Abstract
Saccharomyces cerevisiae Dbr1 is a 405-amino acid RNA debranching enzyme that cleaves the 2′-5′ phosphodiester bonds of the lariat introns formed during pre-mRNA splicing. Debranching appears to be a rate-limiting step for the turnover of intronic RNA, insofar as the steady-state levels of lariat introns are greatly increased in a Δdbr1 strain. To gain insight to the requirements for yeast Dbr1 function, we performed a mutational analysis of 28 amino acids that are conserved in Dbr1 homologs from other organisms. We identified 13 residues (His13, Asp40, Arg45, Asp49, Tyr68, Tyr69, Asn85, His86, Glu87, His179, Asp180, His231 and His233) at which alanine substitutions resulted in lariat intron accumulation in vivo. Conservative replacements at these positions were introduced to illuminate structure–activity relationships. Residues important for Dbr1 function include putative counterparts of the amino acids that comprise the active site of the metallophosphoesterase superfamily, exemplified by the DNA phosphodiesterase Mre11. Using natural lariat RNAs and synthetic branched RNAs as substrates, we found that mutation of Asp40, Asn85, His86, His179, His231 or His233 to alanine abolishes or greatly diminishes debranching activity in vitro. Dbr1 sediments as a monomer and requires manganese as the metal cofactor for debranching.
INTRODUCTION
Lariat RNAs, wherein the 2′-hydroxyl of an internal adenosine nucleotide is linked to the 5′ end of the intron, are formed naturally as splicing intermediates by Group II introns and by the pre-mRNA spliceosome (1–3). Y-shaped branched RNAs with 2′-5′ phosphodiester linkages are generated during trans-splicing in nematodes and trypanosomes (4–6). Bacterial multicopy single-stranded DNAs (msDNAs) are mixed branched structures, wherein the 2′-hydroxyl of an RNA molecule is linked via a phosphodiester bond to the 5′ end of a DNA (7). Branched polynucleotides have also been made synthetically using ribozymes (8), deoxyribozymes (9,10) or solid-phase chemistry (11).
The 2′-5′ phosphodiester linkages in lariat intron RNAs are cleaved by a debranching enzyme, Dbr1, that was discovered in HeLa cell extracts (12). The yeast DBR1 gene was identified in a genetic screen for mutations that affect Ty transposition (13). Chapman and Boeke (13) found that transposition frequency was reduced and the steady-state levels of excised lariat intron RNAs were greatly increased in a dbr1 mutant strain. Whereas the basis for the involvement of Dbr1 in Ty transposition is still not entirely clear (14–17), intron accumulation in a dbr1 mutant strain is a direct consequence of the failure to cleave the 2′-5′ phosphodiester linkage. The debranched linear intron is usually degraded by either the 5′→3′ exonucleases Xrn1 and Rat1 or the 3′→5′ exonucleolytic activities of the exosome (18,19). Some yeast lariat introns are processed by an endonucleolytic pathway involving Rnt1, the yeast ortholog of RNase III (20).
The DBR1 gene is not essential for viability in Saccharomyces cerevisiae and Δdbr1 cells show no discernible growth defect, despite the increased levels of lariat intron RNAs (13). Schizosaccharomyces pombe dbr1+ is also non-essential, however, the S.pombe dbr1null mutant grows more slowly than dbr1+, the mutant cells are significantly elongated and the amounts of lariat intron RNAs are increased (21). The intron accumulation phenotype and the slow growth defect of S.pombe dbr1null cells can be alleviated by expression of Dbr1 homologs from Caenorhabditis elegans, human or mouse (21–23).
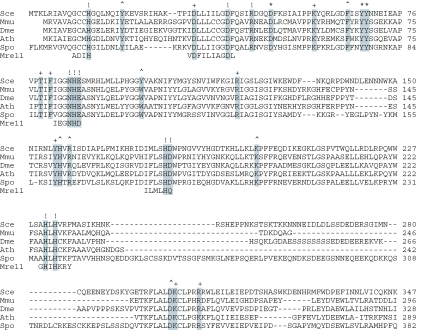
Dbr1 was suggested to be a member of the metallophosphoesterase superfamily that includes λ phosphoprotein phosphatase, calcineurin and Mre11 (24,25). The three signature motifs of this class of enzymes, which contain a dinuclear metal center, are DIH(X)∼25GDYVDR(X)∼27GNHE (25–29). Structural studies of bacteriophage λ phosphatase (a phosphomonoesterase) and Pyrococcus furiosus Mre11 (a phosphodiesterase) showed that conserved His, Asp and Asn side chains are involved in binding of the two manganese ions, whereas conserved His and Asn side chains contact the phosphate in the active site (28,29). Debranching enzymes contain a similar constellation of conserved amino acids (Figure 1).
Figure 1.
Structural conservation among debranching enzymes. The amino acid sequence of S.cerevisae Dbr1 (Sce) is aligned to the homologous proteins of Mus musculus (Mmu), Drosophila melanogaster (Dme), Arabidopsis thaliana (Ath) and Schizosaccharomyces pombe (Spo) and to the metallophosphoesterase motifs of Pyrococcus furious Mre11, a DNA phosphodiesterase for which a crystal structure has been determined. The 28 conserved residues of yeast Dbr1 that were replaced by alanine in the present study are highlighted in grey. The mutational effects of alanine substitutions with respect to accumulation of branched introns are denoted above the sequence as follows: (+) indicates that there is no effect; (^) indicates 14–22% intron accumulation in vivo compared to 100% in Δdbr1 cells; (*) indicates intron accumulation between 40 and 85% of the level in Δdbr1 cells; (!) indicates intron accumulation to ≥90% of the level in Δdbr1 cells.
Most metallophosphoesterases require transition metal ions (e.g. Mn2+ or Ni2+) for catalysis and are inhibited by zinc (26,30–32). The metal-requirement of yeast Dbr1 has not been assessed in detail because the debranching activity of purified recombinant Dbr1 was independent of added metals (33). In contrast, the debranching activity purified from HeLa cell extracts required added divalent cations and magnesium, calcium and manganese were effective cofactors for activity (34). Biochemical studies of yeast Dbr1, which routinely include magnesium in the reaction mixture, showed that a 20- to 100-fold molar excess of enzyme over substrate is typically required to obtain efficient debranching (33,35).
Here we show that Dbr1 is metal-dependent and acts catalytically only when manganese is included in the reaction mixture. We performed an alanine scan of yeast Dbr1 targeting 28 amino acids that are conserved among Dbr1 homologs from other species (Figure 1) and thereby identified 13 residues important for intron debranching activity in vivo. Structure–activity relationships were determined via conservative mutagenesis and selected mutant proteins were characterized with respect to their ability to catalyze debranching in vitro. Our findings indicate that Dbr1 is indeed a member of the metallophoshoesterase family, with an active site similar to that of Mre11.
