Abstract
A folate enzyme, FDH (10-formyltetrahydrofolate dehydrogenase; EC 1.5.1.6), is not a typical tumour suppressor, but it has two basic characteristics of one, i.e. it is down-regulated in tumours and its expression is selectively cytotoxic to cancer cells. We have recently shown that ectopic expression of FDH in A549 lung cancer cells induces G1 arrest and apoptosis that was accompanied by elevation of p53 and its downstream target, p21. It was not known, however, whether FDH-induced apoptosis is p53-dependent or not. In the present study, we report that FDH-induced suppressor effects are strictly p53-dependent in A549 cells. Both knockdown of p53 using an RNAi (RNA interference) approach and disabling of p53 function by dominant-negative inhibition with R175H mutant p53 prevented FDH-induced cytotoxicity in these cells. Ablation of the FDH-suppressor effect is associated with an inability to activate apoptosis in the absence of functional p53. We have also shown that FDH elevation results in p53 phosphorylation at Ser-6 and Ser-20 in the p53 transactivation domain, and Ser-392 in the C-terminal domain, but only Ser-6 is strictly required to mediate FDH effects. Also, translocation of p53 to the nuclei and expression of the pro-apoptotic protein PUMA (Bcl2 binding component 3) was observed after induction of FDH expression. Elevation of FDH in p53 functional HCT116 cells induced strong growth inhibition, while growth of p53-deficient HCT116 cells was unaffected. This implies that activation of p53-dependent pathways is a general downstream mechanism in response to induction of FDH expression in p53 functional cancer cells.
Keywords: apoptosis, folate, 10-formyltetrahydrofolate dehydrogenase, p53, RNA interference (RNAi), tumour suppressor
Abbreviations: Act D, actinomycin D; FDH, 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6); GFP, green fluorescent protein; Mdm2, murine double minute clone 2 oncoprotein; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; PI, propidium iodide; PUMA, Bcl2 binding component 3; RNAi, RNA interference; TS, thymidylate synthase
INTRODUCTION
Activation of the p53 tumour suppressor protein, through its elevation and post-translational modification in response to various types of cellular stress, induces either cell cycle arrest or apoptosis [1,2]. The p53 protein is usually present at low levels but it accumulates in genotoxic stress-induced cells, resulting in transcriptional activation of cell cycle checkpoint genes and/or apoptotic genes [3,4]. It can also be activated by a non-genotoxic stress [4]. Among the factors that activate p53 in the absence of DNA damage are: expression of oncoproteins [5], non-genotoxic drugs [6], ribonucleotide pool depletion [7] and hypoxia [8]. In more than 50% of tumours, the function of p53 is impaired by mutations of the p53 gene [9,10]. Moreover, even in cells with the wild-type p53 phenotype the p53 pathway can still be inactive [10]. Mechanisms that can cause the failure of p53 function include: an enhanced rate of p53 degradation through an Mdm2 (murine double minute clone 2 oncoprotein)-dependent pathway [11,12]; expression of proteins that interact with p53 and prevent its functioning by cytoplasmic sequestration [10]; and the absence of co-activators [13,14]. In addition, defects in pathways upstream or downstream of p53 might also interfere with the wild-type p53 function [10].
Folate deficiency is among the factors that can induce genomic instability and apoptosis, although the underlying mechanisms are largely unknown [15]. Folate coenzymes are crucial for cell integrity because of their involvement in nucleotide biosynthesis and DNA methylation [16]. Therefore, defects in folate metabolism can lead to apoptosis through multiple mechanisms, including: depletion of cellular nucleotide pools [7], misincorporation of uracil into DNA and accumulation of DNA abnormalities [17], and impairment of methylation processes [18]. Several studies demonstrated that apoptosis induced by folate deficiency appears to be p53 dependent [19], although it is not clear at present which of these mechanisms is more likely to trigger a p53-dependent apoptotic cascade in response to folate-related stress. Folates also attract attention because their analogues are potent anticancer drugs [20]. Inhibitory effects of anti-folates on molecular and cellular levels were intensively investigated [20–22], including studies of the role of p53 in these effects [23,24]. These studies led to the conclusion that discrimination between p53-related and p53-unrelated responses of these drugs depends upon the phenotypic/genotypic background [23,24], apparently reflecting different requirements for folate metabolism in different cell types.
It has been demonstrated that a folate-dependent enzyme, TS (thymidylate synthase), directly binds p53 mRNA, preventing its translation and down-regulating p53 levels [25]. Binding of TS to its physiological folate substrate, 5,10-methylenetetrahydrofolate, dramatically decreases the RNA binding activity [26], which should prevent TS-mediated suppression of p53 mRNA translation. This finding suggests an additional mechanism, through regulation of TS function, by which folates could affect p53-dependent pathways. Another folate enzyme, FDH (10-formyltetrahydrofolate dehydrogenase; EC 1.5.1.6), might also be a part of p53-regulating network [27]. We have demonstrated that FDH, which has long being viewed as an ordinary metabolic enzyme [28], possesses properties of a tumour suppressor: it is strongly and ubiquitously down-regulated in tumours, and moderate FDH expression, at below physiological levels, is selectively cytotoxic to FDH-deficient cancer cells [29]. We have also shown that expression of FDH in FDH-deficient human lung carcinoma cell line A549 with wild-type p53 induces G1 arrest and apoptosis that were accompanied by elevation of p53 and its downstream target, p21 [27]. It was not clear, however, whether FDH-induced apoptosis is p53-dependent or not [27]. In the present study, we report that FDH-induced suppressor effects are strictly p53-dependent in A549 cells. We also show that activation of p53 after FDH expression occurs through phosphorylation of Ser-6, Ser-20 and Ser-392 and is followed by expression of a pro-apoptotic protein PUMA (Bcl2 binding component 3).
MATERIALS AND METHODS
Reagents
Reagents and buffers were purchased from Sigma unless otherwise specified. Antibodies against Bax, Bcl2 and BclXL were a gift from Dr Yi-Te Hsu (Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC, U.S.A.).
Cell culture
Cell media and reagents were purchased from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were purchased from Clontech. Tet-On A549/FDH stable cell line for inducible FDH expression was generated in our previous studies [27]. HCT116 p53+/+ and p53−/− cells [a gift from Dr Bert Vogelstein (The Howard Hughes Medical Institute & The Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins Medical Institutions, Baltimore, MD, U.S.A.)] were grown in McCoy 5A medium supplemented with 10% (v/v) fetal bovine serum. Cell viability was assessed by Trypan Blue exclusion or by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] cell proliferation assay as we described previously [27]. MTT cell proliferation assay was performed using a CellTiter 96® kit (Promega) according to the manufacturer's directions.
