Abstract
Neisserial Opa proteins function as a family of adhesins that bind heparan sulfate proteoglycan (HSPG) or carcinoembryonic antigen family (CEACAM) receptors on human host cells. In order to define the CEACAM binding domain on Opa proteins, we tested the binding properties of a series of gonococcal (strain MS11) recombinants producing mutant and chimeric Opa proteins with alterations in one or more of the four surface-exposed loops. Mutagenesis demonstrated that the semivariable domain, present in the first loop, was completely dispensable for CEACAM binding. In contrast, the two hypervariable (HV) regions present in the second and third loops were essential for binding; deletion of either domain resulted in loss of receptor recognition. Deletion of the fourth loop resulted in a severe decrease in Opa expression at the cell surface and could therefore not be tested for CEACAM binding. Chimeric Opa variants, containing combinations of HV regions derived from different CEACAM binding Opa proteins, lost most of their receptor binding activity. Some chimeric variants gained HSPG binding activity. Together, our results indicate that full recognition of CEACAM receptors by Opa proteins requires a highly coordinate interplay between both HV regions. Furthermore, shuffling of HV regions may result in novel HSPG receptor binding activity.
The obligate human pathogens Neisseria meningitidis and Neisseria gonorrhoeae colonize mucosal tissues in the respiratory and genitourinary tracts, respectively. The bacteria are thought to adhere to the mucosal epithelia by using multiple adhesins in a sequential manner. Primary attachment is mediated by the pilus, followed by a more intimate interaction via the opacity (Opa) proteins (35) present in the bacterial outer membrane (28, 40). The Opa proteins recognize distinct receptors present on epithelial cells. Two types of Opa receptors have been discovered so far. Certain Opa proteins bind to the cell surface heparan sulfate proteoglycans (HSPG) syndecan-1 and -4 (11, 15, 39), while other Opa proteins bind to members of the carcinoembryonic antigen (CEA) or CD66 family, recently renamed the CEACAM family (1, 9, 14, 42). CEACAMs can be found on epithelial cells and neutrophils (38), two cell types that are targeted by neisserial strains during natural infection. Some Opas are capable of recognizing both receptor types (7, 10). The interaction between Opa proteins and the CEACAM family members is highly specific; i.e., each Opa variant demonstrates a particular tropism for only certain members of the CEACAM receptor family (7, 10, 17). The binding of Opa protein is based on protein-protein interactions in the N-terminal part (N domain) of CEACAM glycoproteins (8). Some CEACAM family members, such as CEACAM4, -7, and -8, do not bind any Opa protein (30), although their N domains are up to 80% identical to the domains of the Opa binding family members CEACAM1, -3, and -6 and CEA (for historical reasons, the protein encoded by the CEACAM5 gene is still called CEA). We recently mapped the domain in CEA critically involved in Opa binding by homologue scanning mutagenesis. We found that a serine at position 32 in the CEA N domain was the major determinant for Opa protein binding, with less critical contributions of F29 and G41 (6). The importance of S32 was also demonstrated for Opa binding by CEACAM6 (30). In addition, the conserved residues Y34 and I91 in the CEACAM1 N domain were found to be necessary for Opa protein binding (43). While the CEACAM sequences required for Opa recognition have now been well characterized (reviewed in references 5 and 41), the CEACAM binding domain on Opa proteins remains to be elucidated.
Multiple opa genes can be found in the chromosomes of N. gonorrhoeae (11 or 12 loci) and N. meningitidis (3 or 4 loci) (4, 34). Many distinct opa alleles have been identified, since each neisserial strain contains a different repertoire of opa genes (23). The expression of Opa proteins is phase variable as a result of variation in the number of pentameric repeats within the signal peptide-encoding region (27, 33). This on-and-off switching of Opa protein expression may result in the presence of multiple Opa variants within one culture. Opa proteins are predicted to form an eight-stranded β-barrel in the outer membrane with four extracellular loops. Three of those loops contain variable sequence domains termed semivariable (SV) and hypervariable (HV-1 and HV-2) domains, while the fourth loop is highly conserved (23). The overall conservation of sequence and predicted structure outside the variable domains suggests that the different biological characteristics of individual Opa proteins are determined by the variable domains in the exposed loops. The involvement of HV regions in CEACAM recognition was indeed indicated by the finding that antibodies directed against the HV-2 region of an Opa protein of gonococcal strain FA1090 inhibited interactions with neutrophils (31). Furthermore, Virji et al. (43) found that two Opa variants of N. meningitidis strain C751 with identical SV and HV-2 regions differed in their interaction with CEACAM3 and CEACAM6, suggesting that HV-1 also is involved in CEACAM recognition.
In order to further understand the molecular basis for Opa-CEACAM recognition, we undertook the present study to define the domains on Opa proteins that mediate binding to CEACAM receptors. We investigated this in a systematic manner by constructing deletion mutant Opa proteins lacking variable domains and constructing chimeric mutant Opa proteins by exchange of variable domains between different Opas. The mutant Opa proteins, expressed in a non-phase-variable manner in their natural host the gonococcus, were then extensively tested to determine the contributions of each variable region to CEACAM recognition.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The gonococcal strain used in this study was MS11mk (37). Gonococci were maintained in a humidified, 5% CO2 atmosphere at 37°C on GC phosphate medium (35) with the appropriate antibiotic when required. For experiments, bacteria were grown for 3 h in 10 ml of HEPES medium (39) (10 mM HEPES, 145 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1 mM CaCl2, 5 mM glucose, and 1.5% Proteose Peptone no. 3 [Difco] [pH 7.4]) in a gyratory shaker at 37°C. Bacterial suspensions were pelleted and resuspended in 1 ml of HEPES buffer (HEPES medium without Proteose Peptone). Escherichia coli DHα was used as the host strain for all plasmid constructions.
Construction of recombinant Opa variants.