MATERIALS AND METHODS
Yeast plasmids and targeted mutagenesis of DBR1
Plasmid p358-Dbr1 (CEN TRP DBR1) was generated by inserting the NcoI–XhoI fragment from pET16b-DBR1 (36) into p358-dbr1(+/−), a plasmid that contains 648 and 562 bp of upstream and downstream yeast genomic DNA flanking the DBR1 open reading frame (ORF), respectively. Mutations were introduced into the DBR1 gene by the two-stage PCR overlap extension method (37). Plasmid pET16b-DBR1 was used for the first amplification step. Residues targeted for substitutions were His13, Tyr20, Asp33, Asp40, Arg45, Asp49, Lys58, Phe65, Tyr68, Tyr69, Thr79, Phe81, Asn85, His86, Glu87, Tyr100, Arg120, Tyr156, His157, Arg159, His179, Asp180, Lys198, His231, His233, Asp300, Lys301 and Arg306. The mutated DNA products of the second-stage amplification were digested with restriction endonucleases and inserted into p358-dbr1(+/−) (CEN TRP1). In the resulting plasmids DBR1 is encoded as a His10 Dbr1 fusion protein. The presence of the desired mutations was confirmed by DNA sequencing and the segments corresponding to the inserted restriction fragments were then sequenced completely to exclude the acquisition of unwanted mutations during amplification or cloning. DBR1 mutants H13A, D33A, D40A, R45A, D49A, K58A, F65A, Y68A, Y69A, N85A, H86A, E87A, Y100A, R120A, H179A, D180A, H231A, H233A, D300A and R306A were also inserted into the pET16b vector for expression in bacteria.
Plasmids p132-DBR1, p132-mDBR1 and p132-SpDBR1 were generated by inserting the DBR1 ORF and the cDNAs for the DBR1 gene from mouse and S.pombe, respectively, into pYX132 (CEN TRP1) (Novagen). In this vector the genes are under the transcriptional control of the constitutive yeast TPI1 promoter. SpDBR1 was amplified from a S.pombe cDNA library (38) by PCR using oligonucleotides that introduced an NdeI site at the start codon and a BamHI site immediately downstream of the stop codon. A full-length cDNA clone for mDBR1 was purchased from ATCC (clone MGC-6384) and restriction sites were introduced during PCR amplification.
RNA isolation and northern blot analysis
The chromosomal DBR1 gene was deleted and replaced by LEU2 (36) in the yeast strain W303 to yield Δdbr1 strain YFK1 (Mat a ura3-52 trp1-63 his3-Δ200 leu2-1 ade2-101 lys2-801 dbr1::LEU2). To test the functionality of DBR1 mutant alleles in vivo, the p358-Dbr1 plasmids were introduced into the YFK1 strain. The empty vector pSE358 (TRP1 CEN) served as a negative control. Trp+ cells were selected and grown to mid-logarithmic phase (A600 = 0.6–0.8). Aliquots (∼5 × 108 cells) were harvested and RNA was isolated using the Qiagen RNA isolation kit according to the vendor's instructions. RNA concentration was calculated on the basis of A260. Equivalent amounts (15 µg) of total RNA from each sample were resolved by electrophoresis through a 1.4% agarose/formaldehyde gel and transferred to a Hybond membrane (Amersham). DNA fragments to be used as probes to detect the introns for ACT1, snR17A, RPS13 and CYH2 were generated by PCR. Radiolabeled probes were prepared using a random priming kit (Roche). Hybridization was performed as described (39) and the hybridization signal was quantified by scanning the membrane using a phosphorimager (Molecular Dynamics) and visualized by autoradiography. In every sample, the values for intron signal was divided by the control signal (ACT1 mRNA or PGK1 mRNA). This value (intron/control) was set to 100% for the Δdbr1 samples. To compare the different samples we used the following formula: % intron accumulation = (intron/control)Dbr1 mutant × 100/(intron/control)Δdbr1. The ratios were similar (+/−15%) for each of the 4 introns tested and the values given are averages of 2 to 4 measurements.
Expression and purification of recombinant Dbr1 proteins
The pET16-based plasmids for expression of wild-type His10-Dbr1 (36) and the His-tagged Dbr1 mutants were transformed into Escherichia coli strain BL21-Codon Plus(DE3) RIL (Stratagene). Cultures were inoculated from single colonies and maintained in logarithmic growth at 37°C in Luria–Bertani medium containing 0.1 mg/ml ampicillin to a final volume of 500 ml. When A600 reached 0.6–0.8, the cultures were chilled on ice for 30 min, and then adjusted to 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 2% ethanol. The cultures were incubated for 16–18 h at 17°C with constant shaking. Cells were harvested by centrifugation and the pellets stored at −80°C.
All subsequent procedures were performed at 4°C. The cell pellets were suspended in 20 ml of buffer A [50 mM Tris (pH 7.4), 250 mM NaCl and 10% sucrose]. Lysozyme was added to 0.2 mg/ml, the suspension was mixed gently for 30 min and then adjusted to 0.1% Triton X-100. The lysate was sonicated to reduce viscosity and insoluble material was removed by centrifugation at 15 000 r.p.m. in a Sorvall SS34 rotor. The soluble lysate was mixed for 1 h with 2 ml of Ni-NTA agarose (50% slurry, Qiagen) that had been equilibrated in buffer A. The resin was recovered by centrifugation, washed with buffer A and then poured into a column and washed with 40 ml of buffer E [50 mM Tris–HCl (pH 7.4), 10% glycerol and 250 mM NaCl] containing 25 mM imidazole. Bound proteins were eluted with buffer E containing 300 mM imidazole. Five 2 ml fractions were collected and their polypeptide compositions assessed by SDS–PAGE. Fractions containing Dbr1 proteins (∼6 ml) were pooled and 4 ml of buffer D [50 mM Tris–HCl (pH 7.4), 10% glycerol, 5 mM DTT and 1 mM EDTA] was added. The protein solutions were mixed for 1 h with 2 ml of heparin Sepharose fast flow (Amersham) that had been equilibrated in buffer D containing 150 mM NaCl. The slurry was then poured into a column, washed with buffer D containing 150 mM NaCl and eluted with buffer D containing 500 mM NaCl. Dbr1 was recovered in the 500 mM NaCl eluate. Protein concentrations were determined by using the BioRad dye-binding reagent with BSA as the standard. The yields of protein recovered from 500 ml cultures varied between 0.3 mg for mutant Y100A and 3 mg for D49A. The yield of wild-type Dbr1 was 2.5 mg.
Where indicated, the purification protocol outlined above was modified to include dialysis and zonal velocity gradient sedimentation steps. Dbr1-containing fractions that were eluted from Ni-NTA resin were pooled and dialyzed twice against 50 volumes of 50 mM Tris–HCl (pH 7.4), 10% glycerol, 5 mM DTT and 3 mM EDTA. The dialyzed preparation was further purified by heparin Sepharose chromatography as described above. An aliquot of the heparin eluate (300 µg of protein) was layered onto a 4.8 ml 15–30% glycerol gradient containing buffer G [50 mM Tris–HCl (pH 7.4), 250 mM NaCl, 2 mM DTT, 1 mM EDTA and 0.1% Triton X-100] and centrifuged in a Sorvall AH650 rotor for 20 h at 47 000 r.p.m.. Aliquots (180 µl) were collected from the top of the gradient and analyzed by SDS–PAGE. Peak fractions were pooled and the protein concentration was determined. The specific activities of Dbr1 proteins purified by the two protocols were identical when assayed under optimal conditions. However, the dialyzed preparation required added divalent cation for activity.
Purification of mDbr1 and SpDbr1 proteins
The cDNAs for mDbr1 and SpDbr1 were introduced into pET16b plasmids and expression of mDbr1 and SpDbr1 was carried out as described for Dbr1, except that the culture volume was increased to 2.5 l. SpDbr1 and mDbr1 were purified using the modified protocol described above for Dbr1.
