Abstract
Acquisition of genomic elements by horizontal gene transfer represents an important mechanism in the evolution of bacterial species. Pathogenicity islands are a subset of horizontally acquired elements present in various pathogens. These elements are frequently located adjacent to tRNA genes. We performed a comparative genome analysis of Salmonella enterica serovars Typhi and Typhimurium and Escherichia coli and scanned tRNA loci for the presence of species-specific, horizontally acquired genomic elements. A large number of species-specific elements were identified. Here, we describe the characteristics of four large chromosomal insertions at tRNA genes of Salmonella spp. The tRNA-associated elements harbor various genes previously identified as single virulence genes, indicating that these genes have been acquired with large chromosomal insertions. Southern blot analyses confirmed that the tRNA-associated elements are specific to Salmonella and also indicated a heterogeneous distribution within the salmonellae. Systematic scanning for insertions at tRNA genes thus represents a tool for the identification of novel pathogenicity islands.
The genome sequences of many important bacterial pathogens as well as of nonpathogenic relatives are now available or will be completed in the near future. These data allow new approaches for the identification of pathogen-specific regions of the genome and thereby may direct the identification of new virulence genes.
Acquisition of genetic information by horizontal gene transfer is considered the most efficient mechanism of bacterial evolution (for a review, see reference 24). The integration of DNA segments containing clusters of virulence genes has lead to the evolution of pathogenic bacteria from nonpathogenic ancestors. These segments of bacterial genomes are also referred to as pathogenicity islands (PAI) (for a review, see reference 11). PAI of various pathogens share several characteristics, such as a base composition different from that of the host genome. A further remarkable feature of PAI and other horizontally acquired elements is their close vicinity to tRNA genes. These genes are highly conserved among bacterial genomes and are known to serve as anchor points for temperate phages (16). This fact supports the idea that PAI could have been laterally transferred by phages into different genomes (for a review, see reference 4).
During evolution, the enteric pathogen Salmonella enterica acquired a large number of virulence genes that are required during various stages of pathogenesis (reviewed in reference 8). Subsets of virulence genes required for the invasion of nonphagocytic eukaryotic cells and enteropathogenesis, as well as for intracellular survival and systemic progression of infection, are clustered in PAI referred to as Salmonella pathogenicity islands (SPI) (reviewed in reference 20). The association with tRNA genes of SPI2 (14), SPI3 (2), SPI4 (27), and SPI5 (28) has been described.
Based on the observation that many PAI are inserted into the genome adjacent to a tRNA gene, we systematically scanned tRNA genes and compared the tRNA-associated regions of a pathogen and its nonpathogenic relative. For this analysis, we compared genome data of two S. enterica serovars, i.e., serovars Typhi and Typhimurium, with genome data of a nonpathogenic strain of Escherichia coli K-12 (3) and enterohemorrhagic E. coli (EHEC) O157:H7 (25). A comparative analysis of the surrounding areas of all tRNA sequences in the genomes of E. coli and Salmonella was performed. Differences in the DNA sequence on at least one side of the tRNA genes have been further analyzed.
Four DNA fragments that show characteristics of PAI have been identified. These regions contain genes that encode proteins already known as virulence factors. However, these virulence genes have not been correlated to horizontally acquired DNA elements. The fact that all of these regions are associated with several phage-related proteins underlines the presence of mobile elements that could have been used as vehicles to transfer these genetic elements.
MATERIALS AND METHODS
Bacterial strains.
S. enterica subspecies I serotype Typhimurium strain 12023 (identical to ATCC 14028) was used as wild-type strain. A spontaneous isolate resistant to nalidixic acid was used for infection studies. Strain HH104 (15), harboring an insertion of a kanamycin resistance cassette in the sseC gene of SPI2, is highly attenuated in systemic pathogenesis in the mouse model. Chromosomal DNA of serovar Typhi was obtained from strain 2 of the Salmonella Reference Collection C (SARC) reference collection (5). The distribution of the genes specific for the putative islands has been analyzed using strains of the SARC reference collection obtained from the Salmonella Genetic Stock Center (Calgary, Canada). The distribution within the gram-negative pathogens was analyzed using genomic DNAs from various patient isolates of the strain collection of the Max von Pettenkofer-Institut, Munich, Germany, and the Institut für Klinische Mikrobiologie, Immunologie und Hygiene, Erlangen, Germany.
Bioinformatics.