The pHermes6a-pTetM shuttle system was used to obtain gonococcal variants expressing recombinant, non-phase-variable Opa proteins. The pHermes6a plasmid contains an Opa promoter and a modified Opa leader sequence resulting in a locked-on expression status of the opa gene cloned behind this leader sequence. This construct is contained within a “shuttle box” that allows homologous recombination into the conjugative Neisseria plasmid pTetM25.2 (21, 22) The nonmodular chimeras were described by Grant et al. (16). The MS11 opaB gene was modified to construct a modular version by subcloning the NotI/HindIII opaB fragment of pEXB (2) into pCRII (Invitrogen) and introducing an SspI site and a ClaI site 3′ of the HV-1 and HV-2 region (16), resulting in plasmid pCRII-mBB. The NotI/HindIII opaB fragment from pEXB was replaced with the corresponding fragment of pCRII-mBB, resulting in plasmid pEX-mBB. HV regions were amplified from the pEXI and pEXC plasmids (2) by PCR using a 3′ primer containing an SspI site (HV-1) or a ClaI site (HV-2) and the 5′ M13 forward primer. HV-2 regions were exchanged in the pEX-mBB plasmid by digestion and ligation via the ClaI sites. Because the pEX-mBB plasmid contains two SspI sites outside the opa region, the HV-1 regions were ligated into SspI-cut pCRII-mBB. The resultant chimeric opa gene was isolated as a NotI/HindIII fragment and ligated into NotI/HindIII-digested pEX-mBB. The resulting plasmids pEX-mXX (where X stands for B, C, or I) were transformed into DH5α and were sequenced through the entire opa gene to verify constructions. The chimeric opa genes were subcloned in the shuttle vector pHermes6a (21) by HindIII and NotI digestion-ligation of pHermes6a-OpaA (16). After transformation into DH10B Max Efficiency cells (Gibco Life Sciences) and selection on 300 μg of erythromycin per ml, transformants were screened for Opa expression by immunoblotting of whole-cell lysates with anti-Opa antibody 4B12 (a generous gift of M. Blake, North American Vaccines). Plasmids from positive transformants were isolated and used for transformation of gonococcal strain MS11 containing the pTetM plasmid plus a deletion in the gene coding for the major HSPG binding Opa of MS11 (OpaA) (16). Gonococcal transformants were selected on GC plates containing 7 μg of erythromycin per ml, and Opa expression was verified by immunoblotting of whole-cell lysates with 4B12 antibody. E. coli variants expressing chimeric Opa proteins were obtained by transformation of the appropriate plasmid pEX-mXX into strain HB101. The pEX-Opa plasmids encode β-lactamase-Opa fusion proteins that correctly insert into the E. coli outer membrane (2).
SV deletions were constructed as described before (16) in the pCRII-mXX plasmids. HV regions were deleted by removal of SspI (HV-1) or ClaI (HV-2) fragments from pCRII-mXX plasmids. The deletion constructs were moved into the E. coli expression plasmid (pEX) or the shuttle vector for gonococcal expression (pHermes6a) as described above. Variants expressing native recombinant Opa proteins were constructed by moving the NotI/HindIII Opa fragment from pEXB, pEXC, or pEXI into pHermes6a, followed by transformation of MS11.
Surface exposure of chimeric and deletion Opa proteins.
Bacteria (5 × 107) were incubated in 0.5 ml of HEPES buffer containing 30 μg of trypsin per ml (Sigma) for 15 min at room temperature. Twenty-five microliters of fetal calf serum was added, and the bacteria were collected by centrifugation (5 min at 2,000 × g), washed once with 1 ml HEPES buffer containing 5% fetal calf serum, and processed for immunoblotting with 4B12 antibody. Correct outer membrane insertion of the Opa protein was inferred from disappearance of the intact Opa band on the blot in the trypsin-treated samples under conditions where the amount of porin in the cell lysate was unchanged as judged by Coomassie brilliant blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Immunoblotting.
Whole-cell lysates were prepared by boiling cells in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. An equivalent of 2 × 107 cells was run on SDS-13% polyacrylamide gels and transferred to nitrocellulose filters. Opa proteins were visualized by incubation with 4B12 antibody followed by protein A-HRP (Sigma, 1:20.000). Blots were developed with the enhanced chemiluminescence protocol (ECL) (Amersham).
Infection experiments.
Stably transfected HeLa cell lines expressing defined CEACAM molecules were a generous gift from F. Grunert (Institut für Immunbiologie, Freiburg, Germany) (3, 13) and were cultured as described previously (7). Gonococci (1.5 × 107) were added to 2 × 105 cells cultured on 12-mm-diameter glass coverslips in 1 ml of RPMI for 3 h at 37°C and 5% CO2. Nonadherent bacteria were removed by three washes with phosphate-buffered saline, and infected cells were fixed with 2% paraformaldehyde in phosphate-buffered saline. Extracellular bacteria were stained by incubation with an antilipopolysaccharide antibody followed by incubation with protein A-gold and silver enhancement (7). This results in a black appearance of bacteria that were accessible to the antilipopolysaccharide antibody and thus not internalized. Intracellular bacteria and HeLa cells were then visualized by counterstaining with 0.005% crystal violet. Numbers of intra- and extracellular bacteria were evaluated by light microscopy for at least 50 cells per coverslip. When appropriate, heparin (ICN Chemicals) was added at a concentration of 50 μg/ml 5 min before addition of the bacteria. For E. coli infections, bacteria were left on the cells for 5 h, after which fixed infected cells were stained with 0.005% crystal violet. Each bacterial suspension used in any infection experiment was tested for expression of the desired Opa protein by electrophoresis and immunoblotting of whole bacterial cell lysates. The presence of a single band of the correct molecular weight, detected with anti-Opa antibody 4B12, was taken as confirmation of usage of the correct Opa variant. The results shown came from at least three different independent experiments.
Binding of soluble receptors to gonococcal Opa variants.
Binding assays were performed as described previously (8). Briefly, 108 bacterial cells were incubated in 200 μl of HEPES buffer for 20 min at 37°C in the presence of the appropriate receptor preparation. Then bacteria were spun, washed with HEPES buffer, pelleted, and boiled in SDS-PAGE sample buffer. Full-length CEA was derived from HeLa-CEA cells that were treated with phosphotidyl inositol-phospholipase C (ICN) to release CEA. The presence of multiple bands reactive with anti-CEA antibodies in this preparation is due to different glycosylation forms of CEA. A cleared lysate of E. coli BL21 cells expressing His6-tagged CEA N domain served as the source for the CEA N domain. Binding of the receptor was measured by immunoblotting of whole-cell lysates with rabbit polyclonal anti-CEA antibodies (Dako) in case of the full-length CEA or with a mouse monoclonal anti-His antibody (Pharmacia) for CEA N-domain detection. After incubation with protein A-HRP, bands were visualized using ECL (Amersham). Binding assays were performed at least twice, and results of representative experiments are shown.
RESULTS
Binding of soluble CEA to chimeric Opa variants.