Debranching assay
Unless otherwise indicated, reaction mixtures (20 µl) contained 50 mM Tris–HCl (pH 7.0), 4 mM MnCl2, 2.5 mM DTT, 25 mM NaCl, 0.01% Triton X-100, 0.1 mM EDTA, 0.15% glycerol, either 10 µg total RNA isolated from yeast Δdbr1 cells or 50–200 fmol of 5′ 32P-labeled synthetic branched oligonucleotides, and Dbr1 as specified. The mixtures were incubated at 22°C or 30°C as indicated and the reactions were halted by adding 20 µl of formamide dye mix (95% formamide, 40 mM Tris-borate, 0.5 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromphenol blue).
The reaction mixtures containing total Δdbr1 RNA were resolved by denaturing PAGE, transferred to membranes and then analyzed by northern blotting with intron-specific probes. Synthetic branched oligonucleotides (11) were 5′-labeled with [γ32P]ATP and T4 polynucleotide kinase (Fermentas) and then purified by electrophoresis in a denaturing 15% polyacrylamide gel. The products of the debranching reactions containing 32P-labeled oligonucleotides were analyzed by denaturing PAGE, visualized by autoradiography and quantified by scanning the gel with a phosphorimager. The extent of debranching was calculated as follows: % debranched = 100 × (linear RNA)/(linear + branched).
RESULTS
Mutational analysis of Dbr1
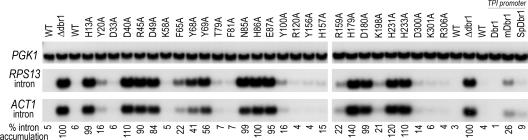
The primary structures of Dbr1 proteins are conserved among fungi, plants and mammals. Their N-terminal segments include putative counterparts of the defining motifs of the metallophosphoesterase superfamily, exemplified by the DNA phoshodiesterase Mre11 (Figure 1). To gain insight to the mechanism of debranching, we initiated a mutational analysis of DBR1 in S.cerevisiae. Guided by the alignment of Dbr1 sequences shown in Figure 1, we selected 28 conserved amino acids for substitution by alanine. Wild-type DBR1 and the DBR1-Ala alleles were cloned into TRP1 CEN plasmids under the control of the native DBR1 promoter. These plasmids, in which the polypeptides are expressed as His10-Dbr1 fusion proteins, were introduced into Δdbr1 cells. None of the mutant strains displayed a growth phenotype. To determine whether the Dbr1-Ala proteins were functional, total RNA was extracted from each of the strains and analyzed by Northern blotting with probes specific for the intron sequences of RPS13, ACT1, CYH2 and snR17A (U3) and the exon sequences of ACT1 and PGK1 mRNAs (Figure 2 and data not shown). As reported previously (13), the steady-state level of branched intron RNAs, a product of mRNA splicing, was greatly increased in Δdbr1 cells compared to wild-type cells (Figure 2, compare lanes Δdbr1 and WT). The levels of unspliced PGK1 and spliced ACT1 mRNAs were unaffected by Dbr1 and they served as loading controls (Figure 2 and data not shown). To quantify the extent of the defects elicited by the Dbr1-Ala mutants, we set the amount of RPS13, CYH2, ACT1 and U3 intron RNA (adjusted for loading) that was detected in Δdbr1 cells to 100%. In wild-type cells, the amounts of branched intron RNAs were between 3 and 6% of the levels measured in Δdbr1 cells. These values were the same whether the cells contained the chromosomal DBR1 gene or the plasmid-borne His10-DBR1 allele. Thus, the N-terminal His10 peptide did not affect the in vivo activity of wild-type Dbr1.
Figure 2.
Effects of Dbr1-Ala mutations on intron levels in vivo. RNA was prepared from Δdbr1 cells containing wild-type DBR1 (WT), the indicated DBR1-Ala mutant alleles, the plasmid vector with no insert (Δdbr1), or plasmids that express Dbr1, mDbr1 and SpDbr1 under the transcriptional control of the constitutive TPI1 promoter. Total RNA (15 µg) was separated on formaldehyde/agarose gels and transferred to membranes. Sequential hybridizations were carried out using 32P-labeled DNA probes specific for PGK1 mRNA and the intron sequences of RPS13 and ACT1. Hybridized 32P-labeled probe was visualized by autoradiography and quantified with a phosphorimager. In each sample, the relative signal measured with the intron probe was divided by the PGK1 signal to control for loading. This number was set to 100% for Δdbr1. The intron level for each of the mutants was normalized to that of Δdbr1. The average of 2–4 measurements (using 4 intron RNAs and 2 control mRNAs; see Materials and Methods) is given as % intron accumulation.
Dbr1 mutants D33A, K58A, T79A, F81A, R120A, Y156A, K301A and R306A showed no debranching defect, as judged from the absence of intron signal above that seen in wild-type cells (Figure 2). We surmise that these eight side chains, denoted by + in Figure 1, are not important for Dbr1 function in vivo. In contrast, H13A, D40A, R45A, N85A, H86A, E87A, H179A, D180A, H231A and H233A appeared to be wholly non-functional, insofar as the steady-state intron levels of ACT1, CYH2, RPS13 and U3 RNAs in these cells were ≥90% of the amount observed in Δdbr1 cells. (The affected amino acids are marked by ! in Figure 1).
Other mutants showed intermediate phenotypes (Figure 2). In Y20A, F65A, Y100A, H157A, R159A, K198A and D300A cells, the levels of branched intron RNAs were between 14 and 22% of the levels in Δdbr1 cells (denoted by ^ in Figure 1). More severe defects were observed in D49A, Y68A and Y69A cells, where each of the four introns tested accumulated to 40–85% of the level seen in Δdbr1 cells (the affected amino acids are denoted by * in Figure 1). We consider intron accumulation that is ≥25% of the level seen in Δdbr1 cells to be indicative of a significant defect in Dbr1 activity in vivo. By this criterion, His13, Asp40, Arg45, Asp49, Tyr68, Tyr69, Asn85, His86, Glu87, His179, Asp180, His231 and His233 are important for Dbr1 function in vivo.
Structure–activity relationships at important Dbr1 residues
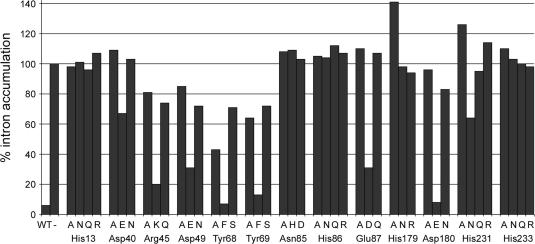
We tested the effects of conservative substitutions at the thirteen positions in Dbr1 that were defined by the alanine scan as important for in vivo activity. Aspartate was replaced by glutamate and asparagine, glutamate by aspartate and glutamine, histidine by asparagine, glutamine and arginine, asparagine by aspartate and histidine, arginine by lysine and glutamine, and tyrosine by phenylalanine and serine. The conservative mutant alleles were introduced into the Δdbr1 strain. Total RNA was isolated from the mutant yeast strains and analyzed by northern blotting in parallel with RNA from the Dbr1-Ala mutants. The level of intron accumulation for each strain is shown in Figure 3.
Figure 3.