The following databases were used for the retrieval of DNA sequence data: the database of the S. enterica serovar Typhi Sequencing Group at the Sanger Centre (ftp://ftp.sanger.ac.uk/pub/pathogens/st/St.dna), the database of the E. coli K-12 and EHEC genomes (University of Wisconsin, Madison [http://www.genome.wisc.edu] and Universität Giessen [http://susi.bio.uni-giessen.de]), and the database of S. enterica serovar Typhimurium (Genome Sequencing Center, Washington University, St. Louis, Mo. [http://genome.wustl.edu/gsc/Projects/bacteria.shtml]). Bacterial genome sequence information was analyzed using the software package Artemis version 3. MacVector version 6 was used for the analysis of the base compositions of various regions of the genome.
Generation of DNA probes and hybridization conditions.
Hybridization probes specific for genes within putative new islands were obtained by PCR using S. enterica serovar Typhimurium 12023 or S. enterica serovar Typhi SARC 2 genomic DNA as the template. Primers used to amplify probes specific for genes within the tRNA-associated elements are listed in Table 1. Additionally, probes specific for gyrB and recA were used to control the hybridization conditions applied. The DNA sequences of gyrB and recA are rather conserved among bacteria, and the genes should therefore be detectable under nonstringent hybridization conditions in all enterobacteria investigated.
TABLE 1.
Oligonucleotides used in this study
| Purpose and gene and/or tRNA | Designation | Sequence |
|---|---|---|
| Hybridization probes | ||
| pgtE (tRNAArgW) | pgtE-for | 5′-ACTTCATCACCTCTCCAG-3′ |
| pgtE-rev | 5′-CTAGAAGCGGTACTGCAAC-3′ | |
| nupC (tRNAArgW) | nupC-for | 5′-ATGGACCGCGTCCTTCATT-3′ |
| nupC-rev | 5′-CAGACTTACAGTACCAGC-3′ | |
| apeE (tRNAArgU) | apeE-for | 5′-GCAATATCTCTGTCGGAT-3′ |
| apeE-rev | 5′-TTGCCGACTGGCGAAATC-3′ | |
| STM0557 (tRNAArgU) | STM0557-for | 5′-GTGCATTGCGACTTCTTG-3′ |
| STM0557-rev | 5′-GGAATAGGTATTCTTGGGG-3′ | |
| sspH2 (tRNAProL) | sspH2-for | 5′-CACGCGGAAGGGGCATC-3′ |
| sspH2-rev | 5′-GCTGGTCAGTTGATTACC-3′ | |
| msgA (tRNAProL) | msgA-for | 5′-TTTCGTCAGGCTTGCATC-3′ |
| msgA-rev | 5′-TCATTTACCTGCCACTGC-3′ | |
| vexA (tRNAProL) | vexE-for | 5′-CCAAATCCCACCGGAAAG-3′ |
| vexE-rev | 5′-CAACCACCCTACTCAAAC-3′ | |
| pilV (tRNAPheU) | pilV-for | 5′-ACATGATGGCGGCTTTGTG-3′ |
| pilV-rev | 5′-GCCAGGTAAGTTCAAACAG-3′ | |
| recA | recA-for | 5′-ATGGCTATCGACGAAAAC-3′ |
| recA-rev | 5′-CGTTAGTTTCTGCTACGC-3′ | |
| gyrB | gyrB-for | 5′-AAAGTCCTGAAAGGGCTG-3′ |
| gyrB-rev | 5′-CGATATTCGCCGCTTTCA-3′ | |
| Deletions | pgtE-Red-Del-for | 5′-TCGGCCGGTTATGACCGATGACAT CCCGATGTGGTCTAGTGTAGGCTGGAGCTGCTTC-3′ |
| pgtP-Red-Del-rev | 5′-AATGTCGGCGCTTCTGTTCCCCAG GAAGGCTAATCGTTTCATATGAATATCCTCCTTAG-3′ | |
| ykgD-Red-Del-for | 5′-CGCCTGGTGTACAACCGAATTC ACGGACAAAAGCTTTGGTGTAGGCTGGAGCTGCTTC-3′ | |
| apeE-Red-Del-rev | 5′-GTCATCAAAATCGGGCGCT AAACCCAACGTTATAACGGGCATATGAATATCCTCCTTAG-3′ | |
| tRNA insertion site detection | ||
| tRNAArgU | tRNA-ArgU-for | 5′-GAGTGACTTTGTCTGCTC-3′ |
| tRNA-ArgU-rev | 5′-GATGTCCCAAATATGTCCC-3′ | |
| tRNAArgW | tRNA-ArgW-for | 5′-CTGGAGCGACTTTCTCTG-3′ |
| tRNA-ArgW-rev | 5′-CGTATTTTGGTGGCGATG-3′ | |
| tRNAProL | tRNA-ProL-for | 5′-CGACAGGTATGGTAATC-3′ |
| tRNA-ProL-rev | 5′-GTAACAGGCTGGTTCTTC-3′ | |
| tRNAPheU | tRNA-PheU-for | 5′-CATAGGCTGGGTTTTCTG-3′ |
| tRNA-PheU-rev | 5′-CCACTCGACACATTACAG-3′ |
PCR fragments were labeled using the digoxigenin DNA labeling kit (Roche). Hybridization was carried out at 50°C overnight in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate [SDS], 1% blocking reagent [Roche]). Two wash steps of 10 min (2× SSC, 0.1% SDS) were performed at room temperature, followed by two wash steps of 30 min at 50°C under nonstringent conditions (0.5× SSC, 0.1% SDS).