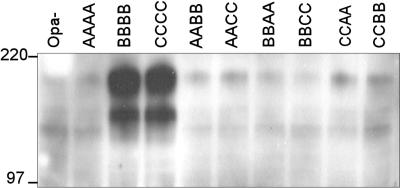
The 11 Opa proteins of gonococcal strain MS11 show different patterns of receptor binding capacities: OpaA is unique in that it strongly binds HSPG but does not bind any CEACAM receptor; OpaB, OpaC, OpaG, and OpaI bind CEACAM1, -3, and -6 and CEA; OpaD, OpaE, OpaF, OpaH, and OpaJ bind CEACAM1 and CEA but not CEACAM3 or -6; and OpaK binds weakly to only CEACAM1. Furthermore, OpaC, OpaF, and OpaH demonstrate low levels of HSPG binding (7, 10, 17). For a previous study where we investigated HSPG binding domains in the OpaA protein of strain MS11, we constructed MS11 variants expressing chimeric Opa proteins. These chimeric Opa proteins consisted of the N-terminal half (SV and HV-1 region) or C-terminal half (HV-2 and conserved-loop region) of the HSPG binding protein OpaA, while the other half of the molecule was derived from a CEACAM binder, i.e., OpaB or OpaC. These variants were referred to using a four-letter designation indicating the origin of their surface-exposed regions, e.g., AABB (16). We tested these chimeric variants for CEA recognition by measuring their ability to bind this receptor in solution. Soluble full-length CEA was obtained by phospholipase C treatment of HeLa-CEA cells. As expected, the native Opa variants BBBB and CCCC readily bound CEA, while the Opa− and AAAA variants did not (Fig. 1). Surprisingly, none of the chimeric variants bound CEA, not even the chimeras BBCC and CCBB, which contained HV regions of two CEA binding Opa proteins. Apparently, combining HV regions of different CEA binding Opas did not preserve CEA receptor binding activity. Furthermore, these results show that an HV region derived from the non-CEA binder OpaA cannot replace the function of the corresponding HV region of a CEA binder such as OpaB or OpaC.
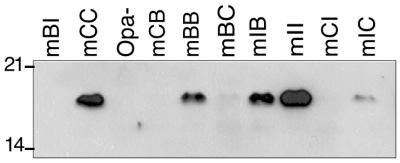
FIG. 1.
Binding of soluble CEA to gonococci expressing chimeric Opa proteins. Bacteria were incubated with CEA in HEPES buffer, spun, and washed. Cell pellets were boiled, separated by SDS-PAGE, blotted onto nitrocellulose, and probed with rabbit polyclonal anti-CEA antibodies, followed by protein A-HRP. Detection was with the ECL kit from Amersham. Numbers on the left are molecular masses in kilodaltons.
Construction of modular Opa variants.
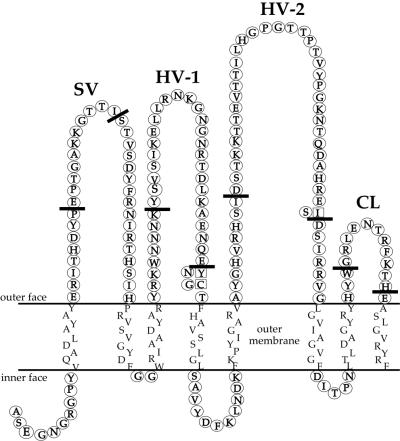
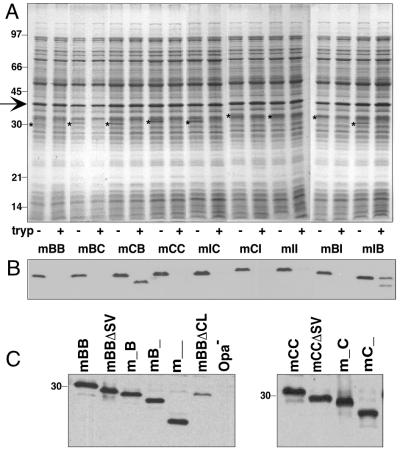
Because of these results, we decided to investigate the CEACAM recognition domain(s) on Opa proteins in more detail. As mentioned above OpaB-, OpaC-, OpaG-, and OpaI-expressing gonococci recognize the largest repertoire of CEACAM family members. Therefore, we considered these the most interesting to use to identify the Opa domains involved in CEACAM binding. Since OpaB and OpaG are virtually identical (4), we used only OpaB for further studies. Mapping of critical Opa protein domains was carried out by the construction and testing of a series of deletion mutants and chimeric Opa proteins. In order to facilitate construction of these recombinant Opa deletions and chimeras we designed a modular opa gene by introducing restriction sites into the native opaB gene to allow for deletion and exchange of loop-encoding regions. These insertions resulted in three amino acid residue changes in the mature OpaB protein (Fig. 2). This slightly altered opaB gene is referred to as modular opaB and served as a backbone to construct new deletion and chimeric variants. All variants based on this backbone are indicated as modular, marked by a lowercase m, followed by uppercase letters indicating the origins of the HV-1 and HV-2 regions, respectively. For instance, mBB indicates that both HV-1 and HV-2 are derived from OpaB; mBI indicates that the HV-1 region came from OpaB while the HV-2 region was derived from OpaI. All Opa proteins described in this study were expressed from the pTetM-Opa expression system, which results in non-phase-variable expression of the recombinant opa gene (21) carried by an MS11 strain in which the chromosomal HSPG binding opaA gene was inactivated (16). The correct insertion of the recombinant Opa proteins in the gonococcal outer membrane was confirmed by their sensitivity to cleavage by trypsin (Fig. 3A and B). The binding activities of the modular Opa variants mBB, mCC, and mII were tested in infection assays with HeLa cells transfected with CEACAM3, -6, or -8 or CEA and the parent HeLa-Neo cells, using the respective parent strains (carrying recombinant native OpaB, OpaC, and OpaI) as controls. The modular variants were indistinguishable in invasion and adherence properties from the native variants; i.e., they adhered to and invaded HeLa-CEACAM3, -CEACAM6, and -CEA cells but did not interact with HeLa-Neo or HeLa-CEACAM8 cells (data not shown). Therefore, we considered the modular variants appropriate tools for determining CEACAM binding domains of Opa proteins.
FIG. 2.
Predicted two-dimensional structure of OpaB protein (23). Thick lines in each loop indicate which portion of the loop was deleted or replaced. Amino acids that are different in the modular OpaB protein compared to native OpaB are in squares, with the native residues shown in circles on the left. Sequences of HV loops that were introduced in the modular OpaB protein are given in Table 3. CL, conserved-loop domain.
FIG. 3.
Expression of deletion and chimeric Opa proteins in gonococcal strain MS11. (A) Expression and surface exposure of chimeric Opa proteins determined by limited trypsinolysis. Whole bacteria were exposed to trypsin as described in Materials and Methods. Cell lysates were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. Molecular mass standards are indicated on the left in kilodaltons. Arrow, porin protein; *, Opa proteins; − and +, absence or presence of trypsin (tryp), respectively. (B) Detection of Opa proteins by immunoblotting with anti-Opa antibody 4B12. Samples are identical to those shown in the gel in panel A. (C) Expression of deletion Opa variants. Whole-cell lysates were separated by SDS-PAGE, and Opa proteins were detected by immunoblotting with anti-Opa antibody 4B12. _, deletions of HV regions (i.e., m_B = mBB with its HV-1 loop deleted; mB_ = mBB with its HV-2 loop deleted).
