Helsinki, Finland) and the intensity of the emitted light was measured using a plate reader assay (Fluostar optima; BMG LabTechnologies, Offenburg, Germany). The amount of ATP produced was determined from the standard curve constructed using 10–100 pmol ATP. Protein content was determined with bicinchoninic acid assay (Pierce, Rockford, IL, U.S.A.).
Oxygen consumption
Cybrids grown in six-well dishes for 48–72 h were washed and placed in a balanced salt solution containing 117 mM NaCl, 5.4 mM KCl, 1.5 mM CaCl2, 0.8 mM MgSO4, 0.9 mM NaH2PO4, 10 mM Hepes and 5.6 mM glucose (pH 7.4). Oxygen consumption of single cells in the absence or presence of 0.5 mM SIN-1 was monitored over 15–30 min at 37 °C by the fully automated system from BioCurrents Research Center (MBL, Wood Hole, MA, U.S.A.) using the self-referencing oxygen electrodes [18]. All measurements were carried out in triplicate.
Exposure of cybrids to nitric oxide-donating drugs
Cybrids were grown to 70% confluence and then exposed to SIN-1 (up to 2.5 mM), DETA-NO (0.25–1.0 mM) (both from Toronto Research Chemical, North York, ON, Canada) or SNP (0.5–1.0 mM; Fisher Scientific, Fair Lawn, NJ, U.S.A.) in complete medium, unless otherwise indicated. In some experiments, the cybrids were placed in GD (glucose-deprivation) medium containing 0.5–1.0 mM SIN-1. Control experiments were performed in the presence of degraded SIN-1 or SNP solution (by exposing to light for 72 h at 25 °C room temperature). Cell viability was measured after 24–48 h of treatments.
Cell viability assays
The CFDA (5-carboxyfluorescein diacetate) assay was used to quantify cell viability [19]. Cells were incubated with 2.5 μg/ml CFDA (Molecular Probes, Eugene, OR, U.S.A.) in Earle's Balanced Salt Solution (Sigma) at 37 °C for 30 min and fluorescence was quantified using a CytoFluor™ 2300/2350 system (Millipore, Bedford, MA, U.S.A.) with λex=480±20 nm and λem=530±25 nm. Morphological assessment of cell viability was determined based on cell staining with CFDA and PI (propidium iodide). Briefly, cells were stained with CFDA/PI and after air-drying the number of dead cells (stained red) and live cells (stained green) were examined under an Olympus BX50 fluorescence microscope using ×40 objective. Flow cytometry was also performed by staining the cells with 5 μg/ml PI. PI excited at 488 nm and the fluorescence were analysed at 610 nm using a Coulter ELITE ESP flow cytometer. The TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) assay was used to visualize 3′-OH breaks in the fragmented DNA.
Measurement of PMOXR (plasma membrane oxidoreductase) activity
PMOXR activity was analysed by the bathophenanthroline-disulphonic acid assay [20]. Cells were washed twice with PBS and incubated for 2 h in a buffer containing 1 mM potassium ferricyanide, 20 mM Tris/HCl (pH 7.4) and 150 mM NaCl. At the end of the incubation, the supernatant and cell pellets were collected by centrifugation at 194 g for 5 min. The supernatant was assayed in duplicate by incubating 700 μl of supernatant with 100 μl of 3 M sodium acetate, 100 μl of 0.2 M citric acid, 50 μl of 3.3 mM ferric chloride (diluted in 0.1 M acetic acid) and 50 μl of bathophenanthroline-disulphonic acid (Sigma). The samples were incubated for 10 min at room temperature and absorbance was measured at 535 nm. The pellets were dissolved in PBS containing 0.2% Triton X-100 and the protein content was determined by the bicinchoninic acid assay.
Measurement of ROS
ROS production was assessed by flow cytometry after cell staining with CMH2DCFDA (5,6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate; acetyl ester, Molecular Probes), a redox-sensitive fluorescent dye. Cells were grown in T-75 flasks to 70% confluence and subjected to nitrative stress (0.5 mM SIN-1+GD) for 24–48 h, washed and resuspended in PBS containing 1% BSA. The cells (1.5×106 cells/0.5 ml) were stained for 20 min at 37 °C with 5 μM CMH2DCFDA. Samples were kept on ice in the dark and a minimum of 20000 events were analysed. CMH2DCFDA was excited at 488 nm and the emitted fluorescence was analysed at 525 nm using a Coulter ELITE ESP flow cytometer and EXPO32™ software (Applied Cytometry Systems, Sheffield, U.K.).
Measurement of mitochondrial membrane potential (Δψm)
Cells were grown in T-75 flasks to 70% confluence and treated with GD+SIN-1. After treatment, cells were washed, resuspended in PBS containing 1% BSA (1.5×106 cells/0.5 ml) and incubated with 20 nM DiOC6 (3,3′-dihexyloxacarbocyanine; Molecular Probes) and 5 μg/ml PI for 15 min at 37 °C in a humidified 5% CO2 incubator. Samples were kept on ice in the dark and a minimum of 20000 events were analysed using a Coulter ELITE ESP flow cytometer and WinMDI version 2.8 software (Purdue University Cytometry Laboratories, West Lafayette, IN, U.S.A.). Mitochondrial Δψm was assessed by measuring the fluorescence using 488 nm excitation and 520 nm emission.
Measurement of caspase activity
Cells were grown (8×104 for cybrids R9 and R921, 1×105 for cybrid R15), harvested, washed once with PBS and the pellet was lysed in 3 vol. of lysis buffer (25 mM Hepes, 5 mM MgCl2, 2 mM dithiothreitol, 1.3 mM EDTA, 1 mM EGTA, 0.1% Triton X-100 and protease inhibitor cocktail) on ice for 30 min. Samples were stored at −80 °C until analysed. Caspase-3 (CPP32) activity was measured using the ApoAlert™ caspase assay according to the manufacturer's instructions. The shift in fluorescence emission following cleavage of DEVD-AFC, a specific substrate for caspase-3, was measured on a CytoFluor™ 2350 plate reader using excitation at 360±40 nm and emission at 460±40 nm. Total protein concentration for each sample was determined using Bio-Rad protein reagent (Bio-Rad Laboratories, Mississauga, ON, Canada).
Immunofluorescence staining
Cells grown on glass coverslips were treated with GD+SIN-1, fixed for 10 min with Genofix™ (DNA Genotek Inc., http://www.DNAGenotek.com), blocked with 1% BSA or 0.15% gelatin in PBS and processed for immunocytochemistry. Double labelling of cultures was carried out by incubation with anti-AIF (apoptosis inducing factor) antibody (1:200, goat polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) followed by detection with rabbit anti-goat IgG-Alexa 488 (1:500; Molecular Probes). Cultures were blocked again and then incubated with anti-hsp60 antibody (1:200, mouse monoclonal, Sigma; where hsp stands for heat-shock protein) followed by detection with goat anti-mouse IgG-rhodamine (1:500; Molecular Probes). Incubation with primary and secondary antibodies was carried out for 1 h at room temperature. The cells were visualized under a Carl Zeiss Axiovert 200M.
Electron microscopy
Cells were fixed with 2.5% (v/v) glutaraldehyde in 100 mM sodium phosphate (pH 7.4) for 1 h at room temperature. They were centrifuged at 900 g and embedded in 22% (w/v) BSA. After washing with 100 mM sodium cacodylate (pH 7.2), the cells were post-fixed with 1% osmium tetroxide in 100 mM sodium cacodylate (pH 7.2) for 1 h at 4 °C. Cell pellets were stained en bloc with 2% (v/v) aqueous uranyl acetate, dehydrated in ethanol and acetone and then embedded in Spurr epoxy resin. Ultrathin sections were stained with lead citrate and photographed on a Hitachi H-7100 electron microscope.
