Abstract
Anaplasma (Ehrlichia) phagocytophila's major immunodominant surface protein antigen, Msp2 (P44, 44-kDa antigen), is encoded by a family of paralogous genes characterized by conserved sequences flanking a hypervariable region. The antigenic profiles of most strains of A. phagocytophila are different, and the differences are principally related to Msp2 expression. To date, multiple unique msp2 gene paralogs have been found in A. phagocytophila isolates, but the overall number in the genome of a single strain is not yet known. Changes in msp2 expression may be related to antigenic variability; thus, we examined the minimal complement of msp2 genes or pseudogenes in two strains of A. phagocytophila and the number of transcriptionally active msp2 gene paralogs during low-passage, steady-state, in vitro propagation. Of 15 BDS strain clones, 1 had a hypervariable region identical to the region in a clone obtained from a BDS strain genomic library previously prepared from organisms after only two horse passages. When 124 Webster strain clones were examined, 18 unique hypervariable regions were identified. Of 64 Webster strain cDNA clones, 56 (87.5%) were derived from a single gene, and transcripts from six additional msp2 genes were also identified. The sequences of several hypervariable regions that were ≥97% similar to regions present in other strains were identified by performing a BLAST analysis of sequences deposited in the GenBank database. These findings suggest that antigenic variability results from transcription of one or a few of the multiple paralogs and not from genetic instability that results in random accumulated mutations, although the possibility that gene recombination plays a role cannot be eliminated. The predominant Msp2 pattern in vitro is determined by transcription from a single gene.
Anaplasma (Ehrlichia) phagocytophila is a causative agent of granulocytic ehrlichiosis. Human infections are acute and usually self-limited, whereas infections of ruminants and rodents may be persistent (7, 24, 25). In addition, strains isolated from different geographic regions are antigenically variable whether they are from humans or animals (2, 6, 31). It is speculated that antigenic variability is related to evasion of immunity and persistent infection (3, 9, 17). The basis for antigenic variability is thought to be differential expression of major immunodominant outer membrane proteins encoded by members of a multigene family (msp2, p44, 44-kDa antigen gene) that is characterized by conserved sequences flanking a hypervariable region (12, 17, 30, 31). The exact role of Msp2 is not understood. However, homologous proteins present in the related bovine pathogen Anaplasma marginale are thought to be adhesins by which the bacterium is attached to the host erythrocytes (16, 19).
To date, multiple unique msp2 gene paralogs have been described (12, 17, 30, 31). However, the total number of gene paralogs in a single strain of A. phagocytophila has not been defined. Thus, we investigated the number of paralogs in the genomes of two low-passage strains and determined which paralogs were transcribed and to what degree each of the paralogs was transcribed during stable in vitro propagation.
(Some of the results were presented at the 101st General Meeting of the American Society for Microbiology, Orlando, Fla., 20 to 24 May 2001.)
MATERIALS AND METHODS
In vitro cultivation of A. phagocytophila.
The Webster and BDS strains of A. phagocytophila (human granulocytic ehrlichiosis agent) were propagated in HL-60 cells in RPMI 1640 medium supplemented with between 1 and 20% fetal bovine serum and 2 mM l-glutamine. Cells were examined for infection by Romanowsky staining three times per week, and the cell concentration was adjusted to between 2 × 105 and 106 cells/ml. When cultures were heavily infected (>70% of the cells were infected), they were harvested by centrifugation and used to prepare nucleic acids. The Webster strain was tested at in vitro passage 7, while the BDS strain was tested after two horse passages and five in vitro passages.
PCR and RT-PCR.
DNA was prepared from infected cells after centrifugation to remove tissue culture supernatant by using a QIAamp DNA mini kit (Qiagen, Valencia, Calif.). RNA was prepared from Webster strain-infected HL-60 cells by using an RNeasy mini kit and a QIAshredder (Qiagen). Genomic DNA contamination was eliminated by using RNase-free DNase (Qiagen) during extraction and again after preparation of the final RNA. The DNA and RNA concentrations of each preparation were determined.