Effects of conservative mutations on intron levels in vivo. RNA was isolated from the indicated strains and analyzed by Northern blotting. The level of intron accumulation (adjusted for loading) is represented in a bar graph and represents the average of two measurements. The value for intron RNA from Δdbr1 cells was set to 100%.
Asn85 and His86 in the putative phosphoesterase motif 84GNHE87 did not tolerate any of the substitutions tested. Lariat intron RNA accumulated in N85H, N85D, H86N, H86Q and H86R cells to the same extents as in N85A and H86A cells. Based on structural studies of λ phosphatase and Mre11 (28,29), we predict that Asn85 is part of the metal-binding site of Dbr1 and that His86 participates in catalysis through contacts with the scissile phosphodiester at the branch (see Discussion). Replacing the side chain of Glu87 by glutamine resulted in a non-functional Dbr1, but aspartate restored the in vivo function partially, i.e. the level of intron in E87D cells was 30% compared to 110% in E87A cells. This finding indicates that a negative charge at position 87 is important, but that optimal debranching activity in vivo requires proper spacing between the main chain and the carboxylate.
A side chain carboxylate was also important at positions 40, 49 and 180 in Dbr1, insofar as D40E, D49E and D180E cells had reduced intron levels compared to Dbr1 strains in which these aspartates were replaced by alanine or asparagine. However, the degree of rescue varied. D180E resembled wild-type cells, with little intron RNA present. Thus, the glutamate substitution at position 180 fully restored debranching activity. In contrast, the intron level in D40E cells was 65%, compared to 110 and 105% for D40A and D40N, respectively. This result suggests that the additional methylene group in the glutamate side chain significantly hinders Dbr1 function. Substitution of Asp49 by glutamate resulted in improved Dbr1 function; the level of intron RNA in D49E cells was 30%, compared to 85% and 70% in D49A and D49N cells.
Dbr1 activity was significantly diminished when Arg45 was replaced by alanine and glutamine; R45A and R45Q cells accumulated 80 and 74% of the intron RNA levels seen in Δdbr1 cells. However, replacing Arg45 by lysine partially restored debranching activity, such that 20% intron RNA accumulated in R45K cells relative to Δdbr1. The positive charge at position 45 is apparently critical for Dbr1 function.
Alanine substitution at Tyr68 and Tyr69 reduced the activity of Dbr1 significantly. The steady-state levels of intron RNA were 40–60% compared to 100% in Δdbr1 cells (Figures 2 and 3). Substituting Tyr68 and Tyr69 with phenylalanine fully restored Dbr1 activity, arguing against the importance of the side chain hydroxyl. The levels of intron RNA were higher in Y68S and Y69S cells (70–75%) compared to the alanine replacements, indicating that serine at these positions had a negative effect on enzyme function.
The strict requirement for histidines at positions 13, 179, 231 and 233 was underscored by the finding that none of the conservative substitutions we tested restored activity (Figure 3). A slight improvement was observed when the histidine at position 231 was replaced by asparagine, yet the level of intron accumulation (65%) remained well above our threshold for a significant loss of debranching activity.
Effects of Dbr1-Ala mutants in vitro
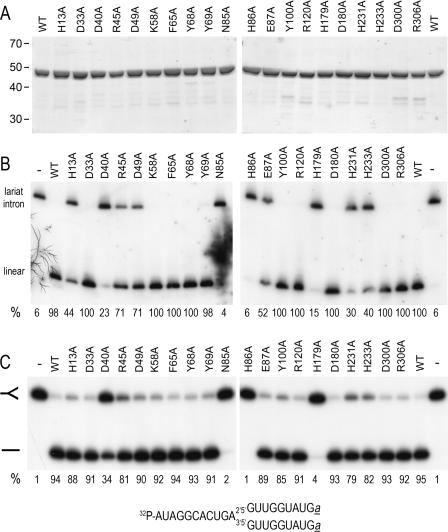
His10-tagged Dbr1 and 20 Dbr1-Ala mutants were purified from soluble bacterial lysates by Ni-NTA agarose and heparin Sepharose chromatography steps. The ∼48 kDa His10-tagged Dbr1 polypeptide was the predominant species in each preparation and the extents of purification were similar for WT and the 20 Ala mutants (Figure 4A).
Figure 4.
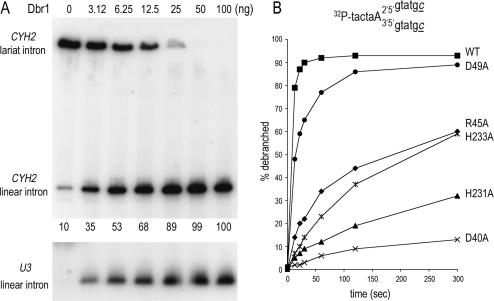
Effect of Dbr1-Ala mutations on debranching in vitro. (A) Aliquots (3 µg) of the heparin Sepharose preparations of wild-type Dbr1 (WT) and the indicated Dbr1-Ala mutants were analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker proteins are indicated at the left. (B) Total Δdbr1 RNA (10 µg) was incubated with 100 ng of the indicated proteins for 1 h at 30°C. The products were analyzed by northern blotting. The membrane was probed for the U3 intron RNA. Hybridized 32P-labeled probe was visualized by autoradiography and quantified with a phosphorimager. The percent linear product is indicated below each lane. (The value for N85A was determined from a different blot.) (C) Reaction mixtures (20 µl) containing 4 ng Dbr1 (WT) and the mutant proteins as indicated and 200 fmol of 5′-labeled RNA oligonucleotide were incubated for 10 min at 22°C. The products were analyzed by denaturing PAGE. The 32P-labeled RNA was visualized by autoradiography. The positions of branched RNA oligonucleotide and the linear product are indicated by the symbols at the left. The amount of linear product as a percentage of total RNA is noted below each lane. The branched RNA substrate is depicted at the bottom. Ribonucleotides are indicated by capital letters; a denotes 3′-terminal L-dA residues (40,41).
Debranching activity of the purified Dbr1 proteins was assayed by incubating the enzyme for 60 min with 4 mM MnCl2 and 10 µg of total RNA isolated from Δdbr1 cells. The reaction products were analyzed by northern blotting using intron-specific probes (Figures 4B and 5A). Inclusion of wild-type Dbr1 in the reaction resulted in near-quantitative conversion of the slowly migrating U3 lariat intron into a more rapidly migrating linear product (Figure 4B, compare lanes − and WT). The CHY2 and ACT1 lariat introns were also quantitatively debranched by wild-type Dbr1 (Figure 5A and data not shown). The extent of debranching was proportional to the level of input Dbr1; 6 ng of Dbr1 debranched ∼50% of the CHY2 intron RNA in 10 µg of total Δdbr1 RNA and 100 ng Dbr1 converted 100% of the CYH2 lariat into linear products (Figure 5A).
Figure 5.
Enzyme-dependence and timecourse analysis of debranching. (A) Aliquots (10 µg) of total RNA isolated from dbr1Δ cells were incubated with increasing amounts of Dbr1 for 1 h at 30°C. The samples were analyzed by northern blotting. The membrane was hybridized with a probe specific for the CYH2 intron (top panel) and U3 intron (bottom panel) and the hybridized 32P-labeled probes were visualized by autoradiography. The percent linear CYH2 intron is indicated below the lanes in the top panel. (B) Wild-type Dbr1 (4 ng) and the indicated Dbr1-Ala mutants were incubated at 22°C with 100 fmol of 5′ 32P-labeled synthetic branched substrate (illustrated on top; deoxyribonucleotides are indicated by small letters, ribonucleotides by capital letters; c denotes 3′-terminal L-dC residues) (40,41). Aliquots were withdrawn at 13, 22, 30, 60, 120 and 300 s. The samples were analyzed by denaturing PAGE. The amount of linear product, as a percentage of total RNA, is plotted as a function of time.