Statistical analysis
Data are expressed as means±S.D. or S.E.M. Data analysis was performed by unpaired t test or ANOVA followed by post-hoc Bonferroni's, Dunnett's or Tukey's multiple comparison tests. A value of P<0.05 was considered statistically significant. Statistical tests were performed using GraphPad Prism version 3.02.
RESULTS
Characterization of the cybrids
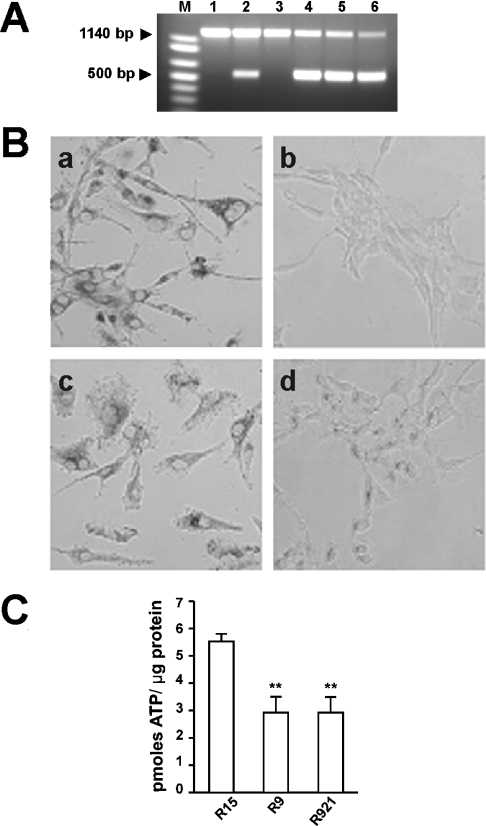
Mitochondria from skin fibroblasts of a MELAS patient were transferred by cytoplast fusion to U87MGρ° cells generated in our laboratory [14]. Several cybrid clones were isolated and heteroplasmy was determined by PCR analysis (Figure 1A). The A3243G MELAS mutation creates a new restriction site for ApaI, hence the enzyme cuts the mutant mtDNA, but not the wt (wild-type) mtDNA, into two fragments of 574 and 566 bp respectively (Figure 1A, lanes 2 and 4–6). The donor fibroblasts carried approx. 30% of mutant mtDNA (lane 2) and the selected cybrids contained different proportions of the wt and mutant mtDNA (lanes 3–6). For example, cybrids R15 (lane 3) and R13 (results not shown) contained only wt mtDNA and are referred to as control cybrids. On the other hand, cybrids R9 and R921 contained both the wt and mutant mtDNA, although at different ratios: R9 (lane 4) harboured approx. 60% mutant and 40% wt, and R921 (lane 6) had approx. 80% mutant and only 20% of wt mtDNA. The cybrids were propagated in a medium containing pyruvate (no uridine), they had similar growth kinetics and an average doubling time of 32.6±3.0 h. The cybrid phenotypes were stable for at least 3 months in culture (results not shown).
Figure 1. Characterization of cybrids.
(A) Restriction enzyme analysis of mtDNA. DNA samples were isolated, PCR amplified and digested with ApaI as described in the Materials and methods section. Lane 1, restriction pattern of mtDNA from U87MGρ+ cells; lane 2, fibroblasts from a MELAS patient; lane 3, mtDNA from a cybrid clone R15 harbouring wt mtDNA; lanes 4 and 5, cybrid clones R9 and R9-6 (not used in this study) harbouring 58.5±0.05% (means±S.D.) mutant mtDNA; lane 6, cybrid clone R921 harbouring 79±0.07% mutated mtDNA; lane M, 1 kb plus molecular mass standard. (B) Assessment of COX activity in U87MGρ+ cells (a), U87MGρ° cells (b), cybrid clone R13 with wt mtDNA (c) and cybrid clone R9 with 58.5% mutant mtDNA (d). COX activity was visualized as a precipitate of diaminobenzidine. Magnification ×400. (C) ATP levels were measured by the luciferase-luciferin assay and are expressed as pmol ATP/μg of protein. Results shown are ATP levels in control and mutant cybrids. Significant differences are shown as **P<0.01.
COX is the terminal metalloprotein of the ETC and transfers electrons from cytochrome c to molecular oxygen. Accordingly, there was no COX activity in U87MGρ° since they lack the mitochondrial-encoded subunits (Figure 1B, panel b) and the impairment of ETC activity by the MELAS mutation was evident in the R9 (Figure 1B, panel d) and R921 (results not shown) cybrids, which showed fewer deposits of diaminobenzidine produced by COX. These cybrids had a significantly lower COX activity compared with the parental U87MGρ+ cells (Figure 1B, panel a) or control cybrids with wt mtDNA (Figure 1B, panel c). Similar levels of COX were observed in the parental U87MGρ+ cells and R13 cybrids with wt mtDNA (Figure 1B, panels a and c). The defective ETC of the MELAS cybrids was clearly reflected in cellular ATP stores (Figure 1C). The mutant cybrids had significantly lower ATP levels than the wt cybrids (Tukey's multiple comparison test, P<0.01 for R15 versus R9 and R15 versus R921, Figure 1C).
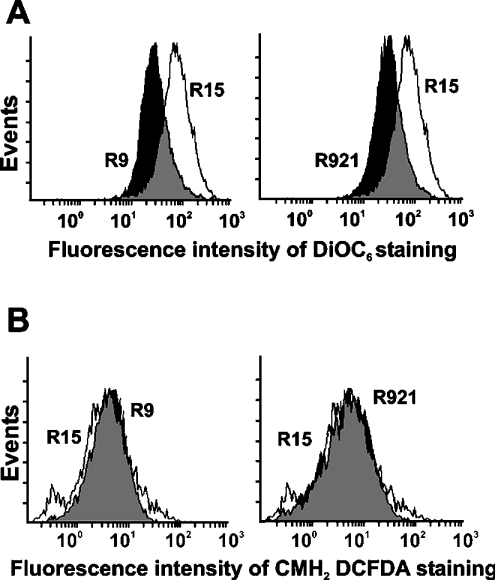
The mitochondrial membrane potential was measured by flow cytometry after staining the cells with DiOC6 dye. As shown in Figure 2(A), the mean DiOC6 fluorescence intensity was lower in the mutant cybrids, i.e. 42.8 for cybrid R9 and 45.3 for cybrid R921 (black histograms) when compared with 66.3 for the control R15 cybrid (clear histogram), indicating a difference in the membrane potential. The intracellular formation of ROS was established based on the oxidation of CMH2DCFDA, which yields an intracellularly trapped fluorescent compound. As shown in Figure 2(B), intracellular ROS levels were not significantly different between the mutants and the control cybrids, although the levels were higher in control cybrids with the functional ETC; the mean CMH2DCFDA fluorescence intensity was 5.7 and 5.8 for R9 and R921 cybrids respectively (black histograms) and 8.8 for control R15 cybrid (clear histogram).
Figure 2. Flow cytometry analysis of mitochondrial function.