Primers for amplification of msp2 were created from an alignment of 10 A. phagocytophila msp2 sequences from GenBank, including two msp2 sequences in λ clones (unpublished data) derived from the BDS strain after two horse passages. These primers were created to anneal with the highly conserved sequences flanking the hypervariable regions of msp2. Primers MSP465f (5′-TGATGTTGTTACTGGACAGA-3′) and MSP980r (5′-CACCTAACCTTCATAAGAA-3′) were used for both PCR and reverse transcription-PCR (RT-PCR) at a final concentration of 1 μM. Amplification from A. phagocytophila nucleic acids was determined by PCR by using DNA from both the BDS and Webster strains; RT-PCR was performed by using RNA from the Webster strain. The specificity of the primers was tested by using Escherichia coli and Ehrlichia chaffeensis DNA templates.
For DNA PCR, each 50-μl reaction mixture contained 1 μg of infected cell DNA template, each deoxynucleoside triphosphate at a concentration of 10 mM, and 1.5 mM MgCl2; a touchdown PCR method was used. Amplification was performed by using the following conditions: 94°C for 2 min, 2 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s, 2 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, 2 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, 2 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s, 26 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s, and finally 72°C for 5 min. Agarose gel electrophoresis and ethidium bromide staining were performed to ensure that an approximately 550-bp DNA fragment was present.
For RT-PCR, we used a one-step PCR protocol with RT-Taq (SUPERSCRIPT One-Step RT-PCR; Life Technologies, Gaithersburg, Md.) and 30 ng of RNA from infected cells as recommended by the manufacturer; touchdown PCR was performed as described above for the DNA PCR. DNA contamination in the RNA preparations was assessed by the same PCR method by substituting Taq for RT-Taq polymerase. Agarose gel electrophoresis and ethidium bromide staining were used to assess the RT-PCR products.
Cloning and sequencing of PCR products.
DNA PCR and RT-PCR products were cloned by ligation into the pCR4-TOPO vector (Invitrogen, Carlsbad, Calif.) and transformation into competent E. coli TOP 10 cells or by using a TA cloning kit (Invitrogen), the pCR2.1 vector, and transformation into competent E. coli DHα2 cells. Transformed cells were plated onto Luria-Bertani agar supplemented with 50 μg of kanamycin per ml. For the BDS strain colonies, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added to the Luria-Bertani agar plates for blue-white color selection. After overnight incubation at 37°C, 15 colonies were selected from the BDS strain DNA transformation preparations and 64 and 63 colonies were selected from the Webster strain RNA and DNA preparations, respectively. The clones were expanded, and the plasmids were purified (Wizard Plus Minipreps DNA purification system; Promega, Madison, Wis.). The plasmids were assessed to determine whether the insert was the correct size after digestion with the EcoRI restriction enzyme.
Sequence analysis of recombinant clones.
To sequence the plasmids obtained from the Webster strain, a BigDye terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.) was used as recommended by the manufacturer; each 20-μl reaction mixture contained 175 ng of plasmid and M13 forward and reverse primers. PCR amplification was performed under the following conditions: 6 cycles of 96°C for 10 s and 65°C for 1 min, 6 cycles of 96°C for 10 s and 64°C for 1 min, 6 cycles of 96°C for 10 s and 63°C for 1 min, 6 cycles of 96°C for 10 s and 62°C for 1 min, 6 cycles of 96°C for 10 s and 60°C for 1 min, 6 cycles of 96°C for 10 s and 59°C for 1 min, 6 cycles of 96°C for 10 s and 58°C for 1 min, 6 cycles of 96°C for 10 s and 57°C for 1 min, 12 cycles of 96°C for 10 s and 55°C for 1 min, and then incubation at 22°C. PCR products were purified by using Centrisep spin columns (Princeton Separations, Adelphia, N.J.). Sequences were determined by automated fluorescent dideoxynucleotide cycle sequencing by using a model 377 sequencer (Applied Biosystems, Foster City, Calif.). BDS strain plasmids were sequenced in a similar fashion except that the T7 and T3 sequencing primers were used in the PCR. All sequence data were assembled by using the Assemgel program (PCGENE; Intelligenetics), and derived sequences were aligned by using the CLUSTALW program, version 1.4 (27). Phylogenetic dendrograms constructed with CLUSTALW were visualized by using the TreeView program, version 1.6.1 (18). Nucleotide variability in the amplicons sequenced was assessed by determining entropy at each position using the Entropy H(x) program in Bioedit Sequence Alignment Editor, version 5.0.7 (10).