Deletion of Salmonella-specific chromosomal elements.
The method of one-step inactivation of chromosomal genes was adapted from that of Datsenko and Wanner (6). Briefly, primers were designed with 38- to 39-nucleotide sequences homologous to the respective flanking regions of the gene of interest followed by 20 nucleotides complementary to the template plasmid pKD4 (Table 1). Primers pgtE-Red-Del-for and pgtP-Red-Del-rev were used in order to delete the pgt gene cluster of the tRNAArgW-associated element, and primers ykgD-Red-Del-for and apeE-Red-Del-rev were used for a deletion in the tRNAArgU-associated element.
PCRs were performed with the Expand High Fidelity PCR system, and the products were gel purified after digestion with DpnI. The PCR products were electroporated into S. enterica serovar Typhimurium 12023 cells carrying the Red recombinase expression plasmid pKD46 and grown with arabinose to induce the Red recombinase. Recombinant clones were selected on Luria-Bertani agar containing 50 μg of kanamycin per ml. Proper chromosomal insertion of the kanamycin cassette in mutant strains was confirmed by PCR using primers k1 and k2 (6) and DNA sequencing of the relevant regions. Finally, mutant alleles were moved into a fresh serovar Typhimurium 12023 strain background by P22 transduction according to standard procedures (19).
Analyses of insertion points.
Conservation of the insertion point of tRNA-associated elements was analyzed by PCR. Forward primers were selected from gene sequences proximal to the tRNA gene and outside the insertion, and reverse primers were complementary to sequences proximal to the tRNA gene inside the insertion (Table 1). PCRs using the Taq polymerase (Gibco) were carried out at annealing temperatures of 47 to 55°C with an elongation time of 2 min at 72°C.
Virulence studies.
Groups of three female C57BL/6 mice (18 to 20 g) were infected by intraperitoneal injection of 200 μl of a mixture consisting of 5,000 CFU of nalidixic acid-resistant wild-type S. enterica serovar Typhimurium 12023 (Nalr Kms) and 5,000 CFU of various mutant strain (Nals Kmr). Mice developed symptoms of severe salmonellosis within 3 days after infection and were sacrificed on day 4 by cervical dislocation. Livers and spleens were dissected aseptically and homogenized in sterile phosphate-buffered saline. Serial dilutions were plated on Luria-Bertani agar containing 50 μg of nalidixic acid per ml or 50 μg of kanamycin per ml for the enumeration of the organ burdens with wild-type Salmonella and the mutant strain. The competitive index (CI) was determined as described previously (1). Animal studies were performed in accordance with national guidelines.
RESULTS
Identification of putative horizontally acquired DNA segments specific to Salmonella spp.
The DNA sequences of about 80 tRNA genes were retrieved from the E. coli K-12 genome database. Individual tRNA gene sequences were aligned to the genome databases of S. enterica serovar Typhimurium LT2 and S. enterica serovar Typhi CT18. DNA sequences of about 2 kb on each side of the tRNA genes were compared between E. coli and Salmonella sequences in order to identify regions with different organization in both species. This screen revealed about 20 chromosomal segments adjacent to tRNA genes that showed different organizations in E. coli K-12 and Salmonella spp. These segments were selected for further analyses. Regions of 6 to 8 kb on each side of the tRNA gene were compared with the National Center for Biotechnology Information database on the DNA level using the BLAST algorithm. Sequences present in Salmonella but absent in E. coli were investigated further.