(A) Measurement of mitochondrial membrane potential. Cells were washed, resuspended in PBS containing 1% BSA with 20 nM DiOC6 and 5 μg/ml PI and incubated for 15 min at 37 °C. A minimum of 20000 events were analysed for each sample as described in the Materials and methods section. (B) Measurement of ROS levels. Cells were washed, harvested and resuspended in PBS containing 1% BSA and 5 μM CMH2DCFDA for 20 min at 37 °C. A minimum of 20000 events were analysed for each sample as described in the Materials and methods section. Results shown are representative of 2–3 separate experiments. Clear histograms show control cybrids and black histograms show mutant cybrids.
Sensitivity of the cybrids to metabolic and nitrative stress
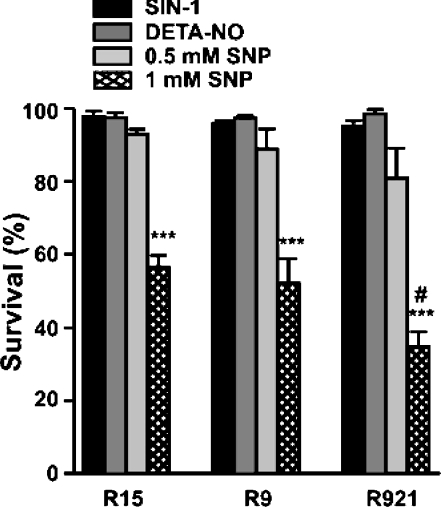
SIN-1 has been previously shown to produce peroxynitrite (ONOO−) by simultaneously generating NO• and O2•− [21]. However, SIN-1 alone even at a concentration of 2.5 mM did not affect the viability of the cybrids (neither control nor mutant); no cell loss occurred over a period of 48 h (Figure 3, black bars refer to cells treated with SIN-1). Similarly, no cell death was observed in either control or mutant cybrids following treatment with 1.0 mM DETA-NO for a period of 24 h (Figure 3, dark grey bars refer to cells treated with DETA-NO; and light grey bars refer to cells treated with 0.5 mM SNP). DETA-NO is a spontaneous donor of NO• with a half-life of 20 h [22]. A clinically used NO•-donor SNP, which requires biotransformation to release NO• [23], was also tested. In contrast with the other two compounds, this drug was highly cytotoxic, especially at a concentration of 1.0 mM (Figure 3, hatched bars refer to cells treated with 1 mM SNP). Percentages of viable cells after the 24 h treatment were: 56.5 for control R15, 52.1 for R9 and 34.7 for R921 (Tukey's multiple comparison test, P<0.001 when compared with untreated controls). Although all cybrids were affected by the treatment, the mutant R921 was clearly the most vulnerable (P<0.05 when compared with treated R15 control). Cell death was eliminated in cultures treated with inactivated SNP (results not shown).
Figure 3. Cell viability following exposure to nitric oxide donors.
Cells grown in a 12-well dish at 5.0×104 cells/well were allowed to attach overnight and treated with SIN-1, DETA-NO or SNP in complete medium for 24 h and cell viability was measured using the CFDA assay as described in the Materials and methods section. Viability of untreated cybrids was used as 100% and the treated cybrids were expressed as percentage of control. Results are expressed as means±S.E.M. from 3–4 separate experiments performed in triplicate. Significant differences are shown as #P<0.05 or ***P<0.001.
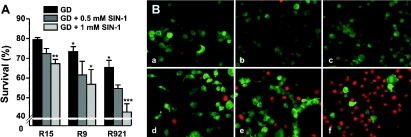
Cytotoxicity was also observed when the cybrids were subjected to a combination of metabolic block caused by GD and nitrative stress produced by SIN-1 (Figure 4). Figure 4(A), clearly shows that the mutant cybrids were more susceptible to killing by ONOO− under GD conditions and the killing was dose-dependent. The metabolic stress itself contributed to cell loss; after 24 h of GD, between 20 and 35% of cybrids were PI-positive. Again, the mutant cybrids were slightly more sensitive (Figure 4A, black bars; Bonferroni's multiple comparison test, P<0.05 when compared with R15 cybrid under GD). However, far greater cell loss was observed in cultures exposed to both stressors, especially to the combination of GD+1 mM SIN-1 (Figure 4A, light grey bars), i.e. 27% under GD alone versus 37% under GD+SIN-1 for R9 cybrid (P<0.05, Bonferroni's multiple comparison test) and 35% under GD versus 57% under GD+SIN-1 for cybrid R921 (P<0.001, Bonferroni's multiple comparison test). Incubation of cells with light-inactivated SIN-1 did not potentiate the effects caused by GD alone (results not shown). Morphological assessment of the cultures subjected to GD+SIN-1 clearly revealed a decrease in the number of viable cells (stained green) and a concomitant increase in the number of dead PI-positive cells (stained red, Figure 4B), especially in the cultures of mutant cybrids.
Figure 4. Analysis of cell viability.
Cells were subjected to GD in the presence or absence of SIN-1 and cell viability was measured using the CFDA assay as described in the Materials and methods section. (A) Effect of metabolic and metabolic plus nitrative stress on cell viability. Viability of untreated cybrids was used as 100% and the treated cybrids were expressed as percentage of control. Results are means±S.E.M. from 3–4 separate experiments performed in triplicate. Significant differences are shown as *P<0.05, **P<0.01 and ***P<0.001. (B) Photomicrographs of cells treated with GD+SIN-1 and stained with CFDA+PI. R15 cybrids untreated (a) or treated (d); R9 cybrids untreated (b) or treated (e); and R921 cybrids untreated (c) or treated (f). Magnification ×200. Images shown are representative of at least three separate experiments.
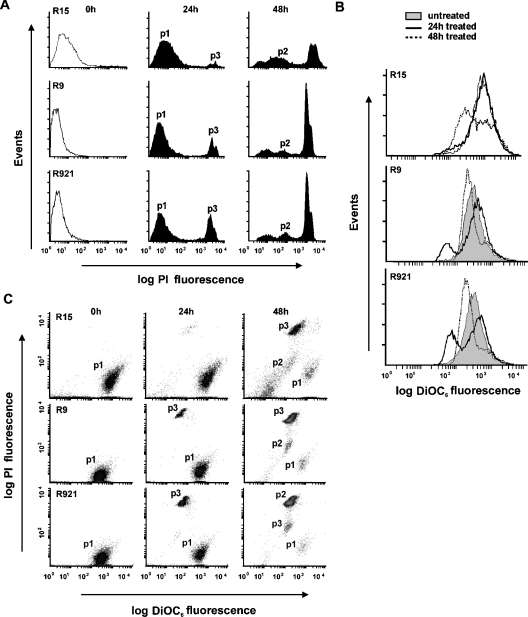
Cell survival and Δψm in GD+0.5 mM SIN-1-treated cells were also measured using flow cytometry after staining the cells with PI and DiOC6 respectively (Figures 5A and 5B). Time-course analysis showed that the loss of viability occurred after 24 h of treatment and further increased by 48 h (Figure 5A). The percentages of cells that were completely permeable to PI (p3) at 24 h were higher in the mutants, 16±2.8 (means±S.E.M.) and 25.4±4.0 for R9 and R921 respectively, when compared with 8.9±1.0 for the R15 control. After 48 h of treatment, the percentages of PI-positive cells (p3) further increased to 60.1±2.5 and 58.9±2.2 in mutant cybrids respectively, when compared with 45.5±2.2 in R15 cybrid. An intermediate population of cells (p2) with increased PI staining was observed at 48 h (Figure 5A). Time-course analysis showed that a noticeable loss of Δψm has occurred in the mutant cybrids after 24 h of treatment (solidline histograms, Figure 5B) with a further loss at 48 h (dotted-line histograms, Figure 5B). At 24 h, the percentage of affected cells was higher in the mutant cybrids, 26±1.8 and 37.5±0.6 for R9 and R921 respectively, when compared with 13.7±1.0 for the R15 cybrid. This was further accentuated at 48 h as 78±3.0% of R9, 82±1.1% of R921 and 57.1±2.0% of R15 cybrids lost Δψm.