Nucleotide sequence accession numbers.
The nucleotide sequences of the Webster strain msp2 hypervariable regions have been deposited in the GenBank database under accession numbers AF443399 (WMSP1), AF443405 (WMSP2), AF443406 (WMSP3), AF443407 (WMSP4), AF443409 (WMSP5), AF443411 (WMSP6), AF443412 (WMSP9), AF443400 (WMSP10), AF443413 (WMSP13), AF443401 (WMSP15), AF443402 (WMSP16), AF443396 (WMSP17), AF443404 (WMSP19), AF443408 (WMSP45), AF443410 (WMSP52), AF443403 (RWMSP11), AF443397 (RWMSP25), and AF443398 (RWMSP57), and the nucleotide sequences of the BDS strain msp2 hypervariable regions have been deposited in the GenBank database under accession numbers AF443419 (MSP1), AF443418 (MSP3), AF443416 (MSP8), AF443415 (MSP9), AF443414 (MSP14), and AF443417 (MSP15).
RESULTS
PCR amplification of msp2 with conserved primers.
A 550-bp PCR product was obtained by using the A. phagocytophila BDS and Webster strains as DNA and RNA templates. DNA amplification was not observed with DNA templates from either E. coli or E. chaffeensis or with any RNA template when Taq polymerase lacking reverse transcriptase activity was used.
BDS strain msp2 partial sequencing.
Fifteen BDS strain amplicon clones from DNA preparations were selected, and 14 of these clones were successfully sequenced and assembled. Six unique paralogs of msp2 were identified from these clones. The sequence of one clone was 99.4% identical to the paralogous part of a gene initially identified in the BDS strain genomic library created 7 years previously after only two horse passages. Hence, since five unique msp2 gene paralogs were identified originally in three λ clones from the BDS genomic library (unpublished data), one of which was represented almost identically in this limited analysis, at least 10 msp2 gene paralogs are present in this strain. The msp2 sequences are characterized by conserved regions flanking a hypervariable, approximately 280-bp region. Thus, the deduced sequence of the hypervariable domain of Msp2 is approximately 95 amino acids long.
Webster strain msp2 partial sequencing.
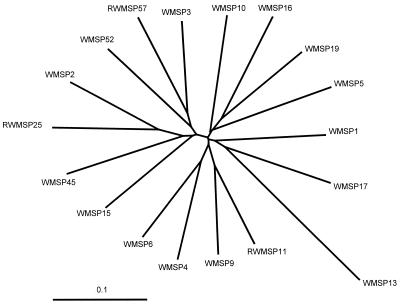
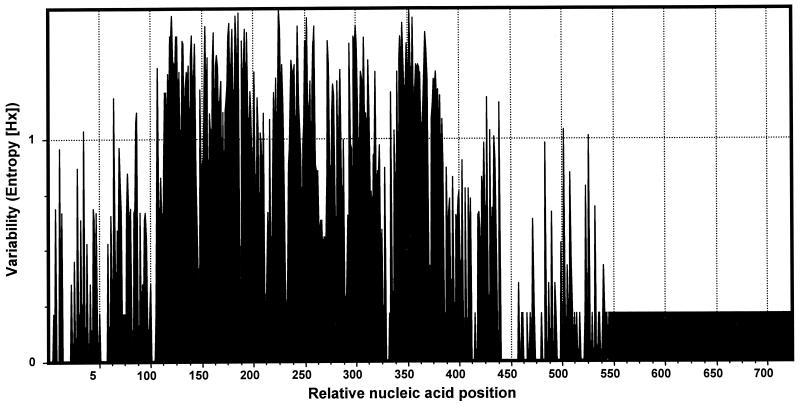
All 64 clones from Webster strain RNA preparations and 60 of 63 clones from the DNA preparations were successfully sequenced. Eighteen unique hypervariable regions were identified in the 124 clones sequenced (Fig. 1); these regions included 11 regions in clones obtained only from DNA preparations, 3 regions in clones obtained only from RNA preparations, and 4 regions in clones obtained from both types of preparations. The majority (87.5%) of the cDNA clones were transcribed from a single msp2 gene, but clones from transcripts of six additional msp2 genes were also identified. The remaining 11 msp2 genes were not transcriptionally active. The 18 msp2 genes were characterized by similar conserved and hypervariable regions (Fig. 2), as observed for the BDS strain, but they included a hypervariable region that was approximately 330 bp long, probably because of the larger number of sequences compared. When the sequences were aligned with predicted open reading frames for fully sequenced msp2, amino acid sequences were determined, and we identified conserved N- and C-terminal peptide sequences flanking a central hypervariable domain that was approximately 109 amino acids long. No hypervariable sequences were found to encode preliminary stop codons.