During the analyses, special attention was given to genes related either to virulence or to mobile elements such as phage proteins or insertion sequences. The complete region with the specific insertion in the Salmonella genome was aligned to the E. coli DNA database to detect sequence differences in both genomes. The analyses of DNA sequences adjacent to tRNA genes in S. enterica serovars Typhi and Typhimurium in comparison with the genomes of E. coli K-12 and EHEC O157:H7 resulted in the identification of various known virulence gene clusters and several genomic regions that may constitute new Salmonella-specific chromosomal insertions.
Using this approach, several PAI that are already known were identified. The tRNASelC-associated PAII of E. coli as well as the Salmonella-specific SPI3 at the corresponding tRNA have been retrieved (2). Furthermore, the sopB gene as part of SPI5 in Salmonella was identified in the neighborhood of the tRNASerX (28) and SPI2 adjacent to the tRNAValV gene (14).
Characterization of new tRNA-associated genomic insertions specific to Salmonella.
Four genomic regions located adjacent to tRNA genes show several characteristics of horizontally acquired elements or possess genes indicative of mobile genetic elements, such as transposases and integrases or genes encoding phage tail fiber proteins. The corresponding tRNA loci have not been associated previously with horizontally acquired genetic elements. The specific characteristics of these regions are summarized in Table 2 and are described in detail below.
TABLE 2.
Characteristics of tRNA-associated elements in Salmonella spp. and E. coli
| Associated tRNA gene | Bacterial strain or serovar | Insertion size (kb) | Localization (centisome) |
|---|---|---|---|
| tRNAProL | S. enterica serovar Typhi | 6.3 | 48 |
| S. enterica serovar Typhimurium | 15.8 | 48 | |
| E. coli K-12 | 4.2 | 49 | |
| E. coli O157:H7 | 3.0 | 24 | |
| tRNAArgU | S. enterica serovar Typhi | 15.6 | 13 |
| S. enterica serovar Typhimurium | 23.4 | 13 | |
| E. coli K-12 | 40.0 | 12-13 | |
| E. coli O157:H7 | 30.1 | 12 | |
| tRNAArgW | S. enterica serovar Typhi | 35.7 | 52 |
| S. enterica serovar Typhimurium | 30.0 | 52 | |
| E. coli K-12 | 58.7 | 53-54 | |
| E. coli O157:H7 | 57.2 | 28 | |
| tRNAPheU | S. enterica serovar Typhi | 146.9 | 91-94 (ViaB region) |
| S. enterica serovar Typhimurium | 15.0 | 94 | |
| E. coli K-12 | 10.9 | 94 | |
| E. coli 0157:H7 | 10.9 | 63 |
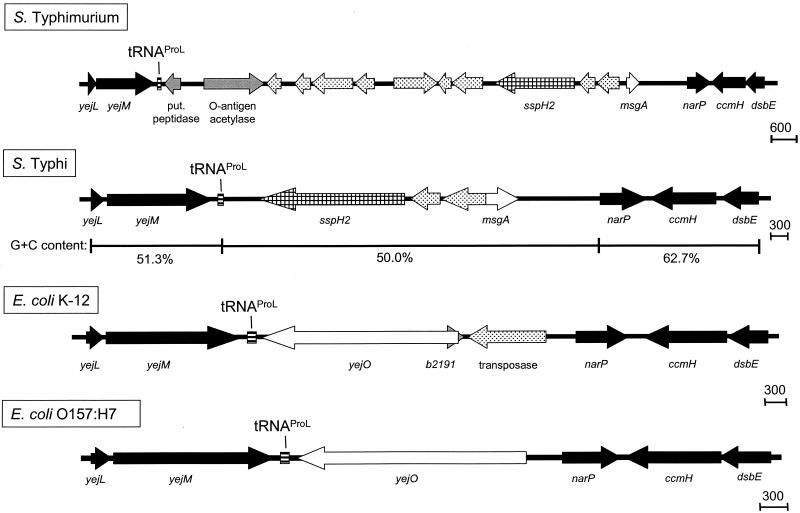
The tRNAProL region.
The gene sspH2 is located in the vicinity of the tRNAProL in the S. enterica serovar Typhi and S. enterica serovar Typhimurium chromosomes (Fig. 1). This gene has recently been described as encoding an effector protein of the type III secretion system encoded by SPI2 of serovar Typhimurium (22). Interestingly, the organizations of the region downstream of sspH2 are different in serovar Typhimurium and serovar Typhi. While the genes upstream of sspH2 in serovar Typhi are similar to those of E. coli K-12 and E. coli O157:H7, a cluster of phage-related genes is located downstream of sspH2 in serovar Typhimurium, followed by the Salmonella-specific gene for O-antigen acetylase and a putative peptidase.