Transient transfection
Tet-On A549/FDH cells were transfected with pCEP4-175 plasmid carrying cDNA for an R175H p53 mutant [a gift from Dr Jennifer Pietenpol (Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN, U.S.A.)] using FuGENE6 reagent (Roche Molecular Biochemicals). Efficiency of transfection was estimated by co-transfection with pEGFP-N3 vector (Clontech) expressing GFP (green fluorescent protein). Transient transfection of HCT116 cells with pcDNA3.1 bearing FDH cDNA was performed essentially as we described previously [29]. In experiments with H1299 cells, the pCEP4 vector bearing the wild-type/mutant p53 cDNA was either co-transfected into the cells with pcDNA3.1 (‘empty’ vector), or co-transfected with pcDNA 3.1 bearing FDH cDNA. Experiments were performed using Lipofectamine 2000. In control experiments co-transfection of corresponding pCEP4 vector with pEGFP-N3 vector was performed to ensure similar transfection efficiency between experiments. Transfection efficiency in these experiments was between 40 and 50%. For each mutant p53 and for the wild-type p53, experiments were performed four times to minimize the influence of deviation of transfection efficiency.
Generation of stable cell line with knocked-down p53 gene using an RNAi (RNA interference) approach
The pSUPER-p53 RNAi System™ (Oligoengine) was used to generate a stable Tet-On A549/FDH cell line with knocked-down p53 gene by an RNAi approach. Tet-On A549/FDH cells at about 80% of confluence were co-transfected with two vectors, pSUPER-p53™ (Oligoengine) and pPUR (BD Dickinson), in the ratio 3:1 using the Superfect® Transfection reagent (Qiagen). After 48 h, cells were passaged at a 1:10 ratio into selection medium with 0.5 μg/ml puromycin. Puromycin-resistant clones were characterized for levels of p53 mRNA by reverse transcriptase-PCR. Total RNA was purified with RNA Easy® Protect Mini Kit (Qiagen). Reverse transcriptase reaction was performed with oligo(dT)18 primer using the BD SMART™ cDNA Synthesis Kit (Clontech). PCR was carried out with p53 sequence-specific primers, 5′-TGGATTGGCAGCCAGACTGCCTTCC-3′ (sense) and 5′-GTGGGAGGCTGTCAGTGGGGAACAA-3′ (antisense), using Pfu polymerase (Stratagene). Samples were normalized for the levels of ribosomal protein S9.
Immunoblot assays
Lysates from approx. 1×106 cells were subjected to SDS/PAGE followed by immunoblotting with the corresponding antibodies. FDH was detected using FDH-specific polyclonal antiserum (1:2000 dilution); p53 and actin were detected using monoclonal antibodies from Calbiochem (0.1 μg/ml and 1:5000 dilution, respectively). Polyclonal antibodies against phosphorylated p53 were from AnaSpec, (Ser-6; 1.5 μg/ml), Cell Signaling (Ser-9, Ser-15, Ser-37, Ser-46 and Ser-392; 1:500) and Santa Cruz Biotechnology (Ser-20; 1:200). In all experiments a Hybond™-ECL® nitrocellulose membrane and an ECL® detection kit (both from Amersham Pharmacia Biotech) were used according to the manufacturer's protocol. In each case membranes were washed and re-probed for the levels of actin.
Assay of apoptosis
Apoptotic cells were detected by annexin V labelling or by assaying caspase-9/6. Cells were labelled with annexin V and PI (propidium iodide) using an Annexin-V-FLUOS Staining Kit (Roche Molecular Biochemicals). Experiments were performed according to the manufacturer's protocol. All cells (attached and floating) were used in these experiments. Annexin V and PI staining was detected by cell flow cytometry. Activity of caspase-9/6 was assayed using ApoAlert™ Caspase Assay Kit (Becton-Dickinson).
Immunofluorescence
Cells grown in Lab-Tek™ II Chamber Slide System (Nalge Nunc International) were treated with doxycycline for the indicated periods of time. Slides were fixed for 5 min with absolute methanol at −10 °C, air-dried and incubated for 30 min in PBS containing 10% (v/v) goat serum. Incubation with primary anti-p53 antibodies (Calbiochem, 2.5 μg/ml) was performed for 1 h followed by incubation with secondary goat anti-mouse antibodies labelled with Alexa Fluor® 488 (Molecular Probes, 5 μg/ml) in a dark chamber for 45 min and counterstaining with TO-PRO®-3 iodide (Molecular Probes). After each step, slides were washed with PBS. Slides were mounted with VECTASHIELD® HardSet™ Mounting Medium (Vector Laboratories). Fluorescent images were captured and processed using a Leica TCS SP2 AOBS scanning confocal microscope.
Site-directed mutagenesis
Mutants of p53 were generated by site-directed mutagenesis using QuikChange kit (Stratagene). The mutated plasmid was sequenced entirely to confirm introduction of a mutation and to ensure the absence of non-specific nucleotide changes.
Nucleotide assays
ATP levels were measured using an ATP Bioluminescence Assay Kit CLSII (Roche) according to the manufacturer's directions. Approximately 105 cells were used in this assay. Luminescence was measured using a Sirius Luminometer (Berthold Detection System). Each experiment was performed in triplicate. GTP, CTP and UTP levels were measured simultaneously by HPLC using a Symmetry C18 column (4.6×250 mm, 5-μm-diam. particles; Waters) and a Waters 515 dual-pump solvent delivery system according to a published procedure [29a]. The elution solvents were (A) 100 mM H3PO4, pH 6.2 (triethylamine), and (B) 100 mM H3PO4, 5 mM MgSO4, pH 6.2 (triethylamine), with a linear gradient of 0–100% solvent B. Peaks were detected at 254 nm using a Waters 2487 dual-wavelength absorbance detector. Calibration of the assay was performed with NTPs obtained from Sigma. A 20 μl aliquot of cell extract prepared according to [29b] (representing approx. 3×106 cells) was used in a chromatography run. Nucleotide concentrations were quantified by peak area against corresponding standards using the Millenium software (Waters). Assays were performed in duplicate. Levels of dNTP were measured by HPLC according to a published procedure [29c].