Effect of deleting extracellular loops on CEACAM recognition.
To investigate which extracellular loop(s) of an Opa protein confers binding to CEACAM receptors, we deleted the major part of each of the four loops separately and in combinations and tested the mutant proteins for recognition of different CEACAM receptors in infection assays. The deleted regions of each loop are indicated in Fig. 2. Western blotting of cell extracts of the different mutants showed that they produced similar amounts of Opa protein, except for the fourth-loop deletion mutant (Fig. 3C). Limited trypsinolysis of intact cells demonstrated that the deletion mutant Opa proteins were exposed on the cell surface (data not shown), similar to what we showed previously for OpaA deletion mutants (16). Results of infection assays performed with mBB and mII deletion variants on CEACAM3-, CEACAM6-, and CEA-transfected HeLa cells are shown in Table 1. Deletion of the SV region in mBB or in mII did not affect recognition of any type of CEACAM receptor. In contrast, deletion of either one of the HV loops in mBB or mII completely abolished invasion into any CEACAM-expressing cell line. The HV deletion variants of mII also lost adherence to the tested cell lines. The mBB mutants with a deletion in either of the HV regions no longer adhered to HeLa-CEACAM3 or -6 cells and demonstrated reduced adherence to HeLa-CEA cells. Recognition of CEA was completely lost when both HV regions were deleted. The mCC deletion mutants demonstrated a pattern of invasion and adherence similar to that of the mBB deletion mutants (data not shown). Deletion of the fourth loop was tested only in variant mBB; it resulted in abolition of all receptor binding. However, the Opa expression level of this deletion mutant was much lower than that of the parental strain (Fig. 3C), as was also found previously for an OpaA fourth-loop deletion mutant (16). This lack of expression may be due to problems with the transport to or stability of the mutant protein in the outer membrane. Hence, we were unable to determine a possible direct role of the fourth loop in CEACAM binding. None of the deletion variants invaded or bound to HeLa-Neo or HeLa-CEACAM8 cells (data not shown), indicating that no alternative, i.e., non-CEACAM-related, binding events were responsible for any of the observed interactions. Together, the data indicate that both HV regions are critically important for CEACAM binding while the SV region does not appear to be required in CEACAM receptor recognition. Moreover, the chimeric variant ABBA, which was constructed during our previous studies on the HSPG binding domain of OpaA, readily bound and invaded HeLa-CEACAM3, -CEACAM6, and -CEA cells, indistinguishably from the behavior of the BBBB variant (data not shown), indicating that even the presence of an SV region of a non-CEACAM binding Opa does not interfere with CEACAM recognition.
TABLE 1.
Infection of HeLa-CEACAM cell lines with Opa deletion mutants
| Receptor type | Infection assay resulta with:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mBB
|
mBB ΔSV
|
mBB ΔHV-1
|
mBB ΔHV-2
|
mBB ΔHV-1+2
|
mII
|
mII ΔSV
|
mII ΔHV-1
|
mII ΔHV-2
|
||||||||||
| in | adh | in | adh | in | adh | in | adh | in | adh | in | adh | in | adh | in | adh | in | adh | |
| CEACAM3 | ++ | ++ | ++ | ++ | − | − | − | − | − | − | ++ | ++ | ++ | ++ | − | − | − | − |
| CEACAM6 | ++ | ++ | ++ | ++ | − | − | − | − | − | − | ++ | ++ | ++ | ++ | − | − | − | − |
| CEA | ++ | ++ | ++ | ++ | − | + | − | + | − | − | ++ | ++ | ++ | ++ | − | − | − | − |
Results of infection assays obtained by microscopic evaluation of invasion (in) and adherence (adh) of bacteria. ++ for CEACAM3, 10 to 15 bacteria per cell (in) or 10 to 15 bacteria per cell (adh); ++ for CEACAM6 and CEA, 4 to 8 bacteria per cell (in) or 40 to 50 bacteria per cell (adh);+, 15 to 20 bacteria per cell;−, 0 bacteria per cell (in) or 0 to 5 bacteria per cell (adh).
Interaction of chimeric Opa proteins with CEACAM receptors expressed by HeLa cells.
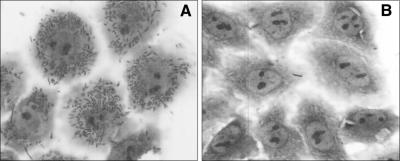
The results with the Opa deletion mutants suggest that both HV loops are important for CEACAM receptor binding. This could imply that each loop directly binds to the receptor, which might be suggested by the low level of binding to HeLa-CEA cells by the single HV region deletions of mBB and mCC, or that one loop contains the binding domain and the other loop acts in support of the binding loop to maintain conformational stability. Neither possibility can be excluded on the basis of the results obtained with the deletion mutants, as in each case they have only one HV loop. To address this question and define the role of HVs in CEACAM binding more precisely, we tested the effect of shuffling HV regions by constructing modular chimeras containing HV-1 and HV-2 loops of OpaB, OpaC, or OpaI. Opa expression levels and surface exposure of these chimeras were similar to those of the modular parental Opas (Fig. 3A and B). The results of infection assays performed with these chimeras on different CEACAM transfectants are shown in Table 2. Infection of the control HeLa-Neo cells, which do not express any CEACAM receptor, surprisingly showed significant binding of the chimeras mIB, mIC, and mCB. Adherence of mIB and mIC was completely inhibited by heparin, indicating that the binding likely occurred through recognition of a proteoglycan (Table 2). Similar heparin-inhibitable binding of mIB and mIC was observed with the human conjunctiva-derived Chang cell line (data not shown), indicating that the binding of mIB and mIC is not mediated by a HeLa cell-specific determinant. Also, when the mIC and mIB proteins were expressed and tested in an E. coli background, heparin-inhibitable binding to HeLa-Neo cells was again observed (Fig. 4), demonstrating that the binding is indeed due to the chimeric Opa protein. Adherence by the mCB-expressing gonococci was very heterogeneous (i.e., these bacteria bound to only about 30% of the HeLa transfectants tested); this binding was not affected by the presence of heparin. The nature of this mCB binding is unknown and is currently under investigation. To exclude any contribution of proteoglycan-mediated binding, further experiments with the CEACAM-expressing cell lines were performed in the presence of heparin.
TABLE 2.