FIG. 1.
Organization of the tRNAProL-associated element. The organizations of the chromosomal regions adjacent to tRNA genes in the two S. enterica serovars Typhi and Typhimurium were compared to those of the corresponding regions of the E. coli K-12 and the E. coli O157:H7 chromosome. Genes shared between the E. coli strains and the Salmonella serovars are depicted by black symbols. Genes that are shared only between E. coli strains or Salmonella serovars are depicted by open symbols, while those genes that are specific to a certain strain or serovar are shown by gray symbols. Genes that are associated with virulence are represented by checked patterns, and tRNA genes and genes with similarity to phage genes are shown by hatched and dotted symbols, respectively. The base composition of the specific region of the serovar Typhi chromosome was analyzed and is expressed as percent G+C.
The insertions between the tRNA gene and the narP gene of Salmonella and E. coli differ in the presence of sspH2. SspH2 is required for virulence of S. enterica serovar Typhimurium in calf models of infection (22). The sspH2 gene is specific for Salmonella and is associated with genes encoding phage proteins. In contrast, at the same tRNA gene in both E. coli strains a gene for a putative ATP-binding component of a transport system (yejO) is present. In addition, in E. coli K-12, but not in E. coli O157:H7, a transposase gene is located adjacent to this gene.
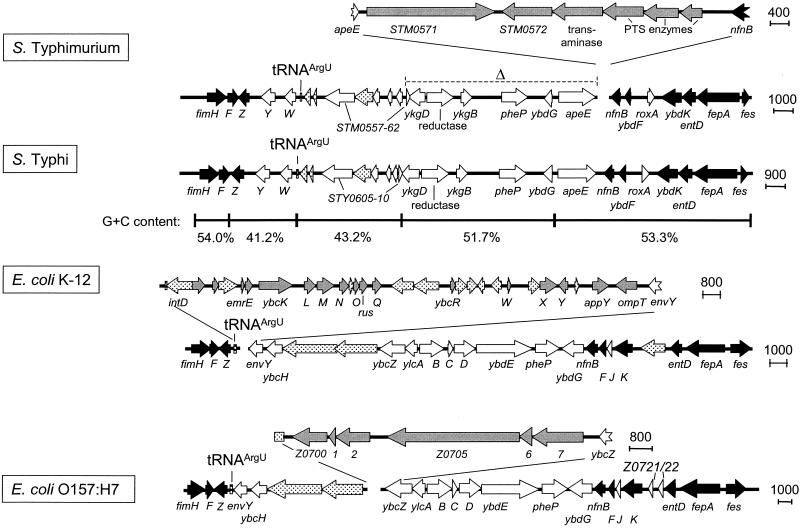
The tRNAArgU region.
The comparative analysis of the vicinity of tRNAArgU indicated that the organization of this region is rather complex (Fig. 2). The regions inserted between the fimbrial operon (fimHFZ) and nfnB gene are very differently organized in Salmonella spp. and E. coli and also exhibit variations within the two E. coli strains and the two Salmonella serovars. In S. enterica serovar Typhimurium, a cluster of genes is inserted between the Salmonella-specific apeE gene and the more globally distributed nfnB. The gene cluster encodes proteins of a phosphotransferase system. This phosphotransferase gene cluster is not present at the same location in the serovar Typhi genome.
FIG. 2.
Organization of the tRNAArgU-associated element. The tRNAArgU-associated element was analyzed as described in the legend to Fig. 1.
A different genetic organization was also detected in the close vicinity of the tRNAArgU gene in both E. coli strains investigated. In addition to many undefined open reading frames (ORFs), the insertion between tRNAArgU and envY in E. coli K-12 contains many genes encoding phage proteins. E. coli O157:H7 also contains a fragment not present at that site in E. coli K-12 (ORFs z0700 to z0707).