Saturating amounts (100 ng) of wild-type Dbr1 and Dbr1-Ala mutants were assayed in the debranching experiment shown in Figure 4B. The D33A, K58A, F65A, Y68A, Y69A, Y100A, R120A, D180A, D300A and R306A proteins completely debranched the lariat intron (≥98% linear product). In contrast, N85A and H86A were inactive; i.e. no increase in linear product was detected. The activities of D40A and H179A proteins were greatly diminished; the mutant proteins debranched 23% and 15% of the total lariat intron RNA, respectively under these conditions. Other mutants were partially active, insofar as 44% (H13A), 71% (R45A and D49A), 52% (E87A), 30% (H231A) and 40% (H233A) of the lariat RNA was debranched (Figure 4B).
Dbr1-Ala mutants were also tested for their ability to cleave a 5′-labeled synthetic branched RNA oligonucleotide (Figure 4C). The proteins (4 ng, ∼80 fmol) were incubated with 4 mM MnCl2 and 200 fmol of substrate for 10 min at 22°C and the products were analyzed by denaturing PAGE. Wild-type Dbr1 converted 95% of the branched RNA into a faster migrating linear product (Figure 4C, compare lanes − and WT). The Dbr1-Ala mutant proteins H13A, D33A, D49A, K58A, F65A, Y68A, Y69A, E87A, Y100A, R120A, D180A, D300A, R306A debranched ≥85% of the synthetic substrate under these conditions. The yield of linear product generated by R45A, H231A and H233A was 79–82%. The efficiency of debranching by D40A and H179A was 34% and 4%, respectively, and N85A and H86A were inactive (Figure 4C).
Assays of the extent of debranching at saturating enzyme levels identify only the most severe mutational effects on Dbr1 activity, but are relatively insensitive to partial losses of function. To better gauge the mutational effects, we measured debranching as a function of time (Figure 5B). In this experiment, 4 ng (∼80 fmol) of Dbr1 was reacted with 100 fmol of branched oligonucleotide. The reaction containing wild-type Dbr1 attained an endpoint at 60 s with 93% of the input substrate converted to linear product; 79% of the substrate was debranched at the earliest time point (13 s). Alanine substitutions at Asp49, Arg45, His233, His231 and Asp40 exerted increasingly severe effects on the rate of debranching (Figure 5B). The initial rates were normalized to the initial rate of wild-type Dbr1 (defined as 100%) as follows: D49A (62%), R45A (17%), H233A (9%), H231A (7%) and D40A (2%).
In summary, the mutational effects on debranching activity in vitro (Figure 4) correlate well with the effects of Dbr1 mutants in vivo (Figure 2), although it appears that the in vivo measurements are more sensitive to defects in Dbr1 function.
Characterization of Dbr1 in vitro
A prior biochemical characterization of recombinant yeast Dbr1 showed that Dbr1 resolved synthetic branched RNA oligonucleotides and msDNA substrates in which a ribonucleotide was linked to the 5′ end of a DNA via a 2′-5′ phosphodiester bond (33). Whereas the debranching activity of recombinant yeast Dbr1 was reported independent of adding divalent cation (33), debranching enzyme purified from HeLa cells was metal-dependent (34).
Our mutational results suggest that Dbr1 is a member of the metallophosphoesterase superfamily. These enzymes have a characteristic binuclear metal cluster and typically require manganese or nickel for catalysis. We evaluated the metal-requirement for debranching by Dbr1 using 5′-labeled branched RNA oligonucleotides as substrate (Figure 6). His10-tagged Dbr1 was isolated from soluble bacterial lysates by Ni-NTA chromatography and then dialyzed against buffer containing EDTA. The protein was further purified by heparin Sepharose chromatography and glycerol gradient sedimentation.
Figure 6.
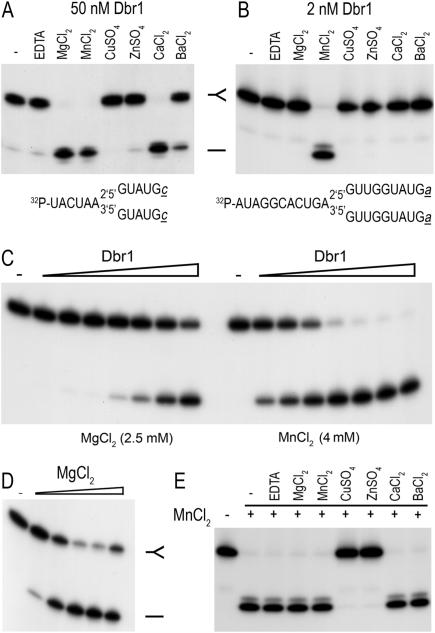
Properties of the in vitro debranching reaction. Debranching of 5′-labeled synthetic branched RNA oligonucleotides was measured during 10 min incubations at 22°C. The reaction mixtures were analyzed by denaturing PAGE; autoradiograms of the gels are shown. (A) Metal specificity at high enzyme concentration. Dbr1 (50 nM) was incubated with 5 nM branched RNA oligonucleotide (depicted at bottom) in the presence of 2.5 mM EDTA or 2.5 mM of the indicated divalent cation. A control reaction lacking EDTA or divalent cation is shown in lane −. (B) Metal specificity at low enzyme concentration. Dbr1 (2 nM) was incubated with 5 nM of branched oligonucleotide (depicted at bottom) for 10 min at 22°C in the presence of 2.5 mM EDTA or 2.5 mM of the indicated divalent cation. (C) Enzyme titration. 10 nM branched substrate (illustrated in panel B) was reacted with increasing concentrations of Dbr1 (0.31, 0.62, 1.25, 2.5, 5, 10 and 20 nM, proceeding from left to right) in the presence of 2.5 mM magnesium (left) or 4 mM manganese (right). Dbr1 was omitted from the control reactions in lanes −. (D) Magnesium dependence. Dbr1 (20 nM) was incubated with 10 nM branched oligonucleotide substrate (depicted in panel B) in the presence of increasing concentrations of MgCl2 (0.5, 1, 2.5, 5 and 10 mM, proceeding from left to right). MgCl2 was omitted from the control reaction in lane −. (E) Mixing of divalent cations. 5 nM 32P-labeled branched oligonucleotide (depicted in panel B) was incubated with 2 nM Dbr1. Reactions mixtures contained 4 mM MnCl2 (+) plus either 2.5 mM EDTA or 2.5 mM of the indicated divalent cations.
Dbr1 required a divalent cation cofactor, insofar as less than 5% of the substrate was debranched when a metal was omitted or when EDTA was added to the reaction mixture (Figure 6). Various metals were tested at 2.5 mM concentration for their ability to support debranching (Figure 6A and B). At 50 nM Dbr1, magnesium, manganese and calcium supported near-quantitative debranching of 5 nM substrate (Figure 6A). No activity was observed with copper or zinc. Magnesium was most effective at 2.5 to 10 mM and higher concentrations inhibited debranching activity (Figure 6D and data not shown).