FIG. 1.
Radial dendrogram of aligned msp2 amplicon sequences from the Webster strain of A. phagocytophila. The tree was constructed from an alignment of 124 sequences by using the neighbor-joining algorithm and was simplified to show only representative members of the 18 unique clades. The names of the sequences were assigned arbitrarily; the prefix RWMSP indicates sequences derived solely from cDNA amplicons. Scale bar = 0.1 nucleotide substitution per site.
FIG. 2.
Variability of nucleotides at each position in 18 aligned msp2 amplicon sequences (lacking the conserved primer sequences) from the Webster strain of A. phagocytophila, determined by using an entropy plot. Greater entropy corresponds to greater nucleotide variability at a position.
One partial msp2 sequence that was identified was substantially longer (681 bp) than other sequences because of the insertion of approximately 125 bp into the 3′ end of the amplicon. The sequence of the insertion was not similar to sequences found in any msp2 gene or in GenBank by a BLAST search (1).
DISCUSSION
Msp2 is a major immunodominant protein expressed in both A. marginale and A. phagocytophila. In both species, msp2 genes are members of multigene families with high degrees of nucleic acid sequence conservation (12, 17, 30, 31). The structure of msp2 is characterized by conserved sequences flanking a hypervariable region that may allow generation of antigenic variation and persistent infection in reservoir hosts (3, 9, 12, 20, 23, 30). The Msp proteins of A. phagocytophila clinical isolates are also heterogeneous, depending in part on geographic origin (2, 31). Preliminary studies have indicated that the Msp2 antigenic profile changes with passage in vitro (unpublished data).
A. phagocytophila animal reservoirs, including small rodents, ruminants, and possibly cervids, can sustain transient or persistent infections without clinical signs (7, 8, 24, 25, 28). The molecular mechanisms that underlie persistence in other bacteria (Borrelia burgdorferi, Neisseria gonorrhoeae) and in protozoan pathogens (Trypanosoma spp., Plasmodium falciparum) involve gene recombination or conversion that allows expression of new antigens and evasion of immunity (4, 21, 22, 26). Persistent infection with A. phagocytophila may occur in part because of the antigenic variability encoded by members of the msp2 gene family.
Our results show that there are at least 18 unique paralogs of the hypervariable region of the msp2 gene in the A. phagocytophila Webster strain and at least 10 paralogs in the BDS strain. Although multiple msp2 paralogs are present in A. marginale, the genes are widely distributed throughout the chromosome (3, 9). Similar proteins and genes have been identified in related genera and species, including E. chaffeensis and Ehrlichia canis, in which the p28 and p30 family genes are tandemly arranged on the chromosome. In E. chaffeensis, each p28 family gene possesses distinct promoter regions, and several regions are simultaneously transcribed (29).