Figure 5. Analysis of cell viability and mitochondrial membrane potential.
Cells treated with GD+0.5 mM SIN-1 were washed, resuspended in PBS containing 1% BSA with 20 nM DiOC6 and 5 μg/ml PI and incubated for 15 min at 37 °C. A minimum of 20000 events were analysed for each sample as described in the Materials and methods section. (A) Viability of cybrids treated with GD+0.5 mM SIN-1. PI fluorescence intensity was used as a measure of cell viability. Shown are untreated PI-negative cybrid cells (clear histograms, starting points), treated PI-negative cybrid cells (p1, surviving cells), PI-positive cells with high PI staining (p3, dead cells) and an intermediate population of cells with increased PI staining (p2). Results shown are representative of three separate experiments carried out in duplicate. (B) Mitochondrial membrane potential of cybrids treated with GD+0.5 mM SIN-1. DiOC6 fluorescence intensity was used as a measure of Δψm. Shown are untreated cybrid cells with high Δψm (grey histograms, starting points) and treated cybrid cells with low Δψm at 24 h (solid-line histograms) or 48 h (broken-line histograms). Surface plots are representative of three separate experiments carried out in duplicate. (C) Scatter plots of PI versus DiOC6. Population p1 represents cells with high Δψm and impermeable to PI, p2 represents cells with low Δψm and increased PI staining and p3 represents cells with low Δψm and high PI staining. Results shown are representative of three separate experiments carried out in duplicate.
Scatter plot of this data clearly showed that cells that had lost their Δψm were also completely permeable to PI, indicating that the loss of Δψm was accompanied by decreased cell viability (Figure 5C). At 24 h, the percentages of normal cells with high Δψm and impermeable to PI (p1) decreased from 86.2±1.5 to 77.0±1.5 for the control R15, from 90.4±0.44 to 71.0±1.4 for R9 and from 86.3±2.2 to 59.0±1.4 for R921. Percentages of healthy cells further declined at 48 h to 31.3±1.4 for R15, and to 20.7±3.0 and 16.2±1.3 for mutants R9 and R921 respectively. This was associated with an increase in the number of dead cells, i.e. cells with low Δψm and high PI staining (p3). At 24 h, the percentages of p3 cells were higher (20.4±2.2 and 31.2±1.1) for the mutants R9 and R921 respectively, when compared with 7.8±0.5 for control R15, which further increased at 48 h to 43.13±0.65 for R15, and to 60.4±2.9 and 59±2.2 for mutants R9 and R921 respectively. At 48 h, the intermediate population of cells (p2) with low Δψm and increased PI staining was detected and the percentages of cells in this population were: 17±2.7 for R921, 13.6±1.1 for R9 and 13.4±2.1 for control R15.
Generation of RNOS
ROS generation in cybrids subjected to GD or GD+SIN-1 treatment was assessed by flow cytometry using a CMH2DCFDA oxidation-based fluorescence assay and reactive nitrogen species generation was determined by measuring the amount of nitrite accumulated and nitrotyrosine staining. As shown in Table 1, variable changes in intracellular ROS levels (i.e. 2.0–3.5-fold increases at 24 h) were observed in all the cybrids. The differences between the control R15 and mutant R9 and R921 cybrids, however, were not statistically significant (P>0.05, Tukey's multiple comparison test). No obvious difference in ROS production was seen at 6 or 24 h in all cybrids subjected to GD alone and the ROS levels were comparable between the control and mutant cybrids (results not shown). Interestingly, the control R15 cybrids with functional ETC produced a slightly higher level of ROS. The basal levels of nitrite were similar in the control and mutant cybrids (means±S.D., 4.75±0.11 μM). A 70-fold increase in nitrite accumulation was found in all cybrids treated with SIN-1 alone or in combination with GD, and anti-nitrotyrosine staining also revealed extensive protein nitration (results not shown).
Table 1. Flow cytometry analysis of ROS production in GD+SIN-1-treated cultures.
After treatments, cells were washed, resuspended in 5 μM CMH2DCFDA in PBS containing 1% BSA and incubated for 20 min at 37 °C. A minimum of 20000 events were analysed for each sample as described in the Materials and methods section and expressed as percentage of cells stained with CMH2DCFDA. Results are shown as means±S.E.M. from three separate experiments.
| CMH2DCFDA-stained cells (%) | |||
|---|---|---|---|
| Cell type | 0 h | 6 h | 24 h |
| R15 | 12.03±2.5 | 27.2±2.1 | 41.6±4.5 |
| R9 | 9.1±0.15 | 17.6±2.8 | 29.0±5.4 |
| R921 | 15.9±1.7 | 26.0±5.5 | 32.3±6.6 |
The effect of RNOS on energy metabolism was studied by measuring oxygen consumption of cybrids. SIN-1 (0.5 mM) did not exert any effect on the rate of oxygen consumption either in the control R15 or mutant cybrids. Oxygen flux was measured to be 0.00086±0.00023 μmol·cm−2·s−1 (means±S.D.) in untreated cybrids compared with 0.00098±0.00014 μmol·cm−2·s−1 in treated R15 cybrids. Similarly, the oxygen flux in mutants was 0.0014±0.00002 μmol·cm−2·s−1 in untreated cybrids compared with 0.0014±0.00011 μmol·cm−2·s−1 in treated R9 cy-brids and 0.0013±0.00029 μmol·cm−2·s−1 in untreated cybrids compared with 0.00096±0.00013 μmol·cm−2·s−1 in treated R921 cybrids, suggesting that ONOO− or a related species did not impair mitochondrial respiration.
ATP content and PMOXR activity
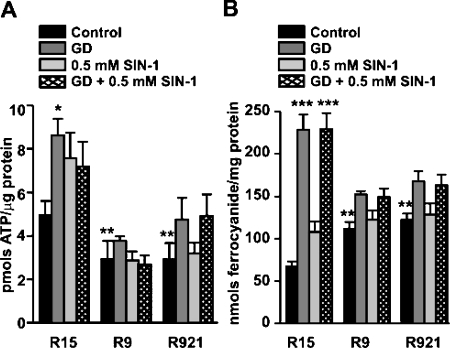
Mitochondria are the main energy-transducing organelles of eukaryotic cells and generate energy in the form of ATP, driven by a substantial Δψm across the mitochondrial inner membrane [24]; therefore it would be expected that cells with defective mtDNA have lower ATP levels. Accordingly, the basal ATP level was lowered by nearly 50% in mutant cybrids when compared with the control (5.52±0.28 pmol/μg for R15 when compared with 2.93±0.57 for R9 and 2.92±0.57 for R921 respectively; Figure 6A, black bars refer to untreated controls). Interestingly, ATP levels were not reduced when the cybrids were subjected to the RNOS-generating treatments (i.e. GD, SIN-1 or GD+SIN-1). In fact, the treatments triggered up-regulation of the ATP-synthesizing pathways, especially in the wt cybrids, in which the ATP stores almost doubled during the 24 h period (Figure 6A, P<0.05, Bonferroni's multiple comparison test). Although no significant up-regulation of ATP stores was observed in the mutant cybrids, they were able to maintain the basal ATP levels unchanged under these conditions.
Figure 6. ATP levels and PMOXR activity.