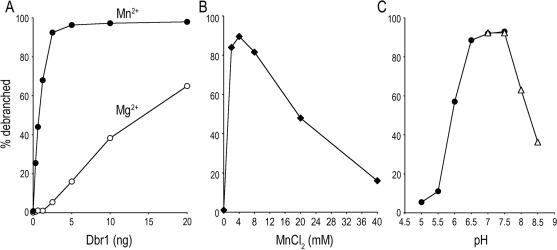
Manganese was the only effective cofactor when the Dbr1 concentration was reduced to 2 nM and the branched substrate concentration was 5 nM. Whereas 2.5 mM MnCl2 supported 98% debranching, little or no linear product was detected with 2.5 mM MgCl2, CuSO4, ZnSO4, CaCl2 or BaCl2 (Figure 6B). The optimal manganese concentration was between 2 and 8 mM; activity was reduced to one-half and one-fifth of the peak value at 20 and 40 mM manganese, respectively (Figure 7B). The extent of debranching was proportional to input Dbr1 (Figures 6C and 7A). From the slopes of the titration curves, we calculated that Dbr1 debranched 0.35 and 7 fmol of substrate per fmol of enzyme in the presence of 2.5 mM magnesium and 4 mM manganese, respectively (Figures 6C and 7A). Thus, Dbr1 acted catalytically in manganese, but not in magnesium. This is consistent with prior studies that routinely used magnesium and a 20- to100-fold excess of enzyme over substrate (33,35).
Figure 7.
Manganese specificity and pH dependence of debranching. (A) Increasing amounts of Dbr1 as specified were incubated with 10 nM of 5′-labeled substrate and either 4 mM MnCl2 or 2.5 mM MgCl2 for 10 min at 22°C. The percentage of the substrate that was debranched is plotted as a function of input Dbr1. (B) Manganese titration. Reaction mixtures containing 50 mM Tris–HCl (pH 7.0), 2.5 mM DTT, 25 mM NaCl, 0.01% Triton X-100, 0.1 mM EDTA, 0.15% glycerol, 1 nM Dbr1 and 10 nM branched substrate and manganese as specified were incubated for 10 min at 22°C. (C) pH dependence. Reaction mixtures containing 50 mM Tris buffer [either Tris–acetate for (pH 5.0, 5.5, 6.0, 6.5, 7.0, 7.5) or Tris–HCl (pH 7.0, 7.5, 8.0, 8.5)], 4 mM MnCl2, 2.5 mM DTT, 25 mM NaCl, 0.01% Triton X-100, 0.1 mM EDTA, 0.15% glycerol 10 nM RNA and 1 nM Dbr1 were incubated for 10 min at 22°C. The extent of debranching is plotted as a function of pH. The branched RNA substrate used for these experiments is depicted in Figure 6B.
An additional experiment was performed in which 4 mM of manganese was mixed with 2.5 mM of each of the other divalent cations (Figure 6D). The debranching activity was abolished in the presence of copper and zinc, but was unaffected by magnesium, calcium or barium. At non-saturating enzyme concentrations, we did not observe stimulation of the debranching activity in 4 mM manganese when the reactions were supplemented with 2.5 to 10 mM magnesium (data not shown). Dbr1 displayed a bell-shaped pH profile with a peak from pH 6.5 to 7.5. Activity declined to one-tenth the peak value at pH 5.5 and less than one-half at pH 8.5 (Figure 7C).
We have used synthetic branched substrates of various lengths and composition (ribonucleotides or deoxyribonucleotides) for in vitro debranching assays (depicted in Figures 4–6 and 9). Enzyme titration experiments, using 100–200 fmol of substrate, showed no difference in the efficiency of debranching of these different substrates by Dbr1 (data not shown). Note that each of the structures contains a branch site adenosine linked to guanosine; this is the naturally occurring branch dinucleotide and prior studies have established that Dbr1 has a preference for purines at these positions (33,34).
Figure 9.
Debranching activities of mouse and S.pombe Dbr1. (A) An aliquot (2 µg) of the mDbr1 preparation was analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown; the positions and sizes of marker polypeptides are indicated at the left. (B) Reaction mixtures (20 µl) contained 200 fmol of the 5′-labeled branched oligonucleotide substrate shown, 2.5 mM of the indicated metal cofactor, 50 mM Tris-acetate (pH 6.5), 2.5 mM DTT, 25 mM NaCl, 0.01% Triton X-100, 0.1 mM EDTA, 0.15% glycerol and 320 ng mDbr1. The products were analyzed by denaturing PAGE. An autoradiogram of the gel is shown. (C) Increasing amounts of mDbr1 were incubated with 200 fmol of the indicated 5′-labeled branched oligonucleotide substrate (shown at bottom) in the presence of either MgCl2 (2.5 mM) or MnCl2 (4 mM) for 10 min at 22°C. The extent of debranching is plotted as a function of input mDbr1. (D) S.pombe Dbr1. Reaction mixtures (20 µl) contained 200 fmol of the indicated 5′-branched oligonucleotide, 4 mM MnCl2 and SpDbr1 as specified. The inset shows the polypeptide composition of the SpDbr1 preparation (2 µg), as gauged by SDS–PAGE and Coomassie blue-staining.
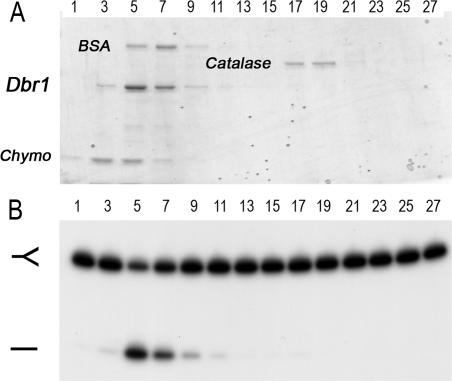
To gauge the native size of Dbr1 (48 kDa), the enzyme was analyzed by sedimentation in a 15–30% glycerol gradient. Catalase (248 kDa), BSA (66 kDa) and chymotrypsinogen (23 kDa) were included as internal standards. Dbr1 sedimented as a single peak between BSA and chymotrypsinogen (Figure 8A). Debranching activity was highest in fraction 5, coinciding with the abundance of the Dbr1 protein (Figure 8B). The sedimentation profile suggests that Dbr1 is a monomer in solution.
Figure 8.
Glycerol gradient sedimentation of Dbr1. Sedimentation was performed as described under Materials and Methods. (A) Aliquots (9 µl) of the odd-numbered gradient fractions were analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown. The positions of the recombinant Dbr1 and the marker proteins chymotrypsinogen (Chymo), BSA and catalase are indicated. (B) Reaction mixtures (20 µl) containing 5 nM of 32P-labeled branched oligonucleotide substrate (depicted in Figure 6B) and 2 µl of a 1:100 dilution of the odd-numbered glycerol gradient fractions were incubated for 10 min at 22°C. The products were analyzed by denaturing PAGE and visualized by autoradiography.