Infection of HeLa-CEACAM cell lines with chimeric Opa variants
| Opa variant | Infection assay resulta with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HeLa-Neo cells
|
Hela-Neo cells + heparin
|
Hela-CEACAM3 cells + heparin
|
Hela-CEACAM6 cells + heparin
|
Hela-CEA cells + heparin
|
||||||
| in | adh | in | adh | in | adh | in | adh | in | adh | |
| mBB | − | − | − | − | ++ | ++ | ++ | ++ | ++ | ++ |
| mCC | − | − | − | − | ++ | ++ | ++ | ++ | ++ | ++ |
| mII | − | − | − | − | ++ | ++ | ++ | ++ | ++ | ++ |
| mBC | − | − | − | − | − | − | − | − | − | + |
| mCB | − | + | − | + | − | + | − | + | − | + |
| mBI | − | − | − | − | − | − | − | + | − | ++ |
| mIB | − | ++ | − | − | − | − | − | − | − | ++ |
| mCI | − | − | − | − | − | − | − | − | − | ++ |
| mIC | − | ++ | − | − | − | − | − | − | − | ++ |
Results of infection assays obtained by microscopic evaluation of invasion (in) and adherence (adh) of bacteria. ++ for HeLa-Neo cells, 20 to 30 bacteria per cell; ++ for HeLa-CEACAM3 cells, 10 to 15 bacteria per cell (in) or 10 to 15 bacteria per cell (adh); ++ for HeLa-CEACAM6 and HeLa-CEA cells, 4 to 8 bacteria per cell (in) or 40 to 50 bacteria per cell (adh); +, 10 to 20 bacteria per cell;−, 0 bacteria per cell (in) or 0 to 5 bacteria per cell (adh).
FIG. 4.
Crystal violet-stained HeLa-Neo cells infected with E. coli expressing the mIB protein in the absence (A) or presence (B) of heparin.
Infection experiments with the chimeric variants in the three HeLa-CEACAM cell lines showed that the chimeric Opa variants lost the ability to invade these cells, since none of these variants were ever found intracellularly, while the parent Opa variants were always readily taken up (Table 2). Furthermore, the chimeras no longer adhered to HeLa-CEACAM3 and -CEACAM6 cells (Table 2). Thus, combining two HV loops from different CEACAM binding Opa proteins did not preserve CEACAM3 or CEACAM6 recognition. In the case of CEA recognition, a more complex picture emerged. All chimeras lost the ability to enter HeLa-CEA cells, indicating a loss of CEA recognition. However, adherence properties of the chimeras were not completely lost. The mBC and mCB chimeras demonstrated a decrease in adherence, while binding by the mBI, mIB, mCI, and mIC chimeras was not significantly different from the binding by the parent Opa variants (Table 2).
Binding of soluble CEA N domain to chimeric Opa variants.
To further investigate the interaction of the chimeric Opa proteins with CEA, we examined whether the chimeric variants were capable of binding the soluble CEA N domain, which carries the Opa binding region of CEA. When Opa-CEA binding is of sufficient avidity, CEA N-domain binding corresponds with adherence to HeLa-CEA cells (8). As expected, the mBB, mCC, and mII variants readily bound the CEA N domain, while the Opa− variant did not (Fig. 5). The mIB- and mIC-producing variants both bound significant although various amounts of CEA N domain, while the mCI, mBI, mBC, and mCB variants did not bind the CEA N domain at all. The lack of binding by the mCB and mBC chimeras agrees with a similar lack of receptor binding by the BBCC and CCBB chimeras (Fig. 1), confirming the appropriateness of the use of modular variants. Apparently, the chimeras mBC, mCB, mBI, and mCI bind CEA with lower affinity than the native Opa proteins, since they adhere to HeLa-CEA cells but do not bind CEA in solution. A lower binding affinity is also suggested by the lack of entry of the chimeras into HeLa-CEA cells, keeping in mind that invasion likely requires higher-affinity interactions than adherence (20). The mIB and mIC variants may bind with intermediate affinity, allowing receptor binding in solution but not entry into receptor-expressing cells.
FIG. 5.
Binding of CEA N domain to gonococci expressing chimeric Opa proteins. Bacteria were incubated in HEPES buffer containing His6-tagged CEA N domain (8) for 20 min at 37°C, spun, and washed. Cell pellets were boiled, separated by SDS-PAGE, blotted onto nitrocellulose, and probed with a mouse monoclonal anti-His6 antibody, followed by protein A-HRP. Detection was with the ECL kit from Amersham. Numbers on the left are molecular masses in kilodaltons.
Overall, our data show that shuffling of HV regions from different CEACAM binding MS11 Opa proteins results in diminished receptor recognition, ranging from a complete loss of interaction for CEACAM3 and CEACAM6 to reduced binding in the case of CEA.
Properties of HV regions of CEACAM binding and nonbinding Opa proteins.
In an attempt to understand why only certain combinations of HV regions result in complete CEACAM recognition, we compared the primary sequences of the HV domains of CEACAM binders to those of nonbinders. Among the 11 MS11 opa genes, seven unique HV-2 regions can be discriminated. There are nine unique HV-1 regions, with two pairs differing in only two residues (4, 21). Allowing this two-residue mismatch, we compared seven unique HV-1 and HV-2 regions in terms of charge, hydrophobicity, and spacing of residues (Tables 3 and 4 ).
TABLE 3.
Sequences of HV regions of MS11 Opa proteinsa
| Region | Proteinb | Sequencec |
|---|---|---|
| HV-1 | 71 OpaB | NNNKYSVSIKELLR--NKGNGNR--TDLK |
| 75 OpaC | HNNKYSVNIKELERKNNKTSGGD-QLNIKYQK | |
| 52 OpaI | NNNKYSVNIENVRI-RKENGIR--IDRK | |
| 73 OpaD | NDNKYSVDIKELE---NKNQNKR---DLK | |
| 84 OpaF | NNNKYSVNIKELLRN DNANSGGNKHLNIKTRK | |
| 77 OpaK | NNSKYSVSIKELGRDDNSTSNSS-HLNIKTQK | |
| 58 OpaA | SDNKYSVSIKNMRV--HKHNSNR--KNLK | |
| HV-2 | 96 OpaB | RHSIDSTKKTTEVTTILHG--PGTTPTVYPGK----NTQNAHRESDSIRRV |
| 76 OpaC | RHGIDSTKKTKNTLTAYHG--AGTKPTYYDDIDSGKNQKNTYRQNRSSRRL | |
| 53 OpaI | RHSIDSTKKTIEVTTVPSNAPNGAVTTYNTDP----KTQNDY-QSNSIRRV | |
| 74 OpaD | RHSIDSTKKTTKFLTSSY---GGLNPTVYTEE----NTQNAHHQSNSIRRV | |
| 85 OpaF | RHQVRSVQQETIAVTTYPQ--NAASSVTTNAP----IRKLPHHESRSISSL | |
| 78 OpaK | KHQVRSVESETTTVTTH----NGAP--VPQGP----TPKPAYHKSRSISSL | |
| 59 OpaA | RHSIDSTKKITGLLTTST---PGIMSGVYKVL----RTPGAHRESDSIRRV |
HV regions of MS11 Opa proteins (sequences are according to reference 4) were defined as in reference 23 and aligned using ClustalW.