RESULTS
Effects of FDH expression on A549 cells transiently transfected with R175H mutant p53
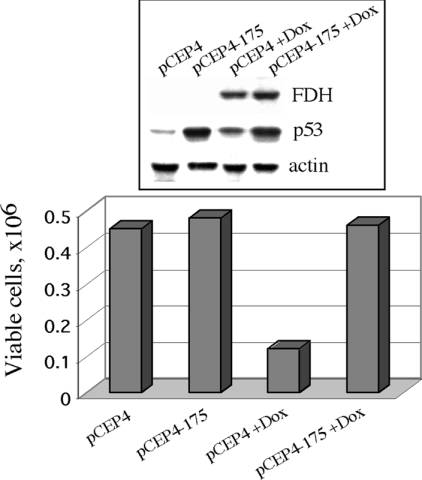
To suppress p53 function we transfected Tet-On A549/FDH cells with a pCEP4-175 vector bearing mutant p53 cDNA. In this mutant p53, Arg-175 is replaced with histidine. This mutant lacks the ability to interact with target DNA sequences and is inactive in induction of both cell cycle arrest and apoptosis [30]. Moreover, expression of this mutant inhibits wild-type p53 function in a dominant-negative manner [31]. We observed significant elevation of p53 protein levels in the cells transfected with pCEP4-175 vector compared to the cells mock-transfected with the ‘empty’ pCEP4 vector (Figure 1, upper panel) that was indicative of mutant p53 expression. Transfection efficiency in these experiments was approx. 40% as estimated from co-transfection with vector expressing GFP. Induction of FDH by doxycycline revealed a four times greater number of viable cells in a population expressing p53 mutant than in the control cells with only wild-type p53 (Figure 1). These experiments suggest that expression of the R175H p53 mutant inhibits the suppressor activity of FDH.
Figure 1. Influence of transient expression of the R175H p53 mutant on FDH growth suppressor effects.
Tet-On A549/FDH cells were transfected with ‘empty’ vector with (pCEP4+Dox) and without (pCEP4) FDH induction; Tet-On A549/FDH cells were transfected with vector expressing the R175H p53 mutant with (pCEP4-175+Dox) and without (pCEP4-175) FDH induction. FDH expression was induced by 2.5 μg/ml doxycycline (Dox). Viable cells were counted 48 h post-transfection using Trypan Blue exclusion. The upper panel shows levels of FDH and p53 assessed by immunoblot techniques. Levels of actin were used as a loading control.
Effects of FDH expression on A549 cells with knocked down p53
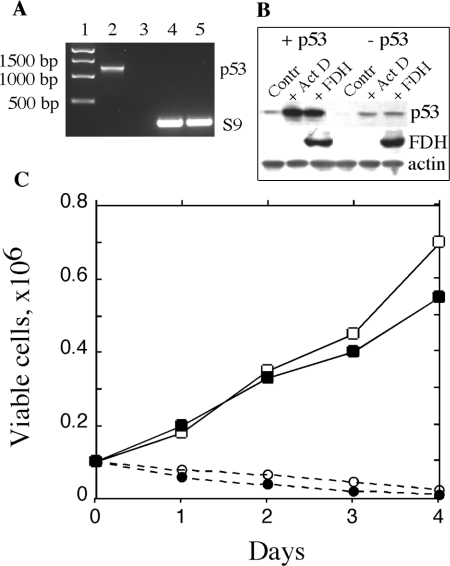
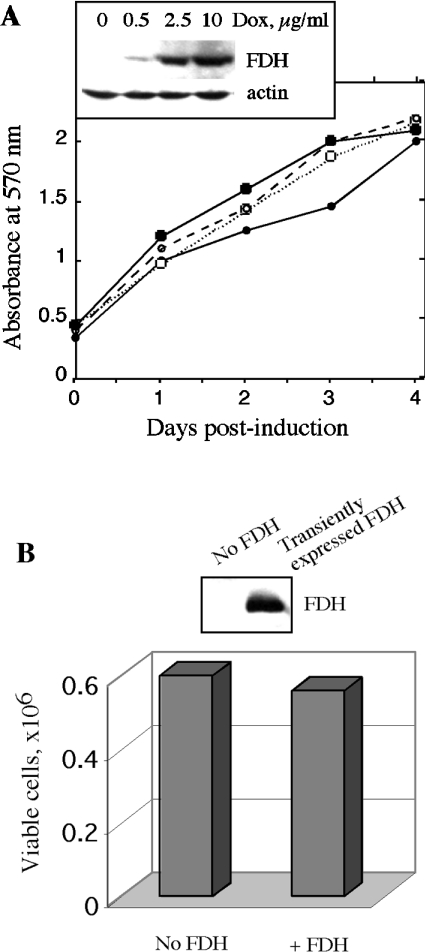
We generated Tet-On A549/FDH cells with knocked down p53 using an RNAi approach [32] with the pSUPER-p53 RNAi System™. We were able to select stable clones that showed almost complete loss of p53 mRNA and p53 protein (results for one of these clones are shown in Figures 2A and 2B). To study the FDH impact on p53 knocked down cells, we induced expression of FDH with 2.5 μg/ml doxycycline in original Tet-On A549/FDH cells and in the cells with knocked down p53. We observed that the two cell lines expressed approximately equal levels of FDH after induction (Figure 2B). After FDH induction, cells with knocked down p53 demonstrated steady-state proliferation (Figure 2C). In contrast, original cells capable of p53 expression revealed complete inhibition of proliferation and strong cytotoxicity after FDH induction (Figure 2C). These results demonstrated that, compared with the p53-proficient cells, the p53-deficient cells were essentially not sensitive to the suppressor effect of FDH. Cells with knocked down p53 were resistant to even higher FDH levels achieved either with higher doxycycline concentrations (Figure 3A) or by transient transfection (Figure 3B). Transient expression in these experiments allows us to produce higher intracellular levels of FDH compared with inducible expression (compare Figures 3A and 3B). These levels, however, still did not suppress proliferation.
Figure 2. Effect of p53 knockdown on proliferation of A549 cells expressing FDH.