It is not possible to ascertain if the 18 paralogs described above are true genes or pseudogenes by the PCR method which we employed. Brayton et al. (5) showed that in the related organism A. marginale, the msp2 and msp3 gene families are composed of local groups of pseudogenes that can be recombined into a single functional expression site to generate new antigenic variants and to presumably evade immunity. While the PCR approach used here is simple and yields abundant information, only statistical predictions can be made regarding the completeness of the analysis. In an attempt to obtain the sequences of all msp2 genes in one strain of A. phagocytophila, we successively sequenced 124 cloned amplicons. On the basis of the Poisson distribution, one would predict that increasing the number of clones sequenced would increase the number of unique sequences found until all sequences have been identified. With 18 unique sequences identified, the number of new clones that would need to be sequenced to identify a single new msp2 hypervariable region is defined by the formula n = log(1 − P)/log[18/(x + 18)], where n is the number of clones to be sequenced, P is the probability of identifying a new sequence, and x is the number of unique msp2 sequences not yet identified. Thus, for 80 and 90% probabilities (P = 0.8 and P = 0.9) that one remaining sequence (x = 1) would be identified, one would need to analyze 30 and 43 new clones, respectively. Since the final analysis was conducted with 34 new clones and identified no new msp2 sequences, the probability that all possible msp2 hypervariable region amplicons that could be identified by this method were found was between 80 and 90%. The depths of the branches in the dendrogram provide evidence of the uniqueness of each sequence and show that the results are not attributable to random errors during amplification. This conclusion is supported by the finding that the minimum number of different nucleotides observed for any two amplicons was 87, which is at least 29 times greater than the predicted maximum number of errors, 3.5 (2.1 × 10−4 error/bp/cycle), for a 550-bp amplicon generated by using Taq polymerase and 35 cycles for the PCR (information found at the U.S. Department of Commerce NOAA/NMFS/NWFSC website http://research.nwfsc.noaa.gov/protocols/taq-errors.html).
The model for antigenic variation in A. marginale implies that recombination of paralogous hypervariable pseudogenes at msp2 and msp3 transcription sites occurs (5). If this model is also applicable to A. phagocytophila, such recombination events might be associated with the rapid emergence of new random sequences and enormous antigenic diversity. However, long-term stability of at least one msp2 gene in A. phagocytophila is evident since it was reidentified in the chromosome of the BDS strain after several in vivo passages and multiple in vitro passages. Moreover, four of the six distinct BDS strain msp2 hypervariable regions were identical to sequences in clones derived from the Webster strain. Since both the Webster and BDS strains were obtained from human patients in adjacent regions of Wisconsin, genetic divergence might be expected to be minimal, a hypothesis confirmed in part by these results. Thus, given the degree of genetic stability demonstrated for these two strains, DNA rearrangement and accumulated mutations that create new gene sequences are probably minor or insignificant mechanisms for generation of antigenic variability. However, these findings are still consistent with the hypothesis that gene conversion occurs, as proposed for the related organism A. marginale (5).
During low-passage, steady-state, in vitro cultivation of A. phagocytophila, the majority of the msp2 transcripts were derived from a single locus. Although low-level transcription of other msp2 loci was detected, we could not determine whether this transcription represented simultaneous transcription from several loci in a homogeneous population, transcription from a single locus in a heterogeneous population dominated by a single clone, or both. Methods to propagate clonal populations of A. phagocytophila have not been devised yet, but when this is accomplished, it should increase our understanding of msp2 gene family transcriptional regulation and its role in Msp2 antigenic variability.