Numbers before Opa designations correspond to the numbers assigned to individual HV regions by Malorny et al. (23).
Basic residues are in boldface and acidic residues are in italic. Spaces (-) were introduced to maximize the alignment.
TABLE 4.
Characteristics of HV regions of MS11 Opa proteins
| Protein | CEACAM recognitiona | HV-1
|
HV-2
|
||||
|---|---|---|---|---|---|---|---|
| No. of residuesb
|
Net chargec | No. of residues
|
Net charge | ||||
| K, R, H | D, E | K, R, H | D, E | ||||
| OpaB | 1,3,6, CEA | 6 | 2 | +4 | 10 | 4 | +4.5 |
| OpaC | 1,3,6, CEA | 8 | 3 | +4.5 | 13 | 4 | +8 |
| OpaI | 1,3,6, CEA | 7 | 3 | +4 | 7 | 4 | +2.5 |
| OpaD | 1, CEA | 6 | 5 | +1 | 9 | 3 | +4.5 |
| OpaF | 1, CEA | 8 | 2 | +5.5 | 8 | 2 | +4.5 |
| OpaK | 1 | 6 | 3 | +2.5 | 8 | 2 | +4.5 |
| OpaA | None | 9 | 1 | +7 | 10 | 3 | +6 |
1,3,6, CEA, recognizes CEACAM1, -3, and -6 and CEA; 1, CEA, recognizes CEACAM1 and CEA; 1, recognizes CEACAM1 weakly; none, no recognition of any CEACAM family member.
Number of basic (K, R, H) or acidic (D, E) residues present in the HV region.
Net charge of the HV region, counting K and R as +1, H as +0.5, and D and E as −1 as described by Swanson (36).
The heparin binding capacity of proteins is determined by spacing of basic residues, often represented by a sequence of 3 basic amino acid residues interrupted by one nonbasic residue (25). The HV-1 region of OpaA, which is the strongest HSPG binder of MS11 Opa proteins, contains the sequence RVHK. Mutation of the arginine residue in this sequence abolishes HSPG binding by OpaA (data not shown), indicating that the RVHK sequence may be the HSPG receptor recognition domain. When all HV domains were checked for a BxBB sequence (where B stands for a basic amino acid residue and x stands for any amino acid residue), we found that the HV-1 domains of OpaF, OpaH, and OpaI and the HV-2 domain of OpaK contained this sequence. A BBxB sequence was found in the HV-2 domain of OpaC (Table 3). OpaF, OpaH, and OpaC have been shown to mediate low levels of HSPG binding, which may be attributed to the presence of a BxBB or BBxB sequence. Interestingly, the HV-1 domain of OpaI contains BxBB, which could possibly explain the observed HSPG recognition mediated by chimeras containing the OpaI HV-1 domain (mIB and mIC). Next, we looked at patterns of charged or hydrophobic residues in combinations of HV regions of CEACAM binders compared to nonbinders; this did not result in any obvious distinction. Overall charge of individual loops was calculated as described by Swanson (36). The total net charge of combinations of HV loops in native, CEACAM binding Opa proteins varied from +6.5 to +12.5, while the net charge of chimeric combinations varied from +6.5 to +12, suggesting that at least the overall charge is not the determining factor for CEACAM recognition.
DISCUSSION
The neisserial Opa protein family, as known today, consists of over 100 sequenced members, each carrying often unique surface-exposed domains. Nevertheless, many Opa proteins are functionally similar; i.e., they recognize the same host cell receptors. This might suggest that the Opa surface domains are functionally conserved and could be constantly exchanged without functional consequences. Our data indicate that this is not the case, at least not for the Opa proteins of strain MS11. Our findings may not necessarily be extrapolated to all Opa proteins, but the fact that we observed loss of function in all possible shuffled variants of three different MS11 Opa proteins suggests that this may be a general phenomenon among Opa proteins.
The Opa variants of strain MS11 that are recognized by the distinct CEACAM receptors contain very dissimilar SV, HV-1, and HV-2 regions within a highly conserved framework sequence. One would therefore possibly expect the CEACAM receptor binding domain to be present in the conserved rather than the variable parts of the different Opa proteins. Yet, our data show that both HV domains are indispensable for receptor binding. Although we cannot exclude the possibility that the HV domains are needed for a correct binding conformation in a conserved part of the Opa protein, we believe it unlikely that only very specific sets of HV regions would allow for this to happen. We consider it more likely that the binding domain resides in the HV regions. The finding that combining HV regions of fully competent CEACAM binders resulted in a decrease or even a loss of CEACAM binding suggests that the HV loops do not contain independent individual binding domains but that they cooperate to create a receptor binding domain. Only the combinations of HV loops present in the native Opa proteins resulted in full receptor recognition, indicating that only restricted sets of HV regions are capable of forming a functional receptor binding domain. The requirements for formation of this domain are not obvious from the primary sequences of Opa proteins, since we could not find distinct similarities in HV regions within the group of CEACAM binding Opa proteins, nor did we find a clear difference in primary sequence between CEACAM binding and nonbinding Opa proteins. Apparently, similar conformational motifs can arise from dissimilar primary sequences. The present work confirms and extends observations from previous studies with meningococci that produce naturally occurring Opa variants sharing HV regions. Two Opa variants of meningococcal strain C751 with similar HV-2 regions but distinct HV-1 regions were bound and taken up by HeLa-CEACAM3 and -6 cells in different amounts (43), indicating that the combination of HV-1 and HV-2 also determines the level of receptor recognition in a different bacterial background.
Full CEACAM recognition, resulting in invasion and adherence to epithelial cells, requires both HV regions of an Opa protein. This appears to be in contrast to HSPG binding, which can be conferred by one HV region (HV-1) in the case of the OpaA protein of strain MS11 (16). It is not unexpected that HSPG and CEACAM binding are mediated through different domains, since the addition of heparin, which completely blocks HSPG recognition, does not interfere with CEACAM recognition of dual-receptor binding Opa proteins (7). The surprising finding that the chimeric Opa variants mIB and mIC demonstrate HSPG binding activity, while the parent mBB, mCC, and mII variants do not, suggests that one or both or a combination of the HV loops forms an HSPG binding domain. This HSPG binding activity is apparently cryptic in the parental Opa protein. The fact that none of the HV deletion mutants bound to HeLa-Neo cells indicates that none of the OpaB, OpaC, or OpaI HV regions by itself is capable of binding HSPG. This implies also that for certain HSPG binding both HV loops are necessary and need to be compatible. In our previous study on HSPG recognition by OpaA, we found only a supportive role for the HV-2 loop of OpaA (16). Although both OpaA and the chimeras mIB and mIC bind HSPG, only binding through OpaA results in substantial invasion of cells, possibly due to a difference in binding affinity. In that respect, the mIB and mIC Opas resemble OpaC, OpaF, and OpaH, which also demonstrate heparin-inhibitable binding but no invasion of epithelial cells (7, 11). Thus, at least among the MS11 Opa proteins, strong HSPG binding can be mediated by only one HV loop, as is the case for OpaA, and alternatively, two compatible HV domains may also form a weaker HSPG binding site.