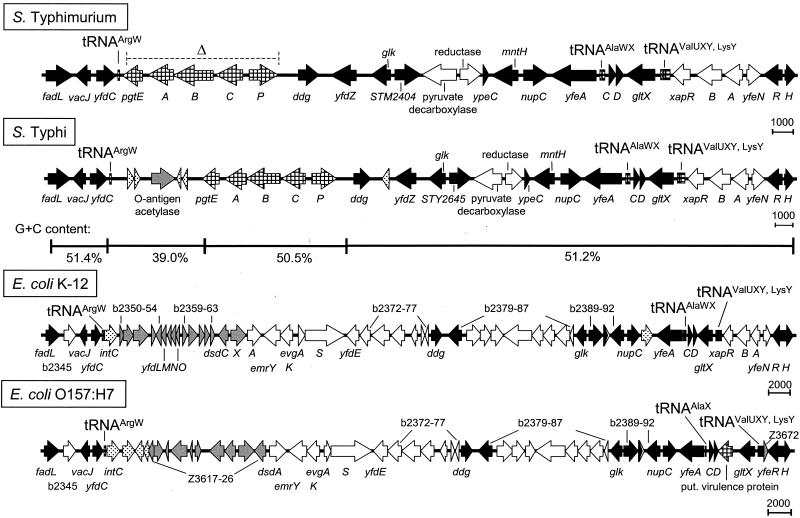
The tRNAArgW region.
The gene yfdC is located adjacent to the tRNAArgW gene in all strains analyzed in this study (Fig. 3). yfeR, the next ORF shared between S. enterica serovar Typhi, S. enterica serovar Typhimurium, E. coli K-12, and E. coli O157:H7, is separated by species-specific insertions of about 31 to 60 kb. The organizations of these insertions are highly different in the four bacterial genomes. A fragment of 35.7 kb is present between the ORFs yfdC and yfeR in serovar Typhi. The gene cluster pgtABCEP containing the gene for the outer membrane protease PgtE is localized in the direct vicinity of the tRNAArgW in serovar Typhimurium. In contrast, four genes encoding phage proteins and a gene for an O-antigen acetylase separate the tRNAArgW gene from the pgt cluster in serovar Typhi. PgtE promotes resistance to alpha-helical antimicrobial peptides and thereby contributes to Salmonella resistance to innate immunity (10). Downstream of pgtP, the same set of genes is inserted in both serovar Typhimurium and serovar Typhi, with the exception of an additional phage gene downstream of ddg in serovar Typhi. Striking differences were seen adjacent to the tRNAArgW gene in both E. coli strains. None contains the pgt cluster, and the genes located between tRNAArgW and dsdA are totally different in E. coli K-12 and E. coli O157:H7. Both gene clusters are associated with phage-related genes. Interestingly, E. coli O157:H7 contains a gene, localized in a small fragment between two tRNA genes (i.e., tRNAAlaX and tRNAValU), which codes for a putative virulence protein that is present neither in the respective region in E. coli K-12 nor in the Salmonella genome.
FIG. 3.
Organization of the tRNAArgW-associated element. The tRNAArgW-associated element was analyzed as described in the legend to Fig. 1. The region of the element that has been deleted and replaced by the kanamycin resistance cassette is indicated by Δ.
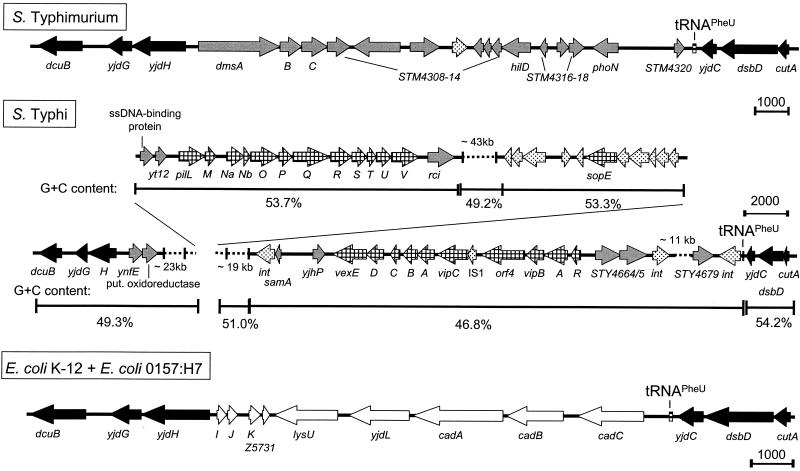
The tRNAPheU region.
An insertion of 147 kb is located adjacent to tRNAPheU gene in the S. enterica serovar Typhi genome. This DNA segment contains several genes and gene clusters whose products are related to Salmonella pathogenicity. Two of these regions have been previously described as residing on the 118-kb major pathogenicity island of the serovar Typhi genome (18, 30). Our data indicate that this major pathogenicity island of serovar Typhi represents a further PAI that is inserted at a tRNA gene.