Msp2 variation may not only be important in evasion of immunity; it may also play a role in propagation of the bacteria in the host and vector. In A. marginale, msp2 remains stable throughout in vitro culture, in acute erythrocyte infections in bovines, and in tick salivary glands; gene transcription varies only during persistent infections (3). Moreover, new data suggest that Msp proteins are adhesins by which the bacterium adheres to the host erythrocytes and granulocytes (15; J. Park, S. Patil, K. Caspersen, and J. S. Dumler, Am. Soc. Rickettsiol. Bartonella Emerg. Pathogen Group 2001 Joint Conf., abstr. 61, 2001). Herron et al. (11) showed that a host ligand for A. phagocytophila is platelet selectin glycoprotein ligand 1 glycosylated by fucose and sialic acid in a specific configuration mediated by α-(1,3)-fucosyltransferase. However, Nelson and Goodman have demonstrated that some cells lacking α-(1,3)-fucosyltransferase but not platelet selectin glycoprotein ligand 1 become infected (C. M. Nelson and J. L. Goodman, Abstr. 15th Meet. Am. Soc. Rickettsiol., abstr. 65, 2000). Thus, one may hypothesize that alterations in Msp2 adhesins may also alter host cell specificity and perhaps biological or even clinical manifestations.
Msp proteins are the major antigenic stimuli between the obligately intracellular bacterium and the host (31). By using a murine model, it has been demonstrated that the histopathology associated with A. phagocytophila is determined not by the quantity of the bacterium present but by a host-mediated response to the organism that results in production of a proinflammatory response, including gamma interferon (13, 14). Whether Msp2 and changes in Msp2 expression are related to immunopathologic injury is not clear. It is also not clear what effects immunologic pressure, prolonged in vitro passage, and in vivo passage have on Msp2 expression. Further study should help us understand the pathogenesis of this disease, including the role of msp2 in persistence and host evasion of immunity, and may facilitate rational design of specific vaccines or therapeutic agents for this unique infection.
Acknowledgments
This study was supported in part by grant RO1-AI44102 from the National Institutes of Allergy and Infectious Diseases (to J.S.D.) and by National Research Service Award training grant RR07002 from the Public Health Service (to K.C.).
We thank the sequencing team of the Molecular Microbiology Laboratory of The Johns Hopkins Hospital for help with sequence analyses, Karl W. Broman, Department of Biostatistics, Johns Hopkins University School of Hygiene and Public Health, for assistance with statistical analyses, and J. S. Bakken for providing clinical samples for microbial isolation.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Asanovich, K. M., J. S. Bakken, J. E. Madigan, M. Aguero-Rosenfeld, G. P. Wormser, and J. S. Dumler. 1997. Antigenic diversity of granulocytic Ehrlichia species isolates from humans in Wisconsin, New York, and a California horse. J. Infect. Dis. 176:1029-1034. [DOI] [PubMed] [Google Scholar]
- 3.Barbet, A. F., J. Yi, A. Lundgren, B. R. McEwen, E. F. Blouin, and K. M. Kocan. 2001. Antigenic variation of Anaplasma marginale: major surface protein 2 diversity during cyclic transmission between ticks and cattle. Infect. Immun. 69:3057-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G., and B. I. Restrepo. 2000. Antigenic variation in vector-borne pathogens. Emerg. Infect. Dis. 6:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumler, J. S., K. M. Asanovich, J. S. Bakken, P. Richter, R. Kimsey, and J. E. Madigan. 1995. Serologic cross-reactions among Ehrlichia equi, Ehrlichia phagocytophila, and human granulocytic ehrlichia. J. Clin. Microbiol. 33:1098-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumler, J. S., and J. S. Bakken. 1996. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J. Infect. Dis. 173:1027-1030. [DOI] [PubMed] [Google Scholar]
- 8.Dumler, J. S., and J. S. Bakken. 1998. Human ehrlichioses. Newly recognized infections transmitted by ticks. Annu. Rev. Med. 49:201-213. [DOI] [PubMed] [Google Scholar]
- 9.French, D. M., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67:5834-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.Herron, M. J., C. M. Nelson, J. Larson, K. R. Snapp, G. S. Kansas, and J. L. Goodman. 2000. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science 288:1653-1656. [DOI] [PubMed] [Google Scholar]