Debranching activity of mDbr1 and SpDbr1
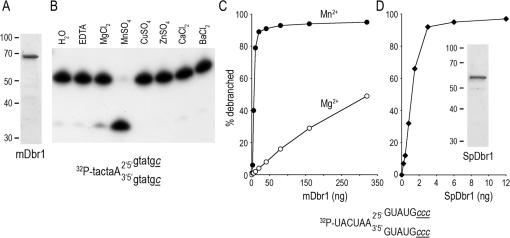
In order to determine whether manganese was an effective cofactor for other debranching enzymes, we expressed the mouse and fission yeast Dbr1 homologs in E.coli and purified soluble His10-tagged mDbr1 and SpDbr1 from bacterial extracts. The sizes of the major polypeptides in the protein preparations were consistent with the calculated molecular masses of recombinant His-tagged mDbr1 and SpDbr1, which are 64 and 57 kDa, respectively (Figure 9A and D). mDbr1 debranched a 5′-labeled synthetic substrate efficiently in the presence of manganese, but not other metals tested (magnesium, copper, zinc, calcium or barium) (Figure 9B). The optimal manganese concentration was 2 to 4 mM (data not shown). The pH optimum for mDbr1 activity was between pH 6 and 7 (data not shown). The extent of debranching was proportional to the amount of mDbr1 and in the linear range of enzyme-dependence, mDbr1 debranched 1.1 fmol of synthetic RNA substrate per fmol of enzyme (Figure 9C). The specific activity of mDbr1 was 40-fold lower in reactions with magnesium than with manganese (Figure 9C). Reaction of increasing amounts of SpDbr1 with 5 nM branched RNA substrate and 4 mM manganese resulted in the formation of 1.8 fmol of linear product per fmol of SpDbr1 (Figure 9D).
DISCUSSION
Here we conducted a genetic and biochemical characterization of yeast Dbr1, the enzyme responsible for debranching the lariat intron products of pre-mRNA splicing. We find that Dbr1 is a monomeric enzyme that acts catalytically in vitro in the presence of manganese. The need for a dialysis step against EDTA-containing buffer in order to reveal the divalent cation dependence of yeast Dbr1 suggests that the enzyme retains a bound metal during purification, which may account for conflicting findings in prior reports concerning the metal-dependence of yeast and human Dbr1 (33,34). The strong preference for manganese at limiting enzyme concentrations is consistent with our mutational results that place Dbr1 as a member of the metallophosphoesterase superfamily.
Although a potential relationship of Dbr1 to the metallophosphoesterases had been proposed 11 years ago based on local amino acid sequence similarities (24,25), no systematic effort had been made to test the validity of this prediction. Since the relationship was posited, a wealth of structural and functional information has been gained for exemplary metallophosphoesterase family members, including calcineurin, λ phosphatase and Mre11 (27–29). The alanine scan of yeast Dbr1 reported here was targeted broadly to 28 amino acids that are conserved in Dbr1 homologs from other organisms. The instructive finding was that seven of the thirteen residues defined by the alanine scan as critical for intron debranching by Dbr1 in vivo (His13, Asp40, Asn85, His86, His179, His231 and His233) correspond to putative counterparts of the side chains that comprise the binuclear metal-binding sites and phosphate-binding site in the crystal structure of Mre11 (29), which, like Dbr1, is a manganese-dependent phosphodiesterase.
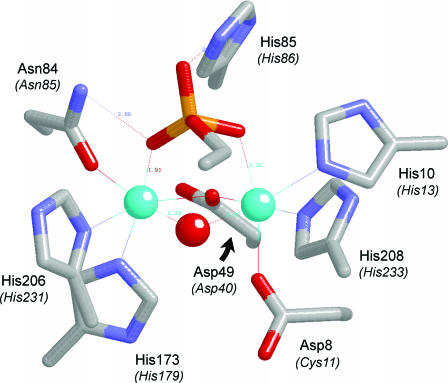
The active site of PfuMre11 (Figure 10) contains two manganese ions coordinated directly by aspartate, histidine and asparagine side chains. Six of the seven metal-binding amino acids in Mre11 (His10, Asp49, Asn84, His173, His206 and His208) are conserved in Dbr1 (as His13, Asp40, Asn85, His179, His231 and His233), essential for debranching in vivo, and intolerant of conservative substitutions. Analysis of the effects of selected Ala mutations on debranching of a synthetic oligonucleotide substrate in vitro highlights important catalytic roles for proposed metal-binding residues Asp40, Asn85, His179, His231 and His233. We surmise that Dbr1 likely contains a binuclear manganese cluster similar to that seen in Mre11.
Figure 10.
Proposed similarity of the active sites of Mre11 and Dbr1. The Figure depicts the active site of the manganese- and 5′-dAMP-bound Pyrococcus furiosus phosphodiesterase Mre11 from the crystal structure (PDB 1II7). The amino acid side chains coordinating the binuclear metal cluster and the nucleotide 5′-phosphate are shown. The corresponding amino acids of yeast Dbr1 are indicated in parentheses. The manganese ions are colored cyan. Water is colored red. For simplicity, only the phosphate and the ribose C4 and C5 atoms of the nucleotide are shown.
The PfuMre11 structure contains 5′-dAMP in the active site (Figure 10). The bridging water between the two manganese ions is a good candidate for the attacking nucleophile, because it is located 3.1 Å from the phosphorus atom and is almost perfectly apical to the phosphate oxygen atom of the leaving group, which is a nucleoside 3′-O in the case of Mre11. As discussed by Voegtli et al. (28), the bridging water ligand between the manganese ions should have a low pKa and thereby be activated by the metals for attack on the phosphate, which, in Mre11, is coordinated by Asn84 and His85 side chains of the signature GNH(E/D) motif. The corresponding Asn85 and His86 side chains of yeast Dbr1 are critical for debranching in vivo and in vitro and intolerant of conservative substitutions. Therefore, we propose that Asn85 and His86 of Dbr1 contact the scissile 2′-5′ phosphodiester of the branched RNA in a manner analogous to that seen in the Mre11-dAMP complex.
Dbr1 residue Glu87 is part of the 84GNHE87 motif and essential for lariat debranching in vivo. Conservative substitution of Glu87 with aspartate resulted in partial restoration of function, but glutamine had no salutary effect. This position is occupied by glutamate in λ phosphatase, but is substituted by an aspartate in Mre11 (Figure 1). The equivalent Glu77 side chain in the λ phosphatase crystal structure (28) makes no contacts with either the metal cluster or the bound sulfate ion (a mimetic of phosphate). Rather, Glu77 appears to play a structural role in ensuring the proper conformation of the active site, via hydrogen bonds from the glutamate to three backbone amide nitrogens, including that of Asp49 (corresponding to Asp40 in Dbr1), which coordinates both of the divalent cations. The equivalent Asp86 side chain in the Mre11 structure (29) also makes hydrogen bonds to backbone amides flanking essential catalytic residues. We therefore invoke a similar structural role for Glu87 in Dbr1. The notion that conformational perturbations in the 84GNHE87 motif can affect Dbr1 activity is consistent with an earlier report that changing Gly84 to alanine resulted in lariat intron accumulation in vivo (15).
Changing Dbr1 residue Arg45 to alanine or glutamine adversely affected Dbr1 activity in vivo, but conservative replacement by lysine ameliorated the defect. The R45A mutation slowed the rate of debranching of a synthetic oligonucleotide substrate by a factor of 6. Although there are no contacts between arginine or lysine residues and the phosphate of 5′-dAMP in the crystal structure of PfuMre11 (29), the structure of λ phosphatase shows that Arg53 makes a bidentate contact with a sulfate ion that likely mimics a phosphate in the active site (28). It is conceivable that Dbr1 Arg45 is analogous to Arg53 of λ phosphatase.