Recombinational reassortment of HV regions among opa genes occurs frequently in vivo (18, 19). Opa proteins are highly immunogenic, as indicated by the reactivity of convalescent-phase sera from meningitis patients with Opa proteins (29). An increase in anti-Opa antibodies is also observed after vaccination with meningococcal outer membrane vesicles (24, 32). Thus, on the one hand there must be strong pressure on the bacteria to change their Opa repertoire in order to evade the host's immune response. On the other hand, the receptor binding function of Opa proteins needs to be maintained. Apparently, the bacteria have found a way to deal with both aspects of survival in the host, at least for HSPG recognition, since shuffling of HV regions can lead to novel HSPG binding activity. For CEACAM recognition, we have found only a loss of binding after shuffling of HVs, but the number of chimeras we looked at may have been too small to conclude that novel CEACAM binding Opas cannot occur. The OpaB protein of meningococcal strain C751 can be regarded as a chimera of the other two C751 Opa proteins (OpaA and OpaD). (It should be noted that the Opa designations OpaA, OpaB, and OpaC, etc., are unrelated among strains; i.e., there is not necessarily any homology between the variable domains in OpaA of strain X versus OpaA of strain Y). In infection assays, higher numbers of OpaB-expressing meningococci than OpaA or OpaD variants were bound or taken up by HeLa-CEACAM3 and -CEACAM6 cells (43). Thus, this is an example where shuffling did not result in a loss of receptor recognition.
It is remarkable that our results on the CEACAM3 and CEACAM6 receptors appear more clear-cut than those concerning the CEA receptor. For CEACAM3 and CEACAM6 we found that any change in the combination of HV regions of three different Opa proteins resulted in a complete loss of recognition. For CEA, however, changes in HV regions had variable effects on recognition. Possibly, binding of CEACAM3 or CEACAM6 requires a high-affinity binding by Opa that is established only with a limited number of HV combinations. CEA binding may be achieved more readily by lower-affinity binding. In that regard, it is noteworthy that of the CEACAM binding Opas so far identified, only a minority bind CEACAM3 or CEACAM6 (7, 17, 26, 43). Recently it was shown that binding of only this CEACAM family member on neutrophils by Opa variants leads to apoptosis of the neutrophil (12). It is not yet clear what role this phenomenon may play in disease, but possibly the course of the natural infection can be affected by the capability of the infecting strain to express a CEACAM3-engaging Opa protein. Detailed understanding of Opa interaction with individual members of the CEACAM family may further elucidate this relevant issue.
In conclusion, we have shown that CEACAM binding by MS11 Opa proteins is dependent on the presence of two, matched HV domains which likely form a binding domain for CEACAM. The structural requirements of these binding domains may be complex, and further characterization will require analyses of purified proteins folded in native conformations. In addition, we found that cryptic HSPG receptor binding activity can be present in Opa proteins, which becomes functionally exposed upon recombinational reassortment of HV domains.
Acknowledgments
We thank Jos van Putten (Utrecht University, Utrecht, The Netherlands) for critical reading of the manuscript.
Editor: B. B. Finlay
REFERENCES
- 1.Beauchemin, N., P. Draber, G. Dveksler, P. Gold, S. D. Gray-Owen, F. Grunert, S. Hammarstrom, K. V. Holmes, A. Karlsson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von Kleist, C. Wagener, S. M. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252:243-249. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., T. Chen, J. Swanson, and S. H. Fischer. 1992. Human neutrophil response to recombinant neisserial Opa proteins. Mol. Microbiol. 6:1729-1737. [DOI] [PubMed] [Google Scholar]
- 3.Berling, B., F. Kolbinger, F. Grunert, J. A. Thompson, F. Brombacher, F. Buchegger, S. von Kleist, and W. Zimmermann. 1990. Cloning of a carcinoembryonic antigen family member expressed in leukocytes of chronic myeloid leukemia patients and bone marrow. Cancer Res. 50:6534-6539. [PubMed] [Google Scholar]
- 4.Bhat, K. S., C. P. Gibbs, O. Barrera, S. G. Morrison, F. Jahnig, A. Stern, E. M. Kupsch, T. F. Meyer, and J. Swanson. 1991. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5:1889-1901. [DOI] [PubMed] [Google Scholar]
- 5.Billker, O., A. Popp, S. D. Gray-Owen, and T. F. Meyer. 2000. The structural basis of CEACAM-receptor targeting by neisserial Opa proteins. Trends Microbiol. 8:258-260. [DOI] [PubMed] [Google Scholar]
- 6.Bos, M. P., D. Hogan, and R. J. Belland. 1999. Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J. Exp. Med. 190:331-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos, M. P., F. Grunert, and R. J. Belland. 1997. Differential recognition of members of the carcinoembryonic antigen family by Opa variants of Neisseria gonorrhoeae. Infect. Immun. 65:2353-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos, M. P., M. Kuroki, A. Krop-Watorek, D. Hogan, and R. J. Belland. 1998. CD66 receptor specificity exhibited by neisserial Opa variants is controlled by protein determinants in CD66 N-domains. Proc. Natl. Acad. Sci. USA 95:9584-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, T., and E. C. Gotschlich. 1996. CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc. Natl. Acad. Sci. USA 93:14851-14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, T., F. Grunert, A. Medina-Marino, and E. C. Gotschlich. 1997. Several carcinoembryonic antigens (CD66) serve as receptors for gonococcal opacity proteins. J. Exp. Med. 185:1557-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T., R. J. Belland, J. Wilson, and J. Swanson. 1995. Adherence of pilus− Opa+ gonococci to epithelial cells in vitro involves heparan sulphate. J. Exp. Med. 182:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, T., S. Bolland, I. Chen, J. Parker, M. Pantelic, F. Grunert, and W. Zimmermann. 2001. The CGM1a (CEACAM3/CD66d)-mediated phagocytic pathway of Neisseria gonorrhoeae expressing opacity proteins is also the pathway to cell death. J. Biol. Chem. 276:17413-17419. [DOI] [PubMed] [Google Scholar]