Cells were grown in T-25 cm2 flasks and subjected to GD in the presence or absence of SIN-1 for 24 h. After washing, cells were collected and analysed for ATP content and PMOXR activity as described in the Materials and methods section. (A) ATP levels were measured by the luciferase-luciferin assay. Shown are changes in ATP levels in cybrids in response to the treatments. Results are expressed as means±S.E.M. from three separate experiments performed in duplicate. (B) PMOXR activity was measured by the bathophenanthroline-disulphonic acid assay and is expressed as nmol ferrocyanide/mg of protein. Results shown are means±S.E.M. from three separate experiments performed in triplicate.
The PMOXR system is a multienzyme complex with at least two distinct activities, NADH-ferricyanide reductase and NADH-oxidase that has been shown to compensate for the defective ETC [25]. Indeed, we observed a marked increase in the NADH-ferricyanide reductase activity in the U87MGρ° cells when compared with the parental U87MGρ+ cells (95±5.9 versus 68±4.7, P=0.013, unpaired t test, n=2 experiments analysed in triplicate). Similarly, the mutant cybrids also had higher PMOXR activity compared with control cybrids [P<0.01, Dunnett's multiple comparison test; Figure 6B, first bar (black)], consistent with their need to rely on the PMOXR system to maintain the redox status. Exposure of cybrids to GD alone (Figure 6B, dark grey bars refer to cells subjected to GD alone; and Figure 6B, light grey bars refer to cells treated with 0.5 mM SIN-1) or in combination with SIN-1 (Figure 6B, hatched bars refer to cells treated with 0.5 mM SIN-1 under GD) triggered a marked increase in PMOXR activity, which was more pronounced in the control cybrid (P<0.001, Bonferroni's multiple comparison test) and correlated with the higher ATP content (Figure 6A) and better cell survival (Figure 4). Interestingly, mutant cybrids failed to up-regulate their PMOXR activity to the same extent as control R15 (Figure 6B) and correlated with lower ATP content (Figure 6A) and poor cell survival (Figure 4). Treatment of the cybrids with SIN-1 alone did not alter the PMOXR activity, consistent with its lack of cytotoxic effect in these cultures (Figure 6B, light grey bars).
Mechanism of cell death
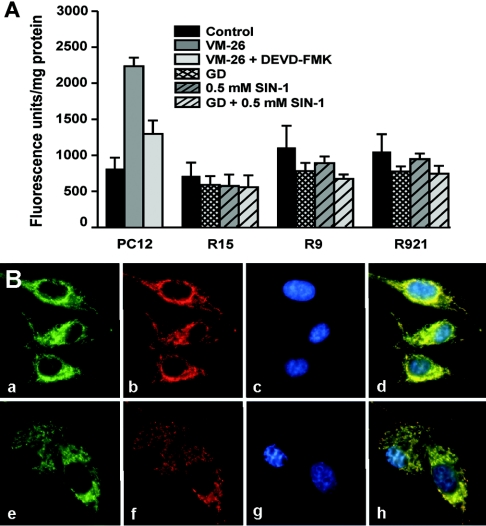
The morphological assessment of GD+SIN-1-treated cultures (Figure 4B) and their further analysis by TUNEL staining or pulsed-field gel electrophoresis (results not shown) did not reveal clear features of apoptotic cell death. Moreover, none of the treatments resulted in the activation of caspase-3, suggesting that the cells were dying by a caspase-independent pathway (Figure 7A). Recently, AIF has been identified as a novel apoptotic-effector protein, which translocates from the mitochondria to the nucleus and induces caspase-independent chromatin condensation [26]. We have examined the behaviour of AIF in cybrids after their treatment with GD+SIN-1 (Figure 7B). The cybrids were double-labelled with anti-AIF (green staining) and anti-hsp60 antibody (red staining), and viewed under a fluorescence microscope. In untreated cybrids, AIF was found in the mitochondria and it clearly co-localized with hsp60 (Figure 7B, panels a–d). A change in cellular morphology evidently occurred after the GD+SIN-1 treatment; however, the AIF signal (green staining) still co-localized with the mitochondria (red staining) and did not translocate to the nucleus (Figure 7B, panels e–h).
Figure 7. Measurements of caspase-3 activity and immunolocalization of AIF.
(A) Cells were grown in T-25 cm2 flasks for 3 days and subjected to GD in the presence or absence of SIN-1. After treatments, cells were collected and analysed for caspase-3 activity using the Clontech kit as described in the Materials and methods section. Caspase activity is expressed as fluorescence units/mg of protein. Results shown are means±S.E.M. from three separate experiments performed in triplicate. Rat pheochromocytoma PC12 cells were treated with 10 μM topoisomerase II inhibitor, VM-26 (teniposide) for 24 h and the cell extract was used as a positive control for caspase-3 activation. (B) R921 cells were grown on glass coverslips and treated with GD+SIN-1. After treatments, cells were processed for double immunofluorescence staining as described in the Materials and methods section. Untreated (a) or treated (b) cells were immunostained using anti-AIF antibody (a, e) and hsp60 antibody (b, f). Nuclei were stained using Hoechst 33258 (c, g). Merged images of AIF and hsp60 showing their co-localization in the mitochondria (d, h) and lack of translocation of AIF to the nucleus. Magnification ×400. Images shown are representative of at least two separate experiments.
Ultrastructural features of cell death
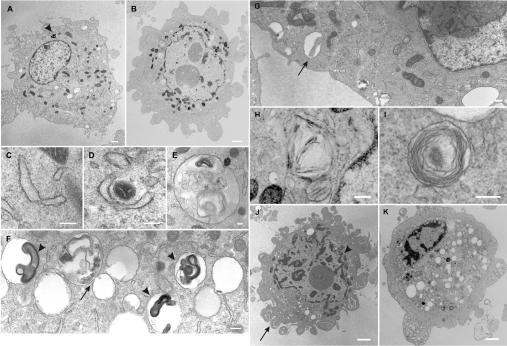
Electron micrographs of untreated mutant cybrids showed normal morphology of all organelles (Figure 8A), with mitochondria scattered homogenously throughout the cell. Ultrastructural analysis of GD+SIN-1-treated cells revealed, at the initiation of cell death, numerous smooth endoplasmic reticulum-like structures, which formed around portions of the cytosol as well as engulfed organelles such as mitochondria (Figures 8C and 8D). These double-membrane structures then resulted in the formation of autophagosomes (Figures 8E–8G) that contained remains of the cytoplasmic material and mitochondria (Figure 8G), which finally fused with the primary lysosomes leading to the formation of single-membrane autolysosomes that contained many myelin figures (Figure 8F). In some cells, the autolysosomes were found close to the plasma membrane where they might be involved in exocytosis (Figure 8G). Consistent with this, myelin figures and remnants of mitochondria were found in the extracellular space of treated cells [27]. At the plasma membrane, increased blebbing was seen in many cells (Figure 8B), chunks of the cytosol were sequestered and extruded from the cell (Figure 8J) resulting in shrinkage. Clumping of the nuclear chromatin was seen only along the inner surface of the nuclear envelope (Figure 8B). Several membranous structures, which appeared crystalline (Figure 8H) or as concentric whorls, were found scattered throughout the cytoplasm (Figure 8I). There was a marked decrease in the number of mitochondria, as they reorganized around the perinuclear area and clustered in groups (Figures 8B and 8J). During the final stages of cell death, nuclear chromatin condensed into small irregular masses of chromatin (Figures 8J and 8K). Cell death was accompanied by loss of the nuclear envelope and extensive vacuolization of the cytoplasm; annulate lamellae, which might represent pores in the nuclear envelope, were seen floating free in the cytoplasm (results not shown). At this stage, most of the mitochondria and other organelles largely disappeared from the cells.