- 12.Ijdo, J. W., W. Sun, Y. Zhang, L. A. Magnarelli, and E. Fikrig. 1998. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect. Immun. 66:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin, M. E., J. E. Bunnell, and J. S. Dumler. 2000. Pathology, immunohistology, and cytokine responses in early phases of HGE in a murine model. J. Infect. Dis. 181:374-378. [DOI] [PubMed] [Google Scholar]
- 14.Martin, M. E., K. Caspersen, and J. S. Dumler. 2001. Immunopathology and ehrlichial propagation are regulated by interferon-γ and interleukin-10 in a murine model of human granulocytic ehrlichiosis. Am. J. Pathol. 158:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGarey, D. J., and D. R. Allred. 1994. Characterization of hemagglutinating components on the Anaplasma marginale initial body surface and identification of possible adhesins. Infect. Immun. 62:4587-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGarey, D. J., A. F. Barbet, G. H. Palmer, T. C. McGuire, and D. R. Allred. 1994. Putative adhesins of Anaplasma marginale: major surface polypeptides 1a and 1b. Infect. Immun. 62:4594-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, C. L., J. R. Storey, J. Recchia, L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, J. S. Bakken, R. T. Coughlin, and G. A. Beltz. 1998. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect. Immun. 66:3711-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 19.Palmer, G. H., G. Eid, A. F. Barber, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62:3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer, G. H., F. R. Rurangirwa, and T. F. McElwain. 2001. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J. Clin. Microbiol. 39:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy, G. R., and C. P. Streck. 1999. Variation in the 28 kDa surface antigen protein multigene locus of isolates of the emerging disease agent Ehrlichia chaffeensis suggests that it plays a role in immune evasion. Mol. Cell. Biol. Res. Commun. 1:167-175. [DOI] [PubMed] [Google Scholar]
- 22.Reddy, G. R., C. R. Sulsona, A. F. Barbet, S. M. Mahan, M. J. Burridge, and A. R. Alleman. 1998. Molecular characterization of a 28 kDa surface antigen gene family of the tribe Ehrlichieae. Biophys. Res. Commun. 247:636-643. [DOI] [PubMed] [Google Scholar]
- 23.Rurangirwa, F. R., D. Stiller, and G. H. Palmer. 2000. Strain diversity in major surface protein 2 during tick transmission of Anaplasma marginale. Infect. Immun. 68:3023-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stafford, K. C., R. F. Massung, L. A. Magnarelli, J. W. Ijdo, and J. F. Anderson. 1999. Infection with agents of human granulocytic ehrlichiosis, Lyme disease, and babesiosis in wild white-footed mice (Peromyscus leucopus) in Connecticut. J. Clin. Microbiol. 37:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuen, S., E. O. Engvall, and K. Artursson. 1998. Persistence of Ehrlichia phagocytophila infection in lambs in relation to clinical parameters and antibody responses. Vet. Rec. 143:553-555. [DOI] [PubMed] [Google Scholar]
- 26.Taylor-Robinson, A., and R. S. Phillips. 1993. Protective CD4+ T-cell lines raised against Plasmodium chabaudi show characteristics of either Th1 or Th2 cells. Parasitol. Immunol. 15:301-393. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls, J. J., K. M. Asanovich, J. S. Bakken, and J. S. Dumler. 1998. Serologic evidence of a natural infection of white-tailed deer with the agent of human granulocytic ehrlichiosis in Wisconsin and Maryland. Clin. Diagn. Lab. Immunol. 5:762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, X. I., J. W. McBride, X. F. Zhang, and D. H. Walker. 2000. Characterization of the complete transcriptionally active Ehrlichia chaffeensis 28 kDA outer membrane protein multigene family. Gene 248:59-68. [DOI] [PubMed] [Google Scholar]
- 30.Zhi, N., N. Ohasi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major membrane proteins are expressed in human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 31.Zhi, N., Y. Rikihisa, H. Y. Kim, G. P. Wormser, and H. W. Horowitz. 1997. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J. Clin. Microbiol. 35:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]