The roles of several other functionally relevant Dbr1 residues defined by the present mutational analysis (e.g. Asp49, Tyr68, Tyr69 and Asp180) cannot be readily inferred, because they do not have clearly identifiable counterparts in the available metallophosphoesterase structures. Conservative mutagenesis shows that the carboxylate is the relevant functional group at positions 49 and 87. The finding that debranching function is restored when phenylalanine is introduced in lieu of tyrosine at positions 68 and 69 is inconsistent with the earlier suggestion that phosphorylation of Tyr68 might be functionally important (15). We envision two potential classes of functionally important residues that would be conserved among Dbr1 proteins, though not necessarily among other metallophosphoesterases: (i) those that stabilize the Dbr1 tertiary structure (e.g. via hydrophobic interactions or buried salt bridges) and (ii) those that contribute to Dbr1 substrate specificity, via interactions with constituents of the branch structure other than the scissile phosphate. Further biochemical, structural and genetic characterization of Dbr1 will be required to address the basis for substrate recognition and specificity.
Acknowledgments
This work was supported by NIH grant GM50288 to B.S., and by a grant from NSERC Canada to M.J.D. We thank Dr Sandra Carriero for branched oligoribonucleotide synthesis. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM50288.
Conflict of interest statement. None declared.
REFERENCES
- 1.Peebles C.L., Perlman P.S., Mecklenburg K.L., Petrillo M.L., Tabor J.H., Jarrell K.A., Cheng H.L. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 2.Padgett R.A., Konarska M.M., Grabowski P.J., Hardy S.F., Sharp P.A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 3.Ruskin B., Krainer A.R., Maniatis T., Green M.R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 4.Murphy W.J., Watkins K.P., Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- 5.Sutton R.E., Boothroyd J.C. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause M., Hirsh D. A trans-spliced leader sequence on actin mRNA in C.elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yee T., Furuichi T., Inouye S., Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984;38:203–209. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]
- 8.Tuschl T., Sharp P.A., Bartel D.P. A ribozyme selected from variants of U6 snRNA promotes 2′,5′-branch formation. RNA. 2001;7:29–43. doi: 10.1017/s1355838201001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Silverman S.K. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 10.Coppins R.L., Silverman S.K. A DNA enzyme that mimics the first step of RNA splicing. Nature Struct. Mol. Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- 11.Damha M.J., Ganeshan K., Hudson R.H., Zabarylo S.V. Solid-phase synthesis of branched oligoribonucleotides related to messenger RNA splicing intermediates. Nucleic Acids Res. 1992;20:6565–6573. doi: 10.1093/nar/20.24.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruskin B., Green M.R. An RNA processing activity that debranches RNA lariats. Science. 1985;229:135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- 13.Chapman K.B., Boeke J.D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 14.Karst S.M., Rutz M.L., Menees T.M. The yeast retrotransposons Ty1 and Ty3 require the RNA Lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem. Biophys. Res. Commun. 2000;268:112–117. doi: 10.1006/bbrc.1999.2048. [DOI] [PubMed] [Google Scholar]
- 15.Salem L.A., Boucher C.L., Menees T.M. Relationship between RNA lariat debranching and Ty1 element retrotransposition. J. Virol. 2003;77:12795–12806. doi: 10.1128/JVI.77.23.12795-12806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Z., Menees T.M. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science. 2004;303:240–243. doi: 10.1126/science.1087023. [DOI] [PubMed] [Google Scholar]
- 17.Coombes C.E., Boeke J.D. An evaluation of detection methods for large lariat RNAs. RNA. 2005;11:323–331. doi: 10.1261/rna.7124405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell P., Tollervey D. mRNA turnover. Curr. Opin. Cell. Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 19.Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 20.Danin-Kreiselman M., Lee C.Y., Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 21.Nam K., Lee G., Trambley J., Devine S.E., Boeke J.D. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol. Cell. Biol. 1997;17:809–818. doi: 10.1128/mcb.17.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J.W., Kim H.C., Kim G.M., Yang J.M., Boeke J.D., Nam K. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 2000;28:3666–3673. doi: 10.1093/nar/28.18.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H.C., Kim G.M., Yang J.M., Ki J.W. Cloning, expression, and complementation test of the RNA lariat debranching enzyme cDNA from mouse. Mol Cells. 2001;11:198–203. [PubMed] [Google Scholar]
- 24.Koonin E.V. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 1994;3:356–358. doi: 10.1002/pro.5560030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo S., Clemens J.C., Stone R.L., Dixon J.E. Mutational analysis of a Ser/Thr phosphatase: identification of residues important in phosphoesterase substrate binding and catalysis. J. Biol. Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]
- 26.Rusnak F., Mertz P. Calcineurin: form and function. Physiol. Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg J., Huang H., Kwon Y., Greengard P., Nairn A.C., Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 28.Voegtli W.C., White D.J., Reiter N.J., Rusnak F., Rosenzweig A.C. Structure of the bacteriophage lambda Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes. Biochemistry. 2000;39:15365–15374. doi: 10.1021/bi0021030. [DOI] [PubMed] [Google Scholar]
- 29.Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 30.Zhuo S., Clemens J.C., Hakes D.J., Barford D., Dixon J.E. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J. Biol. Chem. 1993;268:17754–17761. [PubMed] [Google Scholar]
- 31.Martins A., Shuman S. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′ kinase and polynucleotide 3′ phosphatase activities. J. Biol. Chem. 2004;279:18220–18231. doi: 10.1074/jbc.M313386200. [DOI] [PubMed] [Google Scholar]
- 32.Chen S., Yakunin A.F., Kuznetsova E., Busso D., Pufan R., Proudfoot M., Kim R., Kim S.H. Structural and functional characterization of a novel phosphodiesterase from Methanococcus janaschii. J. Biol. Chem. 2004;279:31854–31862. doi: 10.1074/jbc.M401059200. [DOI] [PubMed] [Google Scholar]
- 33.Nam K., Hudson R.H., Chapman K.B., Ganeshan K., Damha M.J., Boeke J.D. Yeast lariat debranching enzyme. Substrate and sequence specificity. J. Biol. Chem. 1994;269:20613–20621. [PubMed] [Google Scholar]
- 34.Arenas J., Hurwitz J. Purification of a RNA debranching activity from HeLa cells. J. Biol. Chem. 1987;262:4274–4279. [PubMed] [Google Scholar]
- 35.Pratico E.D., Wang Y., Silverman S.K. A deoxyribozyme that synthesizes 2′,5′-branched RNA with any branch-site nucleotide. Nucleic Acids Res. 2005;33:3503–3512. doi: 10.1093/nar/gki656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 37.Ho S.N., Hunt H.D., Horton R.M., Pullen J., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 38.Fikes J.D., Becker D.M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990;346:291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- 39.Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damha M.J., Giannaris P.A., Marfey P. Antisense L/D-oligonucleotide chimeras: nuclease stability, base-pairing properties, and activity at directing ribonuclease H. Biochemistry. 1994;33:7877–7885. doi: 10.1021/bi00191a015. [DOI] [PubMed] [Google Scholar]
- 41.Carriero S., Damha M.J. Inhibition of pre-mRNA splicing by synthetic branched nucleic acids. Nucleic Acids Res. 2003;31:6157–6167. doi: 10.1093/nar/gkg824. [DOI] [PMC free article] [PubMed] [Google Scholar]