Figure 8. Ultrastructural features of autophagic cell death.
R921 cells were treated with 0.5 mM SIN-1 in GD medium for 24 h, fixed and prepared for electron microscopy as described in the Materials and methods section. (A) Untreated control cells showing normal distribution of organelles. Arrowhead shows an autolysosome with a highly electron-dense myelin figure. (B) Features of early autophagic cell death in cells treated with GD+SIN-1. (C, D) Cup-shaped membranous structures in the cytoplasm or occasionally wrapping mitochondria. (E) An autophagosome, containing organelles undergoing degenerative changes and electron-dense myelin figures. (F) An autophagosome fusing with a lysosome (arrow) to form a single membrane vacuole containing phagocytosed cell fragments and myelin figures (arrowheads). (G) Autolysosomes found close to the plasma membrane. Arrow shows a mitochondrion in an autolysosome. (H) Crystalline structures. (I) A vacuole containing membrane whorls. (J) Features of late autophagic cell death showing membrane blebbing and increased number of vacuoles. Arrow shows sequestration of the cytoplasm. The remaining mitochondria seem to organize in the perinuclear area. Arrowhead shows mitochondria in small clusters. Nuclear chromatin is condensed and some detached from the nuclear envelope. (K) Features of very late autophagic cell death. The cell appears highly vacuolated and shows disappearance of all organelles. Nuclear chromatin appears condensed. Scale bars: (A, B), 5 μm; (C, D), 1 μm; (E, F), 1 μm; (G), 1 μm; (H, J and K), 3.3 μm; and (I), 0.83 μm.
DISCUSSION
Mitochondrial diseases are frequently caused by pathogenic mutations (i.e. point mutations and/or rearrangements) in the mtDNA and are associated with a broad spectrum of clinical manifestations. However, despite numerous experimental studies, it is still not clear why only selective cell populations in tissues with high-energy demands, such as the neuromuscular and central nervous system are affected. Here, we studied the vulnerability of MELAS cybrids harbouring A3243G mutation in the mtDNA to exogenous nitrosative and/or nitrative stress and the mechanisms of cell death. The cybrids were made on a nuclear background of glia cells and carried the wt as well as different proportions of defective mtDNA. Indeed, due to impaired mitochondrial protein synthesis and, consequently, defective ETC, the mutant cybrids had a lower Δψm, decreased COX activity and only half of the normal cellular ATP level. A similar decrease in Δψm was also observed in fibroblasts obtained from a MELAS patient [28]. However, despite the defective ETC, the basal ROS levels in the mutant cybrids were similar to those in the wt controls, suggesting that the antioxidant defence system present in these cybrids was sufficient to detoxify the ETC-generated ROS. In general, cells are capable of modulating, to some extent, the activities of antioxidant enzymes in order to adapt to the oxidative stress burden. Accordingly, the activities of superoxide dismutase and catalase were found to be up-regulated in myoblasts obtained from the MELAS patients [6]; similarly we have also found catalase levels to be up-regulated in the mutant cybrids (J. K. Sandhu, A. S. Byrd, C. Sodja, K. McRae, Y. Li, Y. H. Wei, B. Lach, F. Lee and M. Sikorska, unpublished work).
There has been increasing interest in understanding the role of both endogenously and exogenously generated RNOS in the progression of mitochondrial diseases [8,29]. We examined the effect of three different nitric oxide donors (i.e. SIN-1, DETA-NO and SNP) on the viability of cybrids. The amount and duration of NO• released by these drugs is related to their structure and pharmacological properties. DETA-NO is known to generate NO• spontaneously without prior bioactivation [30]. SIN-1 has to be metabolized by enzymatic hydrolysis to release both NO• and O2•− to form ONOO−, which has been generally considered the cytotoxic species. SNP is a complex comprising the Fe2+ ion, five cyanide anions (CN−) and a nitrosonium ion (NO+) and it also requires bioactivation for the liberation of NO• [31]. This is accompanied by the release of CN− and Fe2+ ions, which are known to poison the mitochondrial ETC and to participate in the Fenton reaction to generate more toxic hydroxyl radicals [32]. It has also been reported that SNP treatments cause not only ATP depletion [33] but also inhibition of GSH synthesis [34]. Indeed, in our experiments, only SNP by itself was cytotoxic to these MELAS cybrids; neither SIN-1 nor DETA-NO alone had any adverse effect suggesting that the cytotoxicity of CN− could not be entirely disregarded. Despite the fact that ONOO− and RNOS were produced by the decomposition of SIN-1 resulting in protein nitration, no inhibition of respiration was evident in our experiments. Similarly, no effects of SIN-1 on mitochondrial respiration were reported in HEK-293 (human embryonic kidney 293) cells [35]. Clearly, ONOO− or related species affected neither the ETC nor the intracellular ATP levels and hence were insufficient to trigger cell death. Lack of ONOO− toxicity has been observed in other cell types as well [32,36], suggesting that protein nitration might be a reversible and regulated process. However, we have observed SIN-1 toxicity under GD conditions (Figure 3) indicating the importance of glycolytic pathway in cell survival. The same effects of GD were reported in bovine endothelial cells treated with chloramphenicol or a nitric oxide donor and in human Jurkat cells treated with iodoacetate [37,38].
Taken together, our results clearly demonstrated that drugs affecting energy metabolism were cytotoxic to the cybrids, further strengthening the link between the energy buffers and cell death signals. Accordingly, we observed that cybrids with wt mtDNA were capable of regulating their energy stores and were more resistant to killing by GD+SIN-1 when compared with the MELAS cybrids that could not increase the cellular energy content above the basal level and were more sensitive (Figure 6A). These observations suggested that intracellular pathways involved in energy metabolism, especially oxidative phosphorylation, might play a significant role in defining a threshold of cellular resistance to RNOS. Clearly, the inability of MELAS cybrids to mobilize energy-synthesizing pathways under stress can be attributed to the defective ETC. Upon glucose depletion, cells had to switch to oxidative phosphorylation for ATP synthesis in order to compensate for its diminished supply from the glycolytic pathway. Indeed, it has been shown that many terminally differentiated cells maintain the ability to adjust their oxidative phosphorylation capacity to changing situations [39]. Also, in vitro cells could use glutamine from the medium as an alternative carbon source for oxidative phosphorylation [40].