(A) Levels of p53 mRNA in Tet-On A549/FDH cells before (lane 2) and after (lane 3) p53 knockdown evaluated by reverse transcriptase-PCR. Samples were normalized by levels of ribosomal protein S9 mRNA (lanes 4 and 5, respectively). Lane 1, standards. (B) Levels of p53 and FDH in Tet-On A549/FDH cells, before (+p53) and after (−p53) p53 knockdown, assessed by immunoblot techniques. Contr, control (untreated cells); +Act D, 48 h after beginning of treatment with 50 nM Act D (treatment was carried out for 24 h); +FDH, after induction of FDH for 48 h. Samples were normalized by levels of actin. (C) Proliferation of p53-proficient Tet-On A549/FDH cells after induction of FDH (○) or treatment with 50 nM Act D (●), and of p53-deficient (knocked down p53) Tet-On A549/FDH cells after induction of FDH (□) or Act D treatment (■). Viable cells were assessed by Trypan Blue exclusion.
Figure 3. Effect of increased FDH levels on proliferation of cells with knocked down p53.
(A) Tet-On A549/FDH cells with knocked down p53 induced with different doxycycline concentrations (■, no doxycycline; □, 0.5 μg/ml; ○, 2.5 μg/ml; ●, 10 μg/ml). Viable cells were assessed by MTT assay. Inset shows levels of FDH 48 h post-induction. Actin is shown as a loading control. (B) Tet-On A549/FDH/pSUPER-p53 cells transiently transfected for FDH expression. Viable cells were assessed by Trypan Blue exclusion. FDH levels were detected 48 h post-transfection by immunoblot assays.
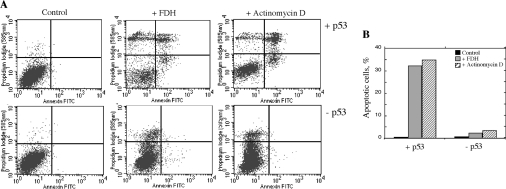
We further treated the p53 functional and p53 knocked down cells with Act D (actinomycin D) (50 nM) for 24 h. Act D is believed to specifically induce apoptosis through a p53-dependent pathway [33]. In the original Tet-On A549/FDH cells, Act D treatment resulted in p53 elevation and strong cytotoxicity (Figures 2B and 2C). On the other hand, in p53-knocked down cells, p53 elevation was significantly lower (Figure 2B) and these cells did not die (Figure 2C). Annexin V assays demonstrated that cells with knocked down p53 did not initiate apoptosis in response to FDH induction or Act D treatment, in contrast to p53-expressing cells (Figure 4A). At 48 h after FDH induction or at the beginning of Act D treatment, in A549 cells with functional p53 the content of apoptotic cells was 32% and 36% respectively compared with 0.5% in non-treated cells (Figure 4B). In contrast, in the cells with knocked down p53 the content of apoptotic cells was at background levels (2.3% and 3.4% respectively compared with 0.7% of non-treated cells, Figure 4B).
Figure 4. Knock-down of p53 prevents FDH-induced apoptosis in A549 cells.
(A) Apoptosis detected by FACS flow cytometry in Tet-On A549/FDH cells (top panels, +p53) and the same cells with knocked down p53 (bottom panels, −p53) after induction of FDH expression or treatment with Act D. Lower left (double negative), live cells; lower right (annexin V positive, PI negative), early apoptosis; upper right (double positive), late apoptosis; upper left (annexin V negative, PI positive), dead cells. Control, non-treated cells. (B) Percentages of apoptotic cells (sum of early and late apoptotic cells) calculated from (A).
Influence of transient FDH expression on HCT116 cells with functional and non-functional p53
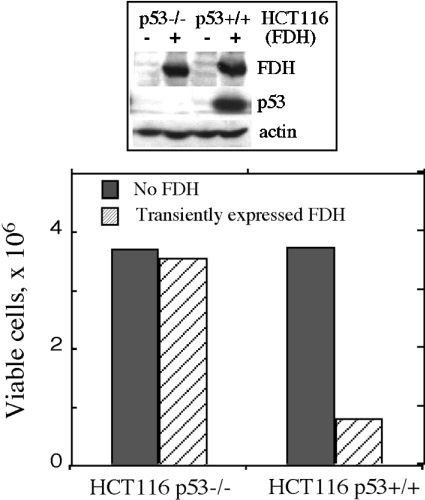
We have also compared effects of FDH expression in isogenic cell lines, HCT116 p53+/+ and p53−/− [34]. We observed that transient FDH expression resulted in accumulation of p53 in p53 functional cells (p53+/+) and caused strong growth inhibition (Figure 5). In contrast, transient expression of FDH in p53-deficient cells (p53−/−) did not inhibit proliferation (Figure 5).
Figure 5. Effect of transient FDH expression on proliferation of p53 functional (HCT116 p53+/+) and p53-null (HCT116 p53−/−) cells.
Viable cells were assessed by Trypan Blue exclusion 48 h post-transfection. Cells were transfected with pcDNA 3.1/FDH vector using Lipofectamine 2000 (hatched bars). Control cells (solid bars, no FDH) were mock-transfected with ‘empty’ vector (no FDH cDNA insert). Efficiency of transfection evaluated by co-transfection with a vector expressing GFP was approx. 35%. Addition of geneticin allowed elimination of non-transfected cells, increasing the accuracy of this approach [29]. Upper panel: FDH levels in control (−) and FDH-transfected (+) cells detected by immunoblot assays.
Intracellular distribution of p53 after FDH expression
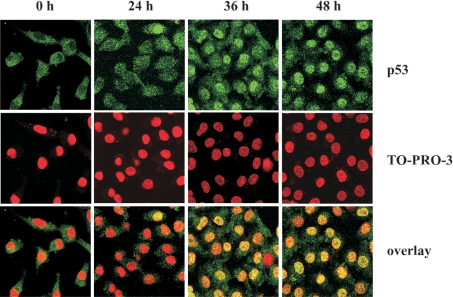
We studied the intracellular redistribution of p53 after induction of FDH expression in Tet-On A549/FDH cells. The p53 protein was monitored by staining with p53-specific first antibodies and fluorescent second antibodies. At 36–48 h post-induction, intense staining of nuclei was observed (Figure 6), suggesting translocation of p53 from the cytoplasm. Counterstaining with the chromatin-specific dye, TO-PRO®-3, confirmed the preferential nuclear localization of p53 at that time (Figure 6).
Figure 6. Accumulation of p53 in nuclei after induction of FDH expression in A549 cells.