The viaB region (vexABCDE-vipABCR) is located adjacent to the tRNAPheU gene, and several phage integrase genes are located at this locus (Fig. 4). The ViaB proteins are necessary for the synthesis and export of the Vi capsular antigen to the bacterial cell surface and thereby contribute to the pathogenicity of S. enterica serovar Typhi. The second virulence-relevant region present on the serovar Typhi-specific insertion is the gene cluster for a type IV pilus (pilL-pilV). The gene cluster encodes a thin pilus required for mating in liquid media (29) and has recently been shown to be involved in the entry of serovar Typhi into human intestinal cells (30). Downstream of the pil operon lies rci, a gene that codes for a site-specific recombinase. The Rci protein was shown to invert DNA segments between two inverted repeats, thereby altering the C-terminal region of the PilV protein (31). A third virulence-associated gene product encoded by this large insertion is SopE. This SPI1-translocated effector protein is known to mediate the entry of Salmonella into host cells via modification of host cell signal pathways. In serovar Typhi, sopE is associated with prophage genes and resides between the two clusters of pil and viaB genes at a distance of about 60 kb from the tRNAPheU gene. sopE of serovar Typhimurium is situated within the temperate prophage φsopE (12, 23).
FIG. 4.
Organization of the tRNAPheU-associated element. The tRNAPheU-associated element was analyzed as described in the legend to Fig. 1. The region of the element that has been deleted and replaced by the kanamycin resistance cassette is indicated by Δ.
In contrast, a far smaller fragment of 15 kb is inserted in the serovar Typhimurium genome at the same tRNAPheU, containing a cluster of anaerobic dimethyl sulfoxide reductase genes (dmsABC) and several ORFs not further characterized. An entirely different insertion was found in the genome of E. coli. Strains K-12 and O157:H7 both contain the same 11-kb fragment, which contains the cad operon (cadABC) for lysine decarboxylase and several ORFs with unknown function.
Base compositions of new tRNA-associated genomic insertions in S. enterica serovar Typhi.
Several PAI are characterized by a base composition that is different from the base composition of the residual genome of the pathogen. Therefore, the base compositions of the four new elements associated with tRNA genes were analyzed. The G+C contents of the elements in serovar Typhi were plotted and compared to that of the surrounding genome. The base composition of the short insertion next to tRNAProL containing sspH2 is not significantly different from the average G+C content of Salmonella of about 52%. However, genes upstream of sspH2 which are homologous to E. coli have a rather high G+C content of 63% (Fig. 1). The regions on both sides of tRNAArgU have very low G+C contents of 41 and 43%, respectively, compared to about 54 and 52% in the adjacent regions (Fig. 2). This observation is consistent with the lower G+C content observed for various SPI. A similar observation was made for the Salmonella-specific insertion at tRNAArgW (Fig. 3). The fragment between the tRNA gene and the pgt cluster, which consists of genes encoding phage-related proteins and an O-antigen acetylase, has a G+C content of only 39%, which is significantly lower than that of the rest of the Salmonella-specific insertion (51%). Finally, the G+C contents of different segments of the insertion at the tRNAPheU gene do not exhibit any significant differences (Fig. 4). Only a small difference was observed for the viaB region, which shows the lowest G+C content of approximately 47%.
Phylogenetic distribution of tRNA-associated genomic insertions.
Southern hybridization analysis was performed to elucidate whether the new tRNA-associated genomic insertions are either specific to Salmonella or distributed among other genera of the Enterobacteriaceae and more distantly related gram-negative pathogens. Furthermore, the distribution of these elements within the salmonellae was analyzed by Southern hybridization with genomic DNAs from strains of the SARC collection representing seven S. enterica subspecies and S. bongori. Genomic DNAs of various bacterial species were isolated and hybridized with probes derived from the tRNA-associated genomic insertions by using gene specific primers (Table 1) and serovar Typhi DNA as a template. Hybridization was performed under nonstringent conditions.
To control that the Southern hybridization conditions were sufficient to identify genes that are conserved among the Enterobacteriaceae and other gram-negative pathogens, recA- and gyrB-specific probes encoding a recombinase and a DNA gyrase, respectively, were generated using E. coli DNA as a template. Under the hybridization conditions applied in this study, both probes hybridized to all of the bacterial genomes investigated, proving that the hybridization conditions allowed interspecies hybridization (data not shown).