MELAS cybrids have the capability to compensate for the decrease in ATP levels by up-regulating the PMOXR system and hence maintain the NADH/NAD+ ratio. Others have also suggested that, indeed, the PMOXR acts as a backup mechanism to maintain the redox status of the cell when the mitochondrial ETC is impaired [41,42]. Although PMOXR is not directly coupled with ATP synthesis, it reoxidizes excess of cytosolic NADH to NAD+, thereby maintaining an appropriate NAD+/NADH ratio essential for cell viability [41]. However, although the basal activity of the PMOXR system of the MELAS cybrids was significantly higher than that in the wt controls (Figure 6), the mutants were unable to derive enough redox capacity from the PMOXR system and hence succumbed to the GD+SIN-1-triggered authophagic cell death. In experiments where U87MGρ° cells were grown in the presence of 10 μg/ml coenzyme Q10 for 7 days, PMOXR activity could be further up-regulated (M. Sikorska, C. Sodja, K. McRae, Y. Li, Y. H. Wei, B. Lach, F. Lee and J. K. Sandhu, unpublished work), suggesting that supplementation with coenzyme Q10 may be useful therapeutically in patients harbouring mutant mtDNA. The exact mechanism linking the PMOXR system to the increased cell survival remains unknown. However, it is plausible that up-regulation of PMOXR activity would result in increased reoxidation of NADH to NAD+, which, in turn, could be utilized by NAD-linked dehydrogenases localized in the mitochondrial matrix. They would then reduce NAD+ to NADH and provide the flow of electrons into the respiratory chain required for the formation of ATP. Our results are consistent with this sequence of events. Under GD+SIN-1-induced stress, PMOXR activity was clearly up-regulated in cybrids with wt mtDNA (Figure 6B), which correlated with higher ATP content and better cell survival. On the other hand, in the MELAS cybrids, the PMOXR activation and ATP increases were much less pronounced and consequently cell survival was poorer. The MELAS mutation affects mitochondrial protein synthesis and impairs ETC complexes containing the mitochondrially encoded subunits, especially complex I. This, in turn, must affect the shuttle systems, which deliver the reducing equivalents from the cytosolic NADH to complex I, hence in the mutant cybrids the PMOXR activation could not be fully utilized for ATP production as required to support cell survival.
In summary, our results showed that cellular ability to utilize different metabolic pathways in support of energy production is critical for survival under stress, and if compromised, the cybrids activated the cell death programme and died by autophagy; a phenomenon whereby cells can digest themselves from within. Autophagic cell death has been documented in several cell types [43], including fibroblasts from MELAS patients harbouring bioenergetically incompetent mitochondria [44]. The reason why the cybrids treated with GD+SIN-1 opted to die by autophagy rather than apoptosis is not clear. One possibility could be that activation of PMOXR might also contribute to the up-regulation of DNA repair mechanism via NAD+-linked repair pathways and, thereby, preventing the initiation of DNA fragmentation [45]. Consistent with this, cybrids subjected to GD+SIN-1 showed numerous autophagic vacuoles and lysosomes (Figure 8), but had TUNEL-negative nuclei.
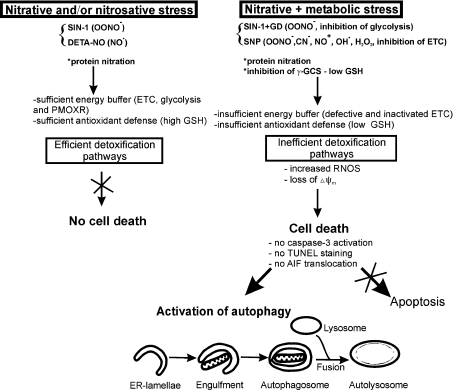
A model depicting some of the pathways, which could be inactivated due to energy crisis, but which, otherwise might modulate the cellular sensitivity to RNOS is presented in Scheme 1. The model predicts that as long as the ATP-synthesizing capability (ETC, glycolysis and PMOXR) and the antioxidant defence systems (i.e. GSH) are sufficient, the cells can detoxify RNOS and/or their targets (i.e. nitrated proteins) and survive under the stress conditions. However, when the energy-buffering capacities of the cells are diminished by metabolic injury (i.e. inhibition of glycolysis) and/or inactivation of ETC (i.e. MELAS mutation), the antioxidant defence also fails and the cells unable to detoxify RNOS succumb to cell death.
Scheme 1. A model depicting some of the cellular events contributing to the activation of cell death pathway in cybrid cells.
Acknowledgments
This research was supported by a joint grant from the National Research Council Canada and National Science Council of the Republic of China to M.S. and Y.-H. Wei. We are very thankful to J. LeBlanc for the help with flow cytometry and M. Boivin (Department of Pathology and Laboratory Medicine, The Ottawa Hospital-Civic Campus) for technical assistance with the electron microscopy.
References
- 1.Pavlakis S. G., Phillips P. C., DiMauro S., De Vivo D. C., Rowland L. P. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann. Neurol. 1984;16:481–488. doi: 10.1002/ana.410160409. [DOI] [PubMed] [Google Scholar]
- 2.DiMauro S., Moraes C. T. Mitochondrial encephalomyopathies. Arch. Neurol. 1993;50:1197–1208. doi: 10.1001/archneur.1993.00540110075008. [DOI] [PubMed] [Google Scholar]
- 3.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu) (UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature (London) 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 4.Goto Y., Nonaka I., Horai S. A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) Biochim. Biophys. Acta. 1991;1097:238–240. doi: 10.1016/0925-4439(91)90042-8. [DOI] [PubMed] [Google Scholar]
- 5.Ciafaloni E., Ricci E., Shanske S., Moraes C. T., Silvestri G., Hirano M., Simonetti S., Angelini C., Donati M. A., Garcia C. MELAS: clinical features, biochemistry, and molecular genetics. Ann. Neurol. 1992;31:391–398. doi: 10.1002/ana.410310408. [DOI] [PubMed] [Google Scholar]
- 6.Rusanen H., Majamaa K., Hassinen I. E. Increased activities of antioxidant enzymes and decreased ATP concentration in cultured myoblasts with the 3243A→G mutation in mitochondrial DNA. Biochim. Biophys. Acta. 2000;1500:10–16. doi: 10.1016/s0925-4439(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 7.Kirkinezos I. G., Moraes C. T. Reactive oxygen species and mitochondrial diseases. Semin. Cell Dev. Biol. 2001;12:449–457. doi: 10.1006/scdb.2001.0282. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie M., Liolitsa D., Hanna M. G. Mitochondrial disease: mutations and mechanisms. Neurochem. Res. 2004;29:589–600. doi: 10.1023/b:nere.0000014829.42364.dd. [DOI] [PubMed] [Google Scholar]
- 9.Elfering S. L., Sarkela T. M., Giulivi C. Biochemistry of mitochondrial nitric-oxide synthase. J. Biol. Chem. 2002;277:38079–38086. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- 10.Ghafourifar P., Bringold U., Klein S. D., Richter C. Mitochondrial nitric oxide synthase, oxidative stress and apoptosis. Biol. Signals Recept. 2001;10:57–65. doi: 10.1159/000046875. [DOI] [PubMed] [Google Scholar]
- 11.Radi R., Cassina A., Hodara R., Quijano C., Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radical Biol. Med. 2002;33:1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 12.Lacza Z., Snipes J. A., Zhang J., Horvath E. M., Figueroa J. P., Szabo C., Busija D. W. Mitochondrial nitric oxide synthase is not eNOS, nNOS or iNOS. Free Radical Biol. Med. 2003;35:1217–1228. doi: 10.1016/s0891-5849(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 13.Brookes P. S. Mitochondrial nitric oxide synthase. Mitochondrion. 2004;3:187–204. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Tarnopolsky M. A., Simon D. K., Roy B. D., Chorneyko K., Lowther S. A., Johns D. R., Sandhu J. K., Li Y., Sikorska M. Attenuation of free radical production and paracrystalline inclusions by creatine supplementation in a patient with a novel cytochrome b mutation. Muscle Nerve. 2004;29:537–547. doi: 10.1002/mus.20020. [DOI] [PubMed] [Google Scholar]