- 13.Daniel, S., G. Nagel, J. P. Johnson, F. M. Lobo, M. Hirn, P. Jantscheff, M. Kuroki, S. von Kleist, and F. Grunert. 1993. Determination of the specificities of monoclonal antibodies recognizing members of the CEA family using a panel of transfectants. Int. J. Cancer 55:303-310. [DOI] [PubMed] [Google Scholar]
- 14.Dehio, C., S. D. Gray-Owen, and T. F. Meyer. 1998. The role of neisserial Opa proteins in interactions with host cells. Trends Microbiol. 6:489-495. [DOI] [PubMed] [Google Scholar]
- 15.Freissler, E., A. Meyer auf der Heyde, G. David, T. F. Meyer, and C. Dehio. 2000. Syndecan-1 and syndecan-4 can mediate the invasion of OpaHSPG-expressing Neisseria gonorrhoeae into epithelial cells. Cell. Microbiol. 2:69-82. [DOI] [PubMed] [Google Scholar]
- 16.Grant, C. C. R., M. P. Bos, and R. J. Belland. 1999. Proteoglycan receptor binding by Neisseria gonorrhoeae MS11 is determined by the HV-1 region of OpaA. Mol. Microbiol. 32:233-242. [DOI] [PubMed] [Google Scholar]
- 17.Gray-Owen, S. D., D. R. Lorenzen, A. Haude, T. F. Meyer, and C. Dehio. 1997. Differential Opa specificities for CD66 receptors influence tissue interactions and cellular response to Neisseria gonorrhoeae. Mol. Microbiol. 26:971-980. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs, M. M., A. Seiler, M. Achtman, and J. G. Cannon. 1994. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol. Microbiol. 12:171-180. [DOI] [PubMed] [Google Scholar]
- 19.Hobbs, M. M., B. Malorny, P. Prasad, G. Morelli, B. Kusecek, J. E. Heckels, J. G. Cannon, and M. Achtman. 1998. Recombinational reassortment among opa genes from ET-37 complex Neisseria meningitidis isolates of diverse geographical origins. Microbiology 144:157-166. [DOI] [PubMed] [Google Scholar]
- 20.Isberg, R. R. 1991. Discrimination between intracellular uptake and surface adhesion of bacterial pathogens. Science 252:934-938. [DOI] [PubMed] [Google Scholar]
- 21.Kupsch, E. M., B. Knepper, T. Kuroki, I. Heuer, and T. F. Meyer. 1993. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 12:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupsch, E. M., D. Aubel, C. P. Gibbs, A. F. Kahrs, T. Rudel, and T. F. Meyer. 1996. Construction of Hermes shuttle vectors: a versatile system useful for genetic complementation of transformable and non-transformable Neisseria mutants. Mol. Gen. Genet. 250:558-569. [DOI] [PubMed] [Google Scholar]
- 23.Malorny, B., G. Morelli, B. Kusecek, J. Kolberg, and M. Achtman. 1998. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of neisserial Opa proteins. J. Bacteriol. 180:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandrell, R. E., and W. D. Zollinger. 1989. Human immune response to meningococcal outer membrane protein epitopes after natural infection or vaccination. Infect. Immun. 57:1590-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margalit, H., N. Fischer, and S. A. Ben-Sasson. 1993. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J. Biol. Chem. 268:19228-19231. [PubMed] [Google Scholar]
- 26.Muenzner, P., C. Dehio, T. Fujiwara, M. Achtman, T. F. Meyer, and S. D. Gray-Owen. 2000. Carcinoembryonic antigen family receptor specificity of Neisseria meningitidis Opa variants influences adherence to and invasion of proinflammatory cytokine-activated endothelial cells. Infect. Immun. 68: 3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, G. L., T. D. Connell, D. S. Barritt, M. Koomey, and J. G. Cannon. 1989. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell 56:665-692. [DOI] [PubMed] [Google Scholar]
- 28.Nassif, X., C. Pujol, P. Morand, and E. Eugene. 1999. Interactions of pathogenic Neisseria with host cells. Is it possible to assemble the puzzle? Mol. Microbiol. 32:1124-1132. [DOI] [PubMed] [Google Scholar]
- 29.Poolman, J. T., C. T. P. Hopman, and H. C. Zanen. 1983. Immunogenicity of meningococcal antigens as detected in patient sera. Infect. Immun. 40:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popp, A., C. Dehio, F. Grunert, T. F. Meyer, and S. D. Gray-Owen. 1999. Molecular analysis of neisserial Opa protein interactions with the CEA family of receptors: identification of determinants contributing to the differential specificities of binding. Cell. Microbiol. 1:169-181. [DOI] [PubMed] [Google Scholar]
- 31.Rest, R. F., and W. M. Shafer. 1989. Interactions of Neisseria gonorrhoeae with human neutrophils. Clin. Microbiol. Rev. 2(Suppl.):S83-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenqvist, E., A. Hoiby, E. Wedege, B. Kusecek, and M. Achtman. 1993. The 5C protein of Neisseria meningitidis is highly immunogenic in humans and induces bactericidal antibodies. J. Infect. Dis. 167:1065-1073. [DOI] [PubMed] [Google Scholar]
- 33.Stern, A., M. Brown, P. Nickel, and T. F. Meyer. 1986. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 47:61-71. [DOI] [PubMed] [Google Scholar]
- 34.Stern, A., and T. F. Meyer. 1987. Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol. Microbiol. 1:5-12. [DOI] [PubMed] [Google Scholar]
- 35.Swanson, J. 1978. Studies on gonococcus infection. XII. Colony color and opacity variants of gonococci. Infect. Immun. 19:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson, J. 1994. Effects of Opa proteins and lipooligosaccharides on surface charge and biological behavior of gonococci, p. 109-125. In C. I. Kado and J. H. Crosa (ed.) Molecular mechanisms of bacterial virulence. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 37.Swanson, J., O. Barrera, J. Sola, and J. Boslego. 1988. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J. Exp. Med. 168:2121-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. A., F. Grunert, and W. Zimmermann. 1991. Carcinoembryonic antigen family: molecular biology and clinical perspectives. J. Clin. Lab. Anal. 5:344-366. [DOI] [PubMed] [Google Scholar]
- 39.van Putten, J. P. M., and S. M. Paul. 1995. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 14:2144-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Putten, J. P. M., and T. D. Duensing. 1997. Infection of mucosal epithelial cells by Neisseria gonorrhoeae. Rev. Med. Microbiol. 8:51-59. [Google Scholar]
- 41.Virji, M. 2000. The structural basis of CEACAM-receptor targeting by neisserial Opa proteins: response. Trends Microbiol. 8:260-261. [DOI] [PubMed] [Google Scholar]
- 42.Virji, M., K. Makepeace, D. J. Ferguson, and S. M. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22:941-950. [DOI] [PubMed] [Google Scholar]
- 43.Virji, M., D. Evans, A. Hadfield, F. Grunert, A. M. Teixeira, and S. M. Watt. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34:538-551. [DOI] [PubMed] [Google Scholar]