Top row, p53 (green) was detected by immunostaining with monoclonal anti-p53 antibodies (clone DO-1, Calbiochem) and polyclonal anti-mouse IgG2a antibodies conjugated with Alexa Fluor 488 dye (Molecular Probes). Middle row, the nuclei (red) were imaged by chromosomal DNA staining with TO-PRO-3 iodide dye (Molecular Probes). Bottom row, overlay with yellow indicating co-localization. Images were obtained by confocal laser microscopy. Time (h) after induction of FDH expression is shown.
FDH elevation results in p53 phosphorylation and expression of the pro-apoptotic protein PUMA
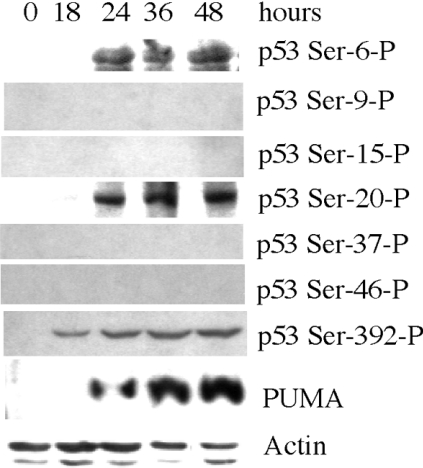
We examined whether elevation of p53 after FDH induction is also accompanied by phosphorylation. Phosphorylation of p53 occurs at multiple sites [35]. In our experiments, phosphorylation at Ser-6, Ser-9, Ser-15, Ser-20, Ser-37 and Ser-46 located in the transactivation domain of p53, and at Ser-392 located in the regulatory domain, were evaluated. FDH induction resulted in phosphorylation of p53 at Ser-6, Ser-20 and Ser-392, while other serines in the transactivation domain remained unphosphorylated (Figure 7). Phosphorylation of Ser-392 was observed as early as 18 h post-induction of FDH, while phosphorylation of serines in the transactivation domain was observed beginning 24 h after FDH induction. Our experiments further revealed expression of the pro-apoptotic protein PUMA, a critical mediator of p53-induced apoptosis [36], in response to FDH induction (Figure 7). We did not observe changes in the protein levels of the other members of the Bax family of proteins, pro-apoptotic Bax and Noxa and anti-apoptotic Bcl2 and BclXL (results not shown).
Figure 7. Phosphorylation of p53 and expression of the pro-apoptotic protein PUMA in A549 cells after FDH induction.
Phosphorylated p53 was detected with antibodies specific for a particular phosphorylated residue. Time (h) after FDH induction is shown. Levels of actin are shown as a loading control.
Mutation of Ser-6 of p53 protects cells from FDH-induced apoptosis
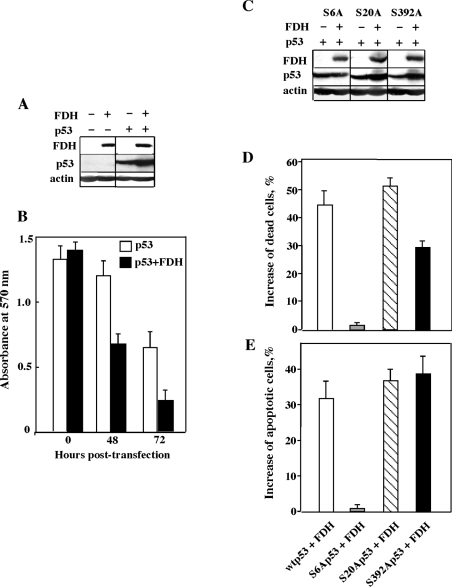
To study whether phosphorylation at multiple Ser residues of p53 is required for the suppressor effects of FDH we changed each of the serine residues that are phosphorylated in response to FDH induction to alanine. Corresponding mutants (S6A, S20A and S392A) were co-expressed with FDH in the H1299 lung carcinoma cell line, which is p53- and FDH-deficient. Expression of the wild-type p53 itself in these cells inhibited proliferation, but the inhibitory effect was enhanced by the presence of FDH (Figure 8B). Likewise, higher levels of p53 in FDH-expressing H1299 cells were observed (Figure 9A). These effects were not changed when FDH was co-expressed with S20A or S392A p53 mutants (Figures 9C–9E): FDH-associated p53 accumulation, enhanced cell death and induction of apoptosis were observed for these mutants. In contrast, mutation of Ser-6 of p53 to alanine prevented p53 accumulation in response to FDH expression and completely abolished p53-related suppressor effects of FDH (Figures 9C–9E).
Figure 8. Effects of transient FDH expression on H1299 cells expressing wild-type and mutant p53.
(A) Levels of FDH and wild-type p53 in H1299 cells after transient transfection. (B) MTT assay of cells transfected with p53 alone (open bars) or in combination with FDH (solid bars). (C) Levels of mutant p53 in H1299 cells in the presence and in the absence of FDH. (D) Changes in number of dead cells (assessed by MTT assays) after co-expression of wild-type (wt) or mutant p53 with FDH (shown as percentages of dead cells observed after expression of p53 alone). (E) Changes in number of apoptotic cells (assessed by caspase-9/6 assays) after co-expression of wild-type or mutant p53 with FDH (shown as percentages of apoptotic cells observed after expression of p53 alone). Each bar represents the mean±S.E.M. for four independent experiments performed in quadruplicate.
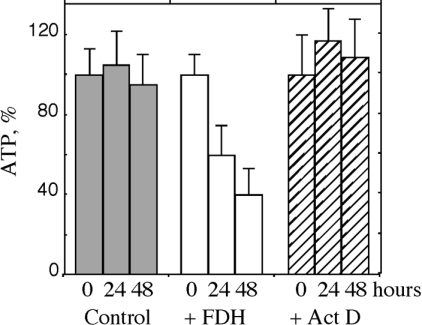
Figure 9. Levels of ATP in Tet-On A549/FDH cells.
Control, untreated cells; +FDH, cells induced with doxycycline for FDH expression; Act D, cells treated with 25 nM Act D. Time (h) after FDH induction or Act D treatment is shown. Levels of ATP are shown as percentages of levels at time 0. Each bar represents the mean±S.E.M. of two independent experiments performed in triplicate.
FDH expression decreases the intracellular ATP/GTP pools
We have hypothesized that FDH exerts its antiproliferative effects on cancer cells through depletion of intracellular purine nucleotide pools. One of the most important nucleotide pools is ATP. In addition to involvement in DNA/RNA biosynthesis and repair, it is required for numerous reactions to supply energy. We observed that expression of FDH resulted in a 2.5-fold decrease in intracellular ATP levels 48 h post-induction (Figure 9A). In contrast, treatment with Act D did not cause ATP depletion (Figure 9A), suggesting that the decrease in ATP levels is a specific characteristic of FDH-induced apoptosis. We also observed significant depletion of GTP after FDH expression, but not of CTP or UTP (Table 1). Overall, these results indicate that FDH elevation causes decreased levels of purine ribonucleoside triphosphates. In contrast, we did not detect decrease in the levels of dNTPs after FDH expression (results not shown).
Table 1. Concentrations of NTPs (mM) in p53-proficient A549 cells after induction of FDH expression.
ATP was measured using a Bioluminescence Assay Kit CLSII (Roche). GTP, UTP and CTP were measured by HPLC techniques as described in the Materials and methods section. The mean±S.E.M. for two independent experiments performed in triplicate is shown for ATP. The average of two measurements is shown for other NTPs.
| Concentration (mM) | ||||
|---|---|---|---|---|
| Time after FDH Induction (h) | ATP | GTP | UTP | CTP |
| 0 | 1.55±0.17 | 0.50 | 0.14 | 0.17 |
| 24 | 0.92±0.21 | 0.15 | 0.18 | 0.28 |
| 48 | 0.60±0.16 | 0.06 | 0.11 | 0.21 |
DISCUSSION
We have previously demonstrated that expression of FDH in p53-proficient A549 cells induces G1 arrest and apoptosis, and is accompanied by elevation of p53 and its downstream target, p21 [27]. In most cases p21 is a negative modulator of apoptosis [37]. Therefore, its up-regulation in FDH-induced A549 cells implies that apoptosis may occur through additional, p53-independent mechanisms. Indeed, it has been reported that p53-independent apoptosis takes place in A549 cells under conditions of p53 accumulation and G1 cell cycle arrest [38]. Our earlier finding that FDH suppresses p53-deficient cells [29] also suggests that p53 might not be required for FDH-induced cell death in these cells. Taken together, these results provide support for the possibility that more than one mechanism of cytotoxicity is operative in different cell lines in response to FDH expression. While the underlying basis for mechanisms of FDH cytotoxicity in p53-proficient and p53-deficient cells is not clear, in the present studies we have shown that functional p53 is required to mediate FDH-induced suppressor effects in A549 cells. Both knockdown of p53 using a RNAi approach [39] and disabling of p53 function by dominant-negative inhibition with mutant p53 [31] prevented FDH-induced cytotoxicity in these cells. The requirement for functional p53 for the suppressor effects of FDH was further confirmed in HCT116 cells, a common model for studies of p53-dependent responses [34]. The finding that elevation of FDH in p53 functional HCT116 cells induced strong growth inhibition, while growth of p53-deficient HCT116 cells was unaffected, implies that activation of p53-dependent pathways could be a general downstream mechanism in response to induction of FDH expression in p53 functional cancer cells. Apparently, ablation of FDH-induced suppressor effects in p53-deficient cells is associated, first of all, with the inability to activate apoptosis in the absence of functional p53.
Accumulation of p53 in response to cellular stress is associated with release from a complex with Mdm2, the protein involved in p53 ubiquitination and degradation [40,41]. This release and consequent stabilization of p53 and its activation as a transcription factor can occur through phosphorylation at several residues within the N-terminal transactivation domain [40,41]. Moreover, phosphorylation at specific residues can cause p53 to initiate transcription of cell cycle checkpoint proteins or apoptotic proteins [1]. FDH-mediated p53 accumulation was accompanied by phosphorylation at Ser-6 and Ser-20, both of which have been implicated in the activation of apoptosis. It has also been shown that phosphorylation at Ser-6 activates p53 by enhancing its tetramerization and stability [42], although phosphorylation at Ser-20 leads to stabilization of p53 by weakening its interaction with Mdm2 [43]. Furthermore, phosphorylation of Ser-20 correlates with induction of p21, a transcriptional target of p53 that is involved in G1 arrest [44]. This is consistent with our studies that demonstrated elevation of p21 associated with FDH expression [27]. Phosphorylation of Ser-392 of the p53 C-terminal domain, known to enhance p53 tetramer formation and to increase its DNA binding ability and transcriptional activity [45,46], was also seen in these experiments. It has been proposed that phosphorylation at Ser-392 might facilitate phosphorylation at the transactivation domain due to a more favourable conformation induced by the tetramer formation [42,47]. Our studies further revealed that Ser-6 is required to mediate suppressor effects of FDH and the two other Ser residues were not important in this model and therefore their phosphorylation could be a secondary effect.
Although the well-established mechanism of p53-induced apoptosis is transcriptional regulation of target genes, it has been recently reported that cytoplasm-localized p53 could exert apoptosis-inducing effects by activating the apoptotic protein Bax through direct interaction [48]. Furthermore, translocation of p53 to mitochondria followed by inhibition of the apoptosis-protective function of anti-apoptotic proteins BclXL and Bcl2 can increase the permeabilization of the mitochondrial outer membrane and result in cytochrome c release [49]. This suggests a potential alternative mechanism for induction of apoptosis by p53. In our experiments, however, translocation of p53 into nuclei after FDH expression suggests the action of p53 occurs through a mechanism of transcriptional activation of apoptotic proteins. Indeed, expression of PUMA, one of the major apoptotic targets of p53 transcriptional activity [36], was seen after FDH induction. Thus the FDH-induced cellular response downstream of p53, that includes expression of the cell cycle-regulating protein p21 and the pro-apoptotic protein PUMA, is consistent with the recent finding that the balance between PUMA and p21 is crucial in determining p53-mediated effects [50].
There are several potential pathways by which FDH could activate p53. Firstly, FDH could activate p53 indirectly through influence on intracellular folate pools. The downstream mechanisms leading to p53 activation could include: altered cellular methylation [18]; nucleotide pool depletion [7]; and accumulation of DNA abnormalities [17]. Secondly, effects of FDH on p53 could be unrelated to its catalytic function. Indeed, there is growing body of evidence that many enzymes, so-called ‘moonlighting’ proteins, possess multiple unrelated functions [51]. The list of enzymes that have a function in apoptosis, perhaps unrelated to their catalytic activities, includes: cytochrome c [52], apoptosis-inducing factor [53], and glyceraldehyde-3-phosphate dehydrogenase [54]. It has been reported that TS, a folate-dependent enzyme, down-regulates p53 at a translational level, through direct interaction with p53 mRNA [25]. Although it is not clear whether FDH could function through a similar mechanism or not, the recently solved crystal structure of FDH N-terminal domain indicates the presence of a potential RNA-binding site [55]. If FDH activates the p53 pathway by a folate-unrelated mechanism it is also not clear whether FDH acts on p53 itself or upstream of p53 through activation of, for example, one of the p53-phosphorylating kinases.
While the direct influence of FDH on p53 or its upstream effectors remains a hypothetical possibility, FDH elevation in our experiments depleted ATP/GTP pools, implying a role for the nucleotide-related mechanism in p53 activation. Activation of p53 by nucleotide pool depletion could potentially proceed through two mechanisms: either purine ribonucleotide depletion can be directly sensed by p53 by an as yet unknown mechanism [7], or insufficient purine deoxyribonucleotide concentrations could initially lead to DNA breakage that would further result in activation of the p53 pathway [56]. In our experiments, dNTP pools were not decreased, suggesting that the latter mechanism does not take part in activation of p53. It would be expected that depletion of ribonucleotide pools should influence, to some extent, cell proliferation even in the absence of p53. Indeed, slowing down of cell proliferation after FDH induction (by about 30%) was observed in p53-deficient cells. However, apoptosis was not initiated in these cells and they were not responsive to the cytotoxic effects of FDH.
Acknowledgments
This work was supported by the National Institutes of Health Grant DK54388. We thank Dr Bert Vogelstein for providing HCT116 cell lines; Dr Jennifer Pietenpol for providing pCEP4-175 vector; Dr Yi-Te Hsu for providing antibodies against Bax, Bcl2 and BclXL; Dr Besim Ogretmen for the helpful discussion; and Margaret Kelly for assistance with confocal microscopy. Confocal microscopy was performed in the Hollings Cancer Center Molecular Imaging Core facility.
References
- 1.Vousden K. H., Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Pietenpol J. A., Stewart Z. A. Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology. 2002;181–182:475–481. doi: 10.1016/s0300-483x(02)00460-2. [DOI] [PubMed] [Google Scholar]
- 3.Burns T. F., El-Deiry W. S. The p53 pathway and apoptosis. J. Cell. Physiol. 1999;181:231–239. doi: 10.1002/(SICI)1097-4652(199911)181:2<231::AID-JCP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Pluquet O., Hainaut P. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 2001;174:1–15. doi: 10.1016/s0304-3835(01)00698-x. [DOI] [PubMed] [Google Scholar]
- 5.Lowe S. W., Ruley H. E. Stabilization of the p53 tumour suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 6.Stewart Z. A., Tang L. J., Pietenpol J. A. Increased p53 phosphorylation after microtubule disruption is mediated in a microtubule inhibitor- and cell-specific manner. Oncogene. 2001;20:113–124. doi: 10.1038/sj.onc.1204060. [DOI] [PubMed] [Google Scholar]
- 7.Linke S. P., Clarkin K. C., Di Leonardo A., Tsou A., Wahl G. M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 8.Koumenis C., Alarcon R., Hammond E., Sutphin P., Hoffman W., Murphy M., Derr J., Taya Y., Lowe S. W., Kastan M., Giaccia A. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.May P., May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B., Lane D., Levine A. J. Surfing the p53 network. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 11.Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 12.Kubbutat M. H., Jones S. N., Vousden K. H. Regulation of p53 stability by Mdm2. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 13.Lill N. L., Grossman S. R., Ginsberg D., DeCaprio J., Livingston D. M. Binding and modulation of p53 by p300/CBP coactivators. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 14.Thornborrow E. C., Manfredi J. J. The tumour suppressor protein p53 requires a cofactor to activate transcriptionally the human BAX promoter. J. Biol. Chem. 2001;276:15598–15608. doi: 10.1074/jbc.M011643200. [DOI] [PubMed] [Google Scholar]
- 15.Li G. M., Presnell S. R., Gu L. Folate deficiency, mismatch repair-dependent apoptosis, and human disease. J. Nutr. Biochem. 2003;14:568–575. doi: 10.1016/s0955-2863(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 16.Wagner C. Biochemical role of folate in cellular metabolism. In: Bailey L. B., editor. Folate in Health and Disease. New York: Marcel Dekker, Inc.; 1995. pp. 23–42. [Google Scholar]
- 17.Blount B. C., Mack M. M., Wehr C. M., MacGregor J. T., Hiatt R. A., Wang G., Wickramasinghe S. N., Everson R. B., Ames B. N. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogribny I. P., Pogribna M., Christman J. K., James S. J. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: possible in vivo relevance during tumorigenesis. Cancer Res. 2000;60:588–594. [PubMed] [Google Scholar]
- 19.Koury M. J., Horne D. W., Brown Z. A., Pietenpol J. A., Blount B. C., Ames B. N., Hard R., Koury S. T. Apoptosis of late-stage erythroblasts in megaloblastic anemia: association with DNA damage and macrocyte production. Blood. 1997;89:4617–4623. [PubMed] [Google Scholar]
- 20.Jackson R. C. Antifolate drugs: past and future perspectives. In: Jackman A. L., editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 1–12. [Google Scholar]
- 21.Chu E., Allegra C. J. Antifolates. In: Chabner B. A., Longo D. L., editors. Cancer Chemotherapy and Biotherapy: Principles and Practice. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 109–148. [Google Scholar]
- 22.Banerjee D., Mayer-Kuckuk P., Capiaux G., Budak-Alpdogan T., Gorlick R., Bertino J. R. Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim. Biophys. Acta. 2002;1587:164–173. doi: 10.1016/s0925-4439(02)00079-0. [DOI] [PubMed] [Google Scholar]
- 23.Lu X., Errington J., Curtin N. J., Lunec J., Newell D. R. The impact of p53 status on cellular sensitivity to antifolate drugs. Clin. Cancer Res. 2001;7:2114–2123. [PubMed] [Google Scholar]
- 24.Zhang C. C., Boritzki T. J., Jackson R. C. An inhibitor of glycinamide ribonucleotide formyltransferase is selectively cytotoxic to cells that lack a functional G1 checkpoint. Cancer Chemother. Pharmacol. 1998;41:223–228. doi: 10.1007/s002800050732. [DOI] [PubMed] [Google Scholar]
- 25.Ju J., Pedersen-Lane J., Maley F., Chu E. Regulation of p53 expression by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3769–3774. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., Schmitz J. C., Lin X., Tai N., Yan W., Farrell M., Bailly M., Chen T., Chu E. Thymidylate synthase as a translational regulator of cellular gene expression. Biochim. Biophys. Acta. 2002;1587:174–182. doi: 10.1016/s0925-4439(02)00080-7. [DOI] [PubMed] [Google Scholar]
- 27.Oleinik N. V., Krupenko S. A. Ectopic expression of 10-formyltetrahydrofolate dehydrogenase in a549 cells induces g(1) cell cycle arrest and apoptosis. Mol. Cancer Res. 2003;1:577–588. [PubMed] [Google Scholar]
- 28.Kisliuk R. L. Folate biochemistry in relation to antifolate selectivity. In: Jackman A. L., editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 13–36. [Google Scholar]
- 29.Krupenko S. A., Oleinik N. V. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumour tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13:227–236. [PubMed] [Google Scholar]
- 29a.Daxecker H., Raab M., Cichna M., Markl P., Muller M. M. Determination of the effects of mycophenolic acid on the nucleotide pool of human peripheral blood mononuclear cells in vitro by high-performance liquid chromatography. Clin. Chim. Acta. 2001;310:81–87. doi: 10.1016/s0009-8981(01)00526-5. [DOI] [PubMed] [Google Scholar]
- 29b.Pizzorno G., Moroson B. A., Cashmore A. R., Beardsley G. P. (6R)-5,10-Dideaza-5,6,7,8-tetrahydrofolic acid effects on nucleotide metabolism in CCRF-CEM human T-lymphoblast leukemia cells. Cancer Res. 1991;51:2291–2295. [PubMed] [Google Scholar]
- 29c.Hwang H. S., Davis T. W., Houghton J. A., Kinsella T. J. Radiosensitivity of thymidylate synthase-deficient human tumour cells is affected by progression through the G1 restriction point into S-phase: implications for fluoropyrimidine radiosensitization. Cancer Res. 2000;60:92–100. [PubMed] [Google Scholar]
- 30.Szak S. T., Pietenpol J. A. High affinity insertion/deletion lesion binding by p53. Evidence for a role of the p53 central domain. J. Biol. Chem. 1999;274:3904–3909. doi: 10.1074/jbc.274.6.3904. [DOI] [PubMed] [Google Scholar]
- 31.Kern S. E., Pietenpol J. A., Thiagalingam S., Seymour A., Kinzler K. W., Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science (Washington, D.C.) 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 32.Paddison P. J., Caudy A. A., Hannon G. J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hietanen S., Lain S., Krausz E., Blattner C., Lane D. P. Activation of p53 in cervical carcinoma cells by small molecules. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8501–8506. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science (Washington, D.C.) 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 35.Saito S., Yamaguchi H., Higashimoto Y., Chao C., Xu Y., Fornace A. J., Jr, Appella E., Anderson C. W. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 2003;278:37536–37544. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 36.Villunger A., Michalak E. M., Coultas L., Mullauer F., Bock G., Ausserlechner M. J., Adams J. M., Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science (Washington, D.C.) 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 37.Dotto G. P. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta. 2000;1471:M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro G. I., Koestner D. A., Matranga C. B., Rollins B. J. Flavopiridol induces cell cycle arrest and p53-independent apoptosis in non-small cell lung cancer cell lines. Clin. Cancer Res. 1999;5:2925–2938. [PubMed] [Google Scholar]
- 39.Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science (Washington, D.C.) 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 40.Appella E., Anderson C. W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 41.Oren M. Decision making by p53: life, death and cancer. Cell. Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 42.Shieh S. Y., Taya Y., Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haupt S., Louria-Hayon I., Haupt Y. P53 licensed to kill? Operating the assassin. J. Cell. Biochem. 2003;88:76–82. doi: 10.1002/jcb.10311. [DOI] [PubMed] [Google Scholar]
- 44.Jabbur J. R., Huang P., Zhang W. DNA damage-induced phosphorylation of p53 at Ser-20 correlates with p21 and Mdm-2 induction in vivo. Oncogene. 2000;19:6203–6208. doi: 10.1038/sj.onc.1204017. [DOI] [PubMed] [Google Scholar]
- 45.Hupp T. R., Lane D. P. Allosteric activation of latent p53 tetramers. Curr. Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 46.Sakaguchi K., Sakamoto H., Lewis M. S., Anderson C. W., Erickson J. W., Appella E., Xie D. Phosphorylation of Ser-392 stabilizes the tetramer formation of tumour suppressor protein p53. Biochemistry. 1997;36:10117–10124. doi: 10.1021/bi970759w. [DOI] [PubMed] [Google Scholar]
- 47.Kapoor M., Hamm R., Yan W., Taya Y., Lozano G. Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene. 2000;19:358–364. doi: 10.1038/sj.onc.1203300. [DOI] [PubMed] [Google Scholar]
- 48.Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science (Washington, D.C.) 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 49.Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U. M. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 50.Yu J., Wang Z., Kinzler K. W., Vogelstein B., Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeffery C. J. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 52.Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell (Cambridge, Mass.) 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 53.Daugas E., Nochy D., Ravagnan L., Loeffler M., Susin S. A., Zamzami N., Kroemer G. Apoptosis-inducing factor (AIF): a ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS Lett. 2000;476:118–123. doi: 10.1016/s0014-5793(00)01731-2. [DOI] [PubMed] [Google Scholar]
- 54.Ishitani R., Tajima H., Takata H., Tsuchiya K., Kuwae T., Yamada M., Takahashi H., Tatton N. A., Katsube N. Proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase: a possible site of action of antiapoptotic drugs. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2003;27:291–301. doi: 10.1016/S0278-5846(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 55.Chumanevich A. A., Krupenko S. A., Davies C. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain. J. Biol. Chem. 2004;279:14355–14364. doi: 10.1074/jbc.M313934200. [DOI] [PubMed] [Google Scholar]
- 56.Chernova O. B., Chernov M. V., Agarwal M. L., Taylor W. R., Stark G. R. The role of p53 in regulating genomic stability when DNA and RNA synthesis are inhibited. Trends Biochem. Sci. 1995;20:431–434. doi: 10.1016/s0968-0004(00)89094-5. [DOI] [PubMed] [Google Scholar]