- 15.Macmillan C., Lach B., Shoubridge E. A. Variable distribution of mutant mitochondrial DNAs (tRNA(Leu[3243])) in tissues of symptomatic relatives with MELAS: the role of mitotic segregation. Neurology. 1993;43:1586–1590. doi: 10.1212/wnl.43.8.1586. [DOI] [PubMed] [Google Scholar]
- 16.King M. P., Attardi G. Mitochondria-mediated transformation of human rho-0 cells. Methods Enzymol. 1996;264:313–334. doi: 10.1016/s0076-6879(96)64030-0. [DOI] [PubMed] [Google Scholar]
- 17.King M. P., Koga Y., Davidson M., Schon E. A. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Land S. C., Porterfield D. M., Sanger R. H., Smith P. J. The self-referencing oxygen-selective microelectrode: detection of transmembrane oxygen flux from single cells. J. Exp. Biol. 1999;202:211–218. doi: 10.1242/jeb.202.2.211. [DOI] [PubMed] [Google Scholar]
- 19.Zigova T., Willing A. E., Saporta S., Daadi M. M., McGrogan M. P., Randall T. S., Freeman T. B., Sanchez-Ramos J., Sanberg P. R. Apoptosis in cultured hNT neurons. Brain Res. Dev. Brain Res. 2001;127:63–70. doi: 10.1016/s0165-3806(01)00107-9. [DOI] [PubMed] [Google Scholar]
- 20.Avron M., Shavit N. A sensitive and simple method for determination of ferrocyanide. Anal. Biochem. 1963;6:549–554. doi: 10.1016/0003-2697(63)90149-0. [DOI] [PubMed] [Google Scholar]
- 21.Hogg N., Darley-Usmar V. M., Wilson M. T., Moncada S. Production of hydroxyl radicals from the simultaneous generation of superoxide and nitric oxide. Biochem. J. 1992;281:419–424. doi: 10.1042/bj2810419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto T., Bing R. J. Nitric oxide donors. Proc. Soc. Exp. Biol. Med. 2000;225:200–206. doi: 10.1046/j.1525-1373.2000.22525.x. [DOI] [PubMed] [Google Scholar]
- 23.Harrison D. G., Bates J. N. The nitrovasodilators. New ideas about old drugs. Circulation. 1993;87:1461–1467. doi: 10.1161/01.cir.87.5.1461. [DOI] [PubMed] [Google Scholar]
- 24.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 25.Larm J. A., Vaillant F., Linnane A. W., Lawen A. Up-regulation of the plasma membrane oxidoreductase as a prerequisite for the viability of human Namalwa rho 0 cells. J. Biol. Chem. 1994;269:30097–30100. [PubMed] [Google Scholar]
- 26.Cande C., Cecconi F., Dessen P., Kroemer G. Apoptosis-inducing factor (AIF): key to the conserved caspase-independent pathways of cell death? J. Cell Sci. 2002;115:4727–4734. doi: 10.1242/jcs.00210. [DOI] [PubMed] [Google Scholar]
- 27.Ghadially F. N. Ultrastructural Pathology of the Cell and Matrix. Oxford: Butterworth-Heinemann; 1997. Mitochondria; pp. 195–342. [Google Scholar]
- 28.Williams A. J., Coakley J. C., Christodoulou J. Flow cytometric evaluation of defects of the mitochondrial respiratory chain. J. Child Neurol. 1999;14:518–523. doi: 10.1177/088307389901400807. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y. H., Lu C. Y., Wei C. Y., Ma Y. S., Lee H. C. Oxidative stress in human aging and mitochondrial disease-consequences of defective mitochondrial respiration and impaired antioxidant enzyme system. Chin. J. Physiol. 2001;44:1–11. [PubMed] [Google Scholar]
- 30.Keefer L. K. Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates. Annu. Rev. Pharmacol. Toxicol. 2003;43:585–607. doi: 10.1146/annurev.pharmtox.43.100901.135831. [DOI] [PubMed] [Google Scholar]
- 31.Bates J. N., Baker M. T., Guerra R., Jr, Harrison D. G. Nitric oxide generation from nitroprusside by vascular tissue. Evidence that reduction of the nitroprusside anion and cyanide loss are required. Biochem. Pharmacol. 1991;42:S157–S165. doi: 10.1016/0006-2952(91)90406-u. [DOI] [PubMed] [Google Scholar]
- 32.Fass U., Panickar K., Williams K., Nevels K., Personett D., McKinney M. The role of glutathione in nitric oxide donor toxicity to SN56 cholinergic neuron-like cells. Brain Res. 2004;1005:90–100. doi: 10.1016/j.brainres.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda M., Fujimori H., Panhou H. NO depletes cellular ATP contents via inactivation of glyceraldehyde-3-phosphate dehydrogenase in PC12 cells. J. Toxicol. Sci. 1998;23:389–394. doi: 10.2131/jts.23.5_389. [DOI] [PubMed] [Google Scholar]
- 34.Niknahad H., O'Brien P. J. Involvement of nitric oxide in nitroprusside-induced hepatocyte cytotoxicity. Biochem. Pharmacol. 1996;51:1031–1039. doi: 10.1016/0006-2952(96)85086-6. [DOI] [PubMed] [Google Scholar]
- 35.Thyagarajan B., Malli R., Schmidt K., Graier W. F., Groschner K. Nitric oxide inhibits capacitative Ca2+ entry by suppression of mitochondrial Ca2+ handling. Br. J. Pharmacol. 2002;137:821–830. doi: 10.1038/sj.bjp.0704949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moro M. A., Darley-Usmar V. M., Goodwin D. A., Read N. G., Zamora-Pino R., Feelisch M., Radomski M. W., Moncada S. Paradoxical fate and biological action of peroxynitrite on human platelets. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6702–6706. doi: 10.1073/pnas.91.14.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran A., Moellering D. R., Ceaser E., Shiva S., Xu J., Darley-Usmar V. Inhibition of mitochondrial protein synthesis results in increased endothelial cell susceptibility to nitric oxide-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6643–6648. doi: 10.1073/pnas.102019899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltran B., Mathur A., Duchen M. R., Erusalimsky J. D., Moncada S. The effect of nitric oxide on cell respiration: a key to understanding its role in cell survival or death. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14602–14607. doi: 10.1073/pnas.97.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiesner R. J. Adaptation of mitochondrial gene expression to changing cellular energy demands. News Physiol. Sci. 1997;12:178–184. [Google Scholar]
- 40.Weber K., Ridderskamp D., Alfert M., Hoyer S., Wiesner R. J. Cultivation in glucose-deprived medium stimulates mitochondrial biogenesis and oxidative metabolism in HepG2 hepatoma cells. Biol. Chem. 2002;383:283–290. doi: 10.1515/BC.2002.030. [DOI] [PubMed] [Google Scholar]
- 41.Lawen A., Martinus R. D., McMullen G. L., Nagley P., Vaillant F., Wolvetang E. J., Linnane A. W. The universality of bioenergetic disease: the role of mitochondrial mutation and the putative inter-relationship between mitochondria and plasma membrane NADH oxidoreductase. Mol. Aspects Med. 1994;15:S13–S27. doi: 10.1016/0098-2997(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 42.Baker M. A., Lawen A. Plasma membrane NADH-oxidoreductase system: a critical review of the structural and functional data. Antioxid. Redox Signal. 2000;2:197–212. doi: 10.1089/ars.2000.2.2-197. [DOI] [PubMed] [Google Scholar]
- 43.Larsen K. E., Sulzer D. Autophagy in neurons: a review. Histol. Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- 44.James A. M., Wei Y. H., Pang C. Y., Murphy M. P. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J. 1996;318:401–407. doi: 10.1042/bj3180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satoh M. S., Poirier G. G., Lindahl T. NAD(+)-dependent repair of damaged DNA by human cell extracts. J. Biol. Chem. 1993;268:5480–5487. [PubMed] [Google Scholar]