The apeE-specific probe representing the tRNAArgU-associated element was detected in all S. enterica subspecies analyzed but not in S. bongori (Table 3). A second probe (ORF 0557) hybridized only with DNAs of isolates of subspecies I. With either probe, no hybridization signals were observed with the Enterobacteriaceae and other gram-negative pathogens (Table 4). Probes for sspH2 and msgA of the tRNAProL-associated element revealed a heterogeneous pattern of distribution within the salmonellae, but the genes were not detected in other species. The distribution of sspH2 within different Salmonella subspecies (SARC) appeared to be heterogeneous, as has been shown previously (26). The use of probes pgtE and nupC indicated the presence of the tRNAArgW-associated element in all Salmonella isolates apart from SARC 9. Further hybridization studies indicated that pgtE is restricted to Salmonella spp., while nupC was detected in most gram-negative bacteria examined (Table 4). The genes pilV and vexE are located on the large insertion of 147 kb in the tRNAPheU-associated element of the serovar Typhi genome. pilV and vexE are restricted to serovar Typhi and are not present in any other species examined (Tables 3 and 4).
TABLE 3.
Distribution of tRNA-associated elements within the salmonellaea
| Salmonella subspecies (strain) | Presence of hybridization signal
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tRNAArgU
|
tRNAArgW
|
tRNAProL
|
tRNAPheU
|
|||||
| apeE | orf0557 | pgtE | nupC | sspH2 | msgA | vexE | pilV | |
| I (SARC 1) | + | + | + | + | + | + | − | − |
| I (SARC 2) | + | + | + | + | + | + | + | + |
| II (SARC 3) | + | − | + | + | − | + | − | − |
| II (SARC 4) | + | − | + | + | − | + | − | − |
| IIIa (SARC 5) | + | − | + | + | + | + | − | − |
| IIIa (SARC 6) | + | − | + | + | + | + | − | − |
| IIIb (SARC 7) | + | − | + | + | − | NDb | − | − |
| IIIb (SARC 8) | + | − | + | + | − | + | − | − |
| IV (SARC 9) | + | − | − | + | + | ND | − | − |
| IV (SARC 10) | + | − | + | + | + | + | − | − |
| V (SARC 11) | − | − | + | + | − | + | − | − |
| V (SARC 12) | − | − | + | + | − | + | − | − |
| VI (SARC 13) | + | − | + | + | − | + | − | − |
| VI (SARC 14) | + | − | + | + | − | + | − | − |
| VII (SARC 15) | + | − | + | + | + | + | − | − |
| VII (SARC 16) | + | − | + | + | + | ND | − | − |
The SARC collection, representing S. enterica subspecies I, II, IIIa, IIIb, IV, VI, and VII and S. bongori, was analyzed for the presence of apeE and orf0557 (tRNAArgU), pgtE and nupC (tRNAArgW), sspH2 and msgA (tRNAProL), and vexE and pilV (tRNAPheU).
ND, not clearly distinguishable.
TABLE 4.
Distribution of tRNA-associated elements within different gram-negative bacteriaa
| Species, serovar, or strainb | Presence of hybridization signal
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tRNAArgU
|
tRNAArgW
|
tRNAProL
|
tRNAPheU
|
|||||
| apeE | orf0557 | pgtE | nupC | sspH2 | msgA | vexE | pilV | |
| S. enterica serovar Typhimurium 12023 | + | + | + | + | + | + | − | − |
| S. enterica serovar Typhi | + | + | + | + | + | + | + | + |
| S. enterica serovar Typhimurium LT2 | + | + | + | + | + | + | − | − |
| E. coli DH5α | − | − | − | + | − | − | − | − |
| EPEC (2) | − | − | − | + | − | − | − | − |
| EHEC (2) | − | − | − | + | − | − | − | − |
| ETEC | − | − | − | + | − | − | − | − |
| EIEC | − | − | − | + | − | − | − | − |
| EAEC | − | − | − | + | − | − | − | − |
| Enterobacter cloacae | − | − | − | + | − | − | − | − |
| Shigella flexneri (2) | − | − | − | + | − | − | − | − |
| Klebsiella pneumoniae (2) | − | − | − | + | − | − | − | − |
| Pseudomonas aeruginosa (3) | − | − | − | − | − | − | − | − |
| Yersinia enterocolitica (2) | − | − | − | + | − | − | − | − |
| Yersinia pseudotuberculosis | − | − | − | + | − | − | − | − |
| Vibrio cholerