Abstract
In over 90% of cervical cancers and cancer-derived cell lines, the p53 tumor suppressor pathway is disrupted by human papillomavirus (HPV). The HPV E6 protein promotes the degradation of p53 and thus inhibits the stabilization and activation of p53 that would normally occur in response to HPV E7 oncogene expression. Restoration of p53 function in these cells by blocking this pathway should promote a selective therapeutic affect. Here we show that treatment with the small molecule nuclear export inhibitor, leptomycin B, and actinomycin D leads to the accumulation of transcriptionally active p53 in the nucleus of HeLa, CaSki, and SiHa cells. Northern blot analyses showed that both actinomycin D and leptomycin B reduced the amount of HPV E6-E7 mRNA whereas combined treatment with the drugs showed almost complete disappearance of the viral mRNA. The combined treatment activated p53-dependant transcription, and increases in both p21WAF1/CIP1 and Hdm2 mRNA were seen. The combined treatment resulted in apoptotic death in the cells, as evidenced by nuclear fragmentation and PARP-cleavage indicative of caspase 3 activity. These effects were greatly reduced by expressing a dominant negative p53 protein. The present study shows that small molecules can reactivate p53 in cervical carcinoma cells, and this reactivation is associated with an extensive biological response, including the induction of the apoptotic death of the cells.
Mutations in the p53 tumor suppressor gene are the most common specific genetic changes in human tumors. They occur only rarely, however, in tumors in which p53 may be inactivated by interaction with cellular or viral proteins. For instance, in cervical cancer, p53 mutation is uncommon, but human papillomavirus (HPV) is present in more than 90% of the tumors. HPV infection is the major risk factor of this disease, which, worldwide, is the second most common form of cancer in women (1). The correlation of HPV infection and p53 mutation has been closely studied in cervical cancer, and, because the HPV E6 protein inactivates p53, it appeared that p53 mutations would be confined to HPV-negative cases. This is, with a few exceptions, found to be the case (2).
The HPV E6 protein complexes with cellular proteins E6-AP and p53 and facilitates p53 degradation via the ubiquitin-dependent proteolytic system (3). E6 proteins of both high risk and low risk HPV types bind to p53 in vitro, but only E6 proteins of oncogenic HPV types can target p53 for degradation (4). Apart from the p53 interaction, evidence is accumulating that E6 has p53-independent transforming and growth-regulating activities (5, 6).
Different stress signals, such as DNA damage, hypoxia, heat shock, and oncogene activation induce an increase in the stability of the wild-type p53 protein. The accumulation of p53 is associated with the transcription of a series of p53-responsive cellular genes, including bax, p21WAF1/CIP1, and gadd45, that mediate p53 induction of growth arrest and apoptosis (7). The activated p53 also induces the transcription of Mdm2/Hdm2, which can directly interact with transactivation domain of p53, inhibiting it's transcription activity and targeting it for degradation (8, 9). Thus, the stability of mutant p53 observed can be explained by its inability to induce the synthesis of Mdm2/Hdm2 (10). The expression of HPV E6 from an ectopic promoter has been shown to inhibit p53 transcriptional activity and cellular responses, such as activation of p21WAF1/CIP1, cell cycle arrest in G1, and induction of apoptosis (11–13). However, when studied in cervical carcinoma cells expressing E6 from their natural promoter, residual wild-type p53 activity has been reported, suggesting that the amount of expressed HPV E6 is proportionate to the inhibition of p53 activity (14, 15). At least in some cervical carcinoma cell lines, the continuous expression of E6-E7 seems to be required for the maintenance of the malignant state (16). Therefore, tilting the balance in favor of p53 might reestablish the tumor-suppressing function of p53 in cervical carcinoma cells.
Leptomycin B (LMB) is a novel anti-tumor antibiotic produced by Streptomyces sp. (17). LMB inhibits the export of nuclear export signal-containing proteins from nucleus to the cytoplasm (18). Mdm2 contains a nuclear export signal that mediates its export from the nucleus (19), and it recently has been reported that LMB can inhibit the degradation of p53 and accumulate active p53 in the nucleus of wild-type (wt) p53 containing cells (20–22). Nuclear accumulation of p53 was also observed in HPV-positive cervical carcinoma cell lines after exposure to LMB, and this was attributed at least in part to nuclear export inhibition (20).
Actinomycin D (AD) (dactinomycin) is also an anti-tumor antibiotic produced by Streptomyces sp. and is currently used against some neoplastic disorders. It intercalates into DNA and interferes with RNA polymerases and DNA topoisomerases I and II. AD is a strong inhibitor of RNA synthesis and induces p53 accumulation in normal cells. The accumulation of p53 has recently been reported to be caused by its inhibition of RNA polymerase II rather than by its DNA damaging effect, which only occurs at higher doses of the drug (23). Our observations show that AD can reduce Mdm2 protein and mRNA levels dramatically (24).
The clinical data relating p53 status and treatment resistance of cancer has been somewhat confusing. Nevertheless, the general view is that tumors harboring p53 mutations are more aggressive and resistant to traditional therapies (25, 26). In the present study, we used LMB and AD to try and reactivate wt p53 in cervical carcinoma cells. We are able to demonstrate that a combination of these agents can induce the activation of the p53 response and p53-dependent apoptotic cell death in HPV-positive human cervical cancer cell lines. Because these results show that the downstream p53-dependant apoptotic pathway is intact in these cells, it encourages the possibility of the development of novel, small-molecule-drug-based treatment protocols for this important disease.
Materials and Methods
Cell Lines and Treatment.
The cervical carcinoma cell lines CasKi, SiHa, and HeLa cells and normal foreskin fibroblasts were maintained in DMEM supplemented with 10% FCS. CasKi and SiHa contain integrated HPV 16, and HeLa cells carry integrated HPV 18 sequences. The dominant negative (dn) p53 expressing SiHa cell line (SiHa DD) was derived by transfecting the parental cell line with a plasmid that expresses a truncated mouse p53 containing amino acid residues 1–14 and 302–390 under the control of the cytomegalovirus (CMV) promoter. The dominant negative effect of this construct involves probably the formation of inactive complex between the truncated protein and the wt p53 (27). Stable transfectants were selected with 0.8 mg/ml G418.
Experiments blocking the interaction between p53 and Mdm2 were carried out expressing a Mdm2 binding peptide in Thioredoxin scaffold in pcDNA 3.1. The expression plasmids STIP and the one containing three alanine substitutions in the Mdm2 binding motif, STIPala, have been described (28). All transfections were carried out with Fugene (Roche).
Leptomycin B was a gift from Patrick Chene and Ying Wang (Novartis, Basel) and was stored as a 10 mM stock in ethanol. Actinomycin D was purchased from Sigma and was stored as a 2-mg/ml stock in ethanol.
Luciferase Assays.
For the p53 transcription studies, 50 ng of the p53 reporter PG13 or the control vector MG15 containing mutated binding site for p53 (29) was cotransfected in 96-well plates with 0.5 ng of Renilla luciferase expressing pRL-SV40 (Promega) as internal standard. Twenty-four hours after transfection, the cells were treated with the drugs. The cells were lysed after 14, 24, and 36 h of treatment with passive lysis buffer, and the luciferase activity was analyzed according to the Dual-Luciferase assay protocol (Promega) with Microplan luminometer LB96U (EG & G Berthold). Variations in cell number and transfection efficiencies were corrected by determining the Renilla luciferase activity of the sample.
Immunofluorescence Analysis.
Cells were grown on Permanox coverslips (Nunc) and were stained as described (21). Primary antibodies were mouse mAb DO-1, which recognizes p53, and polyclonal rabbit anti-thioredoxin antibody (Sigma). Secondary antibodies were FITC- or Texas red-conjugated (Jackson ImmunoResearch). Hoechst 33258 was used for nuclear counterstaining.
Flow Cytometry.
The cells were pulse-labeled with 30 μM BrdUrd for 30 min after which both adherent and detached cells were collected and analyzed as described (30).
Western Blot Analysis.
The cultures were washed twice in ice-cold PBS, were scraped into PBS with a rubber policeman, and were pelleted. In case of PARP analysis, floating cells also were collected. Cells were disrupted in 250 mM Tris (pH 7.8), 60 mM KCl, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF, were frozen and thawed in three cycles, and were centrifuged at 13,000 × g for 15 min. The protein extract in the supernatant was collected, and concentration was determined according to Bradford (Bio-Rad). Equivalent amounts (100 μg) of protein were separated on SDS/polyacrylamide gels and were transferred to Immobilon-P filters (Millipore). Equal loading was confirmed staining the filter with Ponceau S. Proteins were detected with the following antibodies: DO-1 for p53, mouse mAb 118 against p21WAF1/CIP1 (31), mouse mAb 4B2 against MDM2 (32), and polyclonal rabbit anti-PARP H-250 (Santa Cruz Biotechnology). Anti-p73 rabbit sera raised using full length human p73 or a peptide spanning amino acids 9–24 were provided by B. Vojtesek (Masaryk Memorial Cancer Institute, Brno, Czech Republic). Secondary HRP-conjugated rabbit anti-mouse or swine anti-rabbit IgG antibodies were purchased from DAKO. The detection was carried out with enhanced chemiluminescence. Each assay was repeated at least three times with identical results.
Northern Blot Analysis.
Poly(A)+-RNA was isolated according to Rahmsdorf et al. (33). For Northern blot hybridization, 3 μg of mRNA were separated on a 1.4% agarose gel and were transferred onto Hybond N+ membrane (Amersham). Hybridization was performed with random primed cDNA probes: 1,300-bp PstI fragment of mouse GAPDH, 885-bp HindIII fragment of hdm2 provided by B. Vogelstein (Johns Hopkins University, Baltimore), and 1,200-bp EcoRI fragment of human p53 cDNA. The HPV 18 E6-E7 probe was generated by PCR amplification of HPV 18 plasmid with the following primer pair: 5′-AGGGAGTAACCGAAAACG-3′ and 5′-CATAAAACCAGCCGTTAC-3′. HPV 16 E6-E7 transcripts were detected with a probe spanning nucleotide positions 7,454 to 875 and was provided by M. Dürst (DKFZ, Heidelberg). The NdeI-PstI fragment of p21WAF1/CIP1 was provided by E. Warbrick (University of Dundee, Dundee, Scotland). Signal quantitation was done by scanning autoradiograms in the linear exposure range of the film with HP ScanJet 520°C scanner (Hewlett–Packard) and analyzing the data by scion image software (Scion). Hybridization with the mouse GAPDH probe was used as a control for the quantity of the mRNAs.
Clonogenic Survival Assay.
Subconfluent cultures were exposed to drugs for 6 h in 30-mm culture dishes. Then the cells were trypsinized, centrifuged, and resuspended in fresh medium without drugs, and 1/10 000 of the cell suspension was plated to 6-well plates. After 14 days of culture, the colonies were fixed with methanol, were stained with Giemsa, and were counted. Alternatively, cells were plated in 24-well plates and were incubated for 21 days, after which the plates were scanned and analyzed for gray values in individual wells by scion image software. Close correlation of the results were found with these two methods.
Results
p53 Cannot Be Stabilized in Cervical Carcinoma Cells by Inhibiting Hdm2 Interaction with p53.
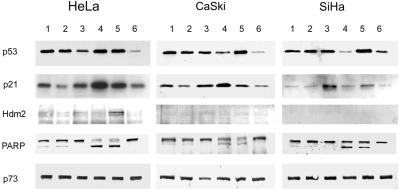
In HPV-transformed cells, it is assumed that p53 is degraded predominantly by the HPV-E6- and HPV E6-AP-mediated ubiquitination, but residual Hdm2 activity may contribute to the low half life of p53 in HPV positive cells. Therefore, we wanted to examine whether this normal cellular p53 degradation pathway is also involved in regulating the p53 levels in cervical cancer cells. Previous experiments from our laboratory have shown that interaction between p53 and Hdm2 can be inhibited by an inhibitory protein (28). This chimeric protein consists of a 12-amino acid peptide that binds to the p53 binding pocket of the Hdm2 protein and is inserted into the active site-loop of Escherichia coli thioredoxin protein (28). We used MCF-7 breast cancer cells expressing wt p53 as a control. Transfection of this peptide construct (STIP) resulted in stabilization of endogenous wt p53 in MCF-7 cells, as we have reported previously (28). The activity is specific because transfection with a construct containing three alanine substitutions in the Hdm2 binding motif (STIPala) did not induce p53 accumulation. However, there was no p53 stabilization in any of the cervical carcinoma cell lines carrying HPV sequences, as analyzed by immunofluorescent staining or Western blots (see supplemental data on the PNAS web site, www.pnas.org). This suggests that the Hdm2 degradation pathway is switched off in HPV-positive cervical carcinoma cells and that the E6-mediated pathway is solely responsible for the low p53 levels in these cells. The very low Hdm2 amount found in the Western blots of HeLa, CasKi, and SiHa (Fig. 4) is also consistent with the fact that, in these E6-positive cells, the p53-Hdm2 feedback loop is interrupted.
Figure 4.
Effect of LMB and AD treatment on the p53, p21, Hdm2, PARP, and p73 proteins. Western blot analysis of cell extracts from cultures treated for 18 h with 20 nM LMB (1), 200 nM (2), 5 nM AD (3), 50 nM AD (4), 20 nM LMB + 5 nM AD (5), and untreated cells (6).
Nuclear Accumulation of p53 Induced by LMB and AD in HPV Positive Cervical Carcinoma Cells.
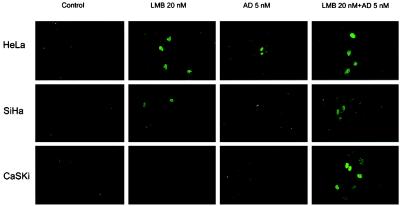
Having established that the low levels of p53 present in HPV-positive cervical cancer cells are independent of the Hdm2 pathway, we examined the effects of p53-activating small molecules on these cells. Immunofluorescence microscopy (Fig. 1) of untreated SiHa, CaSki, and HeLa cell lines with the monoclonal anti-p53 antibody DO-1 showed practically no staining in the nuclei. After 24 h of incubation with leptomycin B at 20 and 200 nM (not shown), nuclear staining of p53 was detected in all three HPV-positive cell lines tested, consistent with the earlier results (20). After treatment with actinomycin D at 5 nM, the p53 staining was also confined to the nucleus in these cell lines. In HeLa cells, AD at 50 nM further increased the nuclear p53 whereas in CasKi cells, the staining was reduced compared with that with 5 nM AD and in SiHa was not distinguishable from the control (not shown). When the two drugs at the lower concentration were combined, even more intense nuclear staining was observed, and this staining pattern was seen in all cell lines. The levels of the p53 staining seen by microscopy were in agreement with the Western blot analyses (Fig. 4).
Figure 1.
Nuclear accumulation of p53 in HeLa, SiHa, and CasKi cells by confocal immunofluorescence microscopy. The cells were treated with indicated concentrations of LMB and AD for 24 h. p53 was detected with DO-1.
Actinomycin D and Leptomycin B Induce Transcriptionally Active p53.
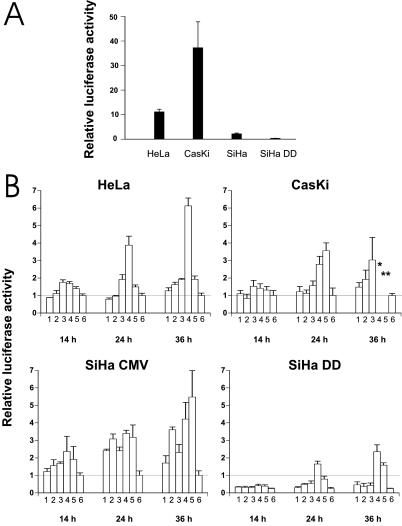
Without drug treatment, HeLa cells showed a 10-fold, CasKi cells a 40-fold, and SiHa cells a 3-fold activation of the PG13-driven reporter vs. the control MG15 reporter (Fig. 2A). SiHa cells that contain only one copy of HPV 16 DNA per cell show less activity than CasKi cells containing 600 HPV 16 or HeLa cells with 25 copies of HPV 18. However, RNA is transcribed only from a minor fraction of the DNA copies (34), and only a small fraction of the early region transcripts are translated to full length E6 protein capable of interfering p53 function (34, 35). Therefore, the HPV DNA copy number does not directly correlate to the activity of p53. The dn p53 SiHa DD cell line showed no activity above control. Therefore, these HPV-positive cells have some basal wt p53 activity despite the presence of HPV E6.
Figure 2.
(A) Endogenous transcriptional activation of the p53 responsive promoter PG13 and MG15 containing mutated binding site for p53. The values represent relative activities of PG13 versus MG15 construct. SiHa DD is a SiHa cell line stably transfected with a dominant negative p53 gene. (B) Transcriptional activation of p53 responsive promoter after treatment with LMB and AD. Twenty-four hours after the transfection, cell lines were treated with the drugs and thereafter analyzed for luciferase activity. The bars represent: 1, 20 nM LMB; 2, 200 nM LMB; 3, 5 nM AD; 4, 50 nM AD; 5, 20 nM LMB + 5 nM AD; 6, untreated cells. Values are expressed relative to the readings of untreated cells arbitrarily set to 1. SiHa DD values are calculated relative to untreated SiHa CMV cells. * and **, not enough living cells for reliable quantitation. SD was calculated from triplicate wells.
LMB showed a small activation of the reporter in SiHa cells carrying the empty vector (SiHa CMV) already at 14 h after drug treatment, but in HeLa and CasKi cells only at 36 h (Fig. 2B). AD induced the promoter already at 14 h in all cell lines. The strongest activator of PG13 was the drug combination of LMB and AD in SiHa CMV and CasKi cells whereas AD alone at 50 nM was the best inducer in HeLa cells. SiHa CMV cells treated with 50 nM AD showed clear reporter activation, although the p53 protein levels were not elevated, suggesting that the activation of p53 had occurred through phosphorylation or other protein modification. Treatment of CasKi with 50 nM AD or the LMB + AD combination for 36 h resulted in almost complete death of the cells. Although the few surviving cells showed strong activation of the p53 reporter, the high rate of cell death meant that there was a very high level of variation between individual assays.
The SiHa DD expressing the dn p53 protein showed clearly lower luciferase activities compared with SiHa CMV, but when stimulated with the drugs a detectable PG13 activation was observed, especially with 50 nM AD or LMB + AD combination (Fig. 2B). This indicates that either the C-terminal miniprotein does not fully inhibit the p53 activity in these cells or there is some other mechanism that activates the PG13 promoter. The former explanation is conceivable based on the original data showing that the transactivation of p53 is not completely inhibited in co-transfection experiments with this construct (27). Nevertheless, one possible candidate to activate p53 responsive promoter is p73, which is a p53-related protein capable of inducing apoptosis and has been reported to be resistant to E6 (36). However, no alterations in the protein levels were observed after LMB or AD treatment (Fig. 4). The same was true with the SiHa DD cell line (not shown).
HPV E6-E7 mRNA Levels Are Down-Regulated by LMB and AD.
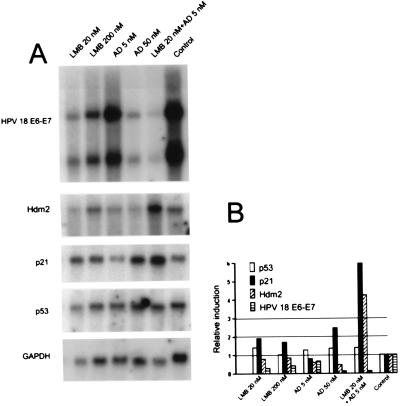
Both LMB and AD decreased HPV E6-E7 mRNA levels in HeLa, CaSki, and SiHa cells (Fig. 3; data not shown). The combination of the two drugs made these mRNAs almost undetectable. Because suitable anti-E6 antibodies for a reliable quantitation of E6 protein levels are not available, no Western blot analyses could be done for the E6 expression.
Figure 3.
(A) Northern blot analysis of HPV 18 E6-E7, Hdm2, p21, and p53 mRNA expression in HeLa cells after 18 h of LMB and AD treatment. Each lane contained 3 μg of polyadenylated RNA. (B) Quantitation of the mRNA levels after normalization for GAPDH mRNA expression. The values are reported as the relative level over untreated cells.
LMB and AD Alter p21WAF1/CIP1 and Hdm2 mRNA and Protein Levels.
To further search for evidence of p53-mediated transactivation, we performed both Northern and Western blot analyses of the products of two p53 target genes, p21WAF1/CIP1 and hdm2. After after 18 h of LMB and 50 nM AD treatment, HeLa cells showed slightly increased p21WAF1/CIP1 mRNA levels. AD treatment reduced the signals of Hdm2, and they were practically unchanged after LMB. The clearest induction of p21WAF1/CIP1 and Hdm2 was obtained by combining 5 nM AD and 20 nM LMB treatments (Fig. 3).
Despite the changes in the mRNA levels of all other studied genes, p53 mRNA levels remained constant (Fig. 3). This means that the elevated protein levels are caused by stabilization of the protein, likely through inhibition of the degradation pathway.
The p21WAF1/CIP1 protein levels were clearly increased in HeLa and CasKi cells after an 18-h treatment with 50 nM AD (Fig. 4). This increase was lower in 5 nM AD and in cells treated with 20 nM LMB + 5 nM AD. However, after a 24-h treatment with 50 nM AD or 20 nM LMB + 5 nM AD, a substantial portion of the cells had lost viability. In these cultures, p21WAF1/CIP1 levels decreased relative to the 5 nM AD (not shown). SiHa cells showed the highest p21WAF1/CIP1 levels with 5 nM AD. After LMB treatment alone, no increase in p21WAF1/CIP1 was detected in SiHa cells and a subtle increase with 20 nM but not 200 nM concentrations in HeLa and CasKi cells.
The amounts of Hdm2 protein in all cell lines were very low and could only be detected in HeLa cells (Fig. 4). In HeLa cells, LMB treatment resulted in slight increase and 20 nM LMB + 5 nM AD induced a clear increase in Hdm2 levels.
Induction of Apoptosis by LMB and AD.
AD at 50 nM and 5 nM AD + 20 nM LMB had a profound cytopathic effect in all of the cell lines. After approximately 10 h (CasKi and HeLa) or15 h (SiHa) of treatment, cells started to show blebbing in the cell membrane followed by rounding up of the cells and cell shrinkage (see the time-lapse video published as supplemental data on the PNAS web site), nuclear fragmentation detected by Hoechst 33258 staining, and accumulation of events in sub-G1 fraction in FACS analyses (Fig. 5). LMB alone also killed the cells, but this occurred after a delay. CaSki cells were most sensitive to LMB and AD, followed by HeLa. SiHa cells were least sensitive, as observed by microscopy and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide cytotoxicity assays (not shown). AD at 5 nM did not cause cytopathic changes, but the cells were inhibited in proliferation. This is consistent with the expression of p21 WAF1/CIP1 protein in these cells.
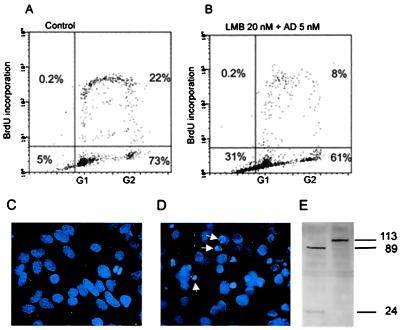
Figure 5.
(A and B) Flow cytometry analysis of apoptosis in HeLa cells after 16 h of treatment with the combination of 20 nM LMB + 5 nM AD. (C) Hoechst 33258 staining of untreated HeLa cells. (D) Nuclear fragmentation of HeLa cells after 18 h of 20 nM LMB + 5 nM AD. (E) Western blot analysis of PARP protein in HeLa cells 35 h after 20 nM LMB + 5 nM AD (first lane) and without treatment (second lane).
Poly(ADP-ribose) polymerase, PARP, is an enzyme that is known to be cleaved by caspases rapidly after induction of apoptosis (37). After 20 nM LMB + 5 nM AD treatment, the typical cleavage pattern was found in which the 113-kDa PARP was cleaved to an 89-kDa catalytic fragment and a 24-kDa DNA binding fragment (Fig. 5). The PARP cleavage was also observed in cultures treated with LMB alone. The blots prepared 20 h after treatment show, however, that the 89-kDa fragment was fainter than with 50 nM AD or 20 nM LMB + 5 nM AD, suggesting that the caspase activity was induced later or was lesser in LMB treated cultures. This is in accordance with the kinetics of the cell death seen after these treatments.
Expression of Dominant Negative p53 Partially Rescues SiHa Cells from the Cytotoxicity of LMB and AD.
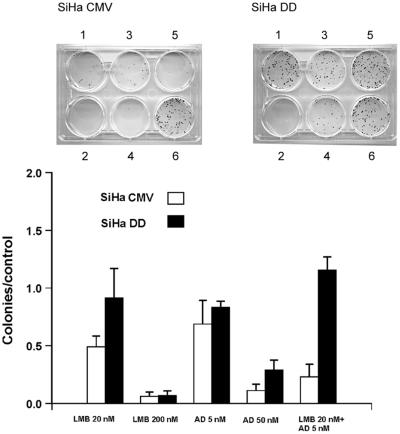
Clonogenic survival assay shows that SiHa DD cells survived better the 20 nM LMB treatment than SiHa CMV cells (Fig. 6). The 5 nM AD reduced the number of colonies slightly for SiHa CMV, but AD at 50 nM killed both cell lines, albeit somewhat less efficiently the SiHa DD cells. When 20 nM LMB and 5 nM AD were combined, SiHa DD survived the treatment with little decrease in the colony formation ability whereas the cells carrying the empty vector formed virtually no colonies. This finding suggests that the effect of LMB and LMB + AD combination on the cell survival is partially mediated by p53.
Figure 6.
Comparison of clonogenic cell survival of SiHa cells either stably transfected with empty vector (SiHa CMV) or with dominant negative p53 (SiHa DD). The treatments were 20 nM LMB (1), 200 nM (2), 5 nM AD (3), 50 nM AD (4), 20 nM LMB + 5 nM AD (5), and untreated cells (6).
Discussion
The tumor suppressor function of p53 is abrogated in human cervical carcinoma cells by the HPV E6 protein. A number of approaches have previously been proposed to control the growth of E6-expressing cancer cells. These include (i) the use of papillomavirus E2 gene to repress E6 (and E7) expression (38, 39), (ii) antisense strategies (40), and (iii) the use of variant forms of E6 that interact both with full length E6 protein and with p53, resulting in suppression of E6-mediated degradation of p53 (35). In the present study, we used LMB and AD and show that these small molecule compounds can activate the p53 response and have a synergistic effect in inducing apoptosis in HPV-positive cervical carcinoma cells.
The basal p53 activity, measured with the reporter assays, in the untreated cervical cancer cells indicates that these cells have residual p53 activity, which can be abrogated by expressing a dn p53 miniprotein. This result is in full agreement with the previous findings by Butz et al. (14, 15) showing that p53 is not completely inactivated in cervical cancer cells. However, they are not in accordance with previous results that p53 is inactive in HPV 16 E6-expressing cells, even after DNA damage (11). In these studies, the expression of E6 was driven by strong constitutive promoters, resulting in huge excess of E6, which explains the different results. Kessis et al. (11) failed to induce p53 in HeLa cells with AD at 0.5 nM, which is 10 times lower than the concentration we used in the present study. In our own experiments, we also observed a similar lack of p53 induction in HeLa cells when we used 0.5 nM and 0.05 nM AD (not shown). It is interesting that CaSki cells had the highest basal p53 activity. The further increase after stimulation by LMB and AD makes the overall level of p53 reporter activity much higher than the other cell lines and might account for the higher sensitivity of this cell line to the drugs. However, direct comparison of the p53 reporter activities between cell lines has limitations as different p53 target promoters may have different reporter activities, depending on the cellular background (41).
The down-regulation of HPV E6 is probably one of the key factors in increasing the function of p53 in the present study. Another mechanism may be the selective inhibition of RNA synthesis by actinomycin D, which has been reported to be the main reason of this agent to cause p53 accumulation (23, 42). Nevertheless, p53 activity does not necessarily depend on its amount (43), but the active portion evolves by posttranslational modifications (44, 45).
Considering potential therapeutic aims, it is interesting to note that the DNA damaging activity of AD has been detected only above a 200 nM concentration, but no significant induction of DNA strand breaks could be detected at 20 nM (23). What makes E6-E7 mRNA synthesis and/or stability so sensitive to both of these compounds remains to be determined, but it is interesting to note that LMB has been shown to accumulate transcription factors in the nucleus (46, 47). Interestingly, p53 mRNA levels remained constant even after strong general transcription suppression reported for AD at higher doses (23), or despite various effects of the drugs to other genes, suggesting a general mechanism may exist to protect it.
In HeLa cells, after treatment of 5 nM AD, the slight decrease in E6-E7 mRNA and Hdm2 mRNA was not sufficient to induce a substantial increase in the PG13 promoter activity. The combination of 20 nM LMB + 5 nM AD promotes a lesser PG13 luciferase activity than AD alone at 50 nM. This may be attributable to observed increases in Hdm2 mRNA and protein levels. Instead, in SiHa and CasKi cells, there was considerable PG13 activation with AD + LMB. Interestingly, in these cells, the levels of Hdm2 protein do not increase after treatment. This correlates well with the fact that the major difference in the clonogenic survival between dn p53 or vector alone carrying SiHa is observed with the combined treatment. With this treatment, the survival seems to be most p53-dependent, and the p53 is most active on the PG13 promoter.
It has been previously demonstrated that Hdm2 protein contains a nuclear export signal and is shuttling between nucleus and cytoplasm, and mutant Hdm2 that is not exported from the nucleus does not target p53 for degradation (19). Blocking nuclear export by LMB results in inhibition of Hdm2-dependent degradation (20, 21). The association of degradation and nuclear export in HPV-positive cells is less clear because it is not known whether E6 or E6-AP harbor nuclear export signals. It has been reported that E6-targeted degradation of p53 also requires nuclear export (20). Our results support the observation that the p53 levels increase in cervical carcinoma cells after LMB treatment, but, because the E6-E7 mRNA levels were found to be reduced by LMB, it is possible that the elevated p53 levels may be attributable to this. Studies showing that p53, HPV E6, and E6-AP are localized to the perinuclear cytoplasmic region would support the view that the degradation at least partially takes place in the cytoplasm (48). E6 may inhibit the nuclear localization of p53, as suggested by a recent study in which proteasome inhibition in HPV positive cells resulted in the increase in the perinuclear p53 staining, but not nuclear staining (49). Taken together, it seems plausible that at least some of the E6 mediated degradation of p53 occurs in the cytoplasm and that this degradation is inhibited by LMB through nuclear export inhibition.
In the present study, the dn p53 in SiHa cells rendered these cells more resistant to the apoptotic effects by AD and LMB than the control cells, indicating that this response was at least partially p53-dependent. However, it is possible that some effect of the drugs is mediated by p53 relatives p73 or p63, known to mediate apoptosis (50). It has been described that cisplatin-induced DNA damage also increases the amount of this protein (51) whereas some other forms of DNA damage do not (50, 52, 53). We did not observe alterations in the levels of p73 in our studies in response to AD or LMB. In addition, it is unlikely that the dn p53 would oligomerize with p63 or p73 (54), and, because the dn p53 partially blocked the biological response, the effect of the agents is clearly partially p53-dependent.
The present study gives evidence that cervical carcinoma cell lines are sensitive to AD and LMB. In cervical cancer, the tumor is confined to the cervix, parametrial tissue, and local lymph nodes until late progression of the disease, when distant metastases develop. Therefore, at early stages, or when disease relapses occur in the vagina, the tumor would be accessible with potential local drug treatment without systemic side effects. An important question that arises from this study is, How would a local drug treatment with LMB + AD act in early cervical neoplasia in which HPV is in an episomal stage in contrast to the situation presented here, where the HPV genome is integrated to the host cell genome?
Supplementary Material
Acknowledgments
We thank Dr. Dan Gibbs for the advice in time-lapse video microscopy, and Dr. Jean-Cristophe Bourdon and Dr. Dimitris Xirodimas for general advice. Sakari Hietanen was recipient of grants from The Finnish Academy, Maud Kuistila, and Turku University Foundations. Sonia Lain and Christine Blattner are recipients of postdoctoral fellowships from the Cancer Research Campaign. David Lane is a Gibb fellow of the Cancer Research Campaign.
Abbreviations
- HPV
human papillomavirus
- LMB
leptomycin B
- AD
actinomycin D
- wt
wild-type
- dn
dominant negative
- CMV
cytomegalovirus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.zur Hausen H. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 2.Park D J, Wilczynski S P, Paquette R L, Miller C W, Koeffler H P. Oncogene. 1994;9:205–210. [PubMed] [Google Scholar]
- 3.Scheffner M. Pharmacol Ther. 1998;78:129–139. doi: 10.1016/s0163-7258(98)00003-5. [DOI] [PubMed] [Google Scholar]
- 4.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 5.Kubbutat M H, Vousden K H. Trends Microbiol. 1998;6:173–175. doi: 10.1016/s0966-842x(98)01267-0. [DOI] [PubMed] [Google Scholar]
- 6.Rapp L, Chen J J. Biochim Biophys Acta. 1998;1378:F1–F19. doi: 10.1016/s0304-419x(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 7.Bates S, Vousden K H. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 10.Midgley C A, Lane D P. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 11.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster S A, Demers G W, Etscheid B G, Galloway D A. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Han S, Baluda M A, Park N H. Oncogene. 1997;14:2347–2353. doi: 10.1038/sj.onc.1201078. [DOI] [PubMed] [Google Scholar]
- 14.Butz K, Shahabeddin L, Geisen C, Spitkovsky D, Ullmann A, Hoppe-Seyler F. Oncogene. 1995;10:927–936. [PubMed] [Google Scholar]
- 15.Butz K, Whitaker N, Denk C, Ullmann A, Geisen C, Hoppe Seyler F. Oncogene. 1999;18:2381–2386. doi: 10.1038/sj.onc.1202557. [DOI] [PubMed] [Google Scholar]
- 16.von Knebel-Doeberitz M, Rittmüller C, Aengeneyndt F, Jansen-Durr P, Spitkovsky D. J Virol. 1992;51:831–834. doi: 10.1128/jvi.68.5.2811-2821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaumberg J P, Hokanso G C, French J C. J Chem Soc Chem Commun. 1984;21:1450–1452. [Google Scholar]
- 18.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 19.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedman D A, Levine A J. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 22.Smart P, Lane E B, Lane D P, Midgley C, Vojtesek B, Lain S. Oncogene. 1999;18:7378–7386. doi: 10.1038/sj.onc.1203260. [DOI] [PubMed] [Google Scholar]
- 23.Ljungman M, Zhang F, Chen F, Rainbow A J, McKay B C. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 24.Blattner C, Sparks A, Lane D. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J M, Wouters B G. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- 26.Bunz F, Hwang P M, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler K W, Vogelstein B. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaulian E, Haviv I, Shaul Y, Oren M. Oncogene. 1995;10:671–680. [PubMed] [Google Scholar]
- 28.Böttger A, Böttger V, Sparks A, Liu W L, Howard S F, Lane D P. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 29.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 30.Renzing J, Lane D P. Oncogene. 1995;10:1865–1868. [PubMed] [Google Scholar]
- 31.Fredersdorf S, Milne A W, Hall P A, Lu X. Am J Pathol. 1996;148:825–835. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahmsdorf H J, Schonthal A, Angel P, Litfin M, Ruther U, Herrlich P. Nucleic Acids Res. 1987;15:1643–1659. doi: 10.1093/nar/15.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smotkin D, Wettstein F O. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pim D, Massimi P, Banks L. Oncogene. 1997;15:257–264. doi: 10.1038/sj.onc.1201202. [DOI] [PubMed] [Google Scholar]
- 36.Prabhu N S, Somasundaram K, Satyamoorthy K, Herlyn M, el-Deiry W S. Int J Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 37.D'Amours D, Desnoyers S, D'Silva I, Poirier G G. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 38.Dowhanick J J, Mcbride A A, Howley P M. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang E S, Naeger L K, DiMaio D. Oncogene. 1996;12:795–803. [PubMed] [Google Scholar]
- 40.Steele C, Cowsert L M, Shillitoe E J. Cancer Res. 1993;53:2330–2337. [PubMed] [Google Scholar]
- 41.Lohrum M, Scheidtmann K H. Oncogene. 1996;13:2527–2539. [PubMed] [Google Scholar]
- 42.An W G, Chuman Y, Fojo T, Blagosklonny M V. Exp Cell Res. 1998;244:54–60. doi: 10.1006/excr.1998.4193. [DOI] [PubMed] [Google Scholar]
- 43.Weinberg W C, Azzoli C G, Chapman K, Levine A J, Yuspa S H. Oncogene. 1995;10:2271–2279. [PubMed] [Google Scholar]
- 44.Jimenez G S, Khan S H, Stommel J M, Wahl G M. Oncogene. 1999;18:7656–7665. doi: 10.1038/sj.onc.1203013. [DOI] [PubMed] [Google Scholar]
- 45.Meek D W. Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 46.Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. J Biol Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez M S, Thompson J, Hay R T, Dargemont C. J Biol Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 48.Daniels P R, Sanders C M, Maitland N J. J Gen Virol. 1998;79:489–499. doi: 10.1099/0022-1317-79-3-489. [DOI] [PubMed] [Google Scholar]
- 49.Mantovani F, Banks L. Oncogene. 1999;18:3309–3315. doi: 10.1038/sj.onc.1202688. [DOI] [PubMed] [Google Scholar]
- 50.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, et al. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 51.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. Nature (London) 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 52.Agami R, Blandino G, Oren M, Shaul Y. Nature (London) 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 53.Yuan Z M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. Nature (London) 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 54.Davison T S, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith C H. J Biol Chem. 1999;274:18709–18714. doi: 10.1074/jbc.274.26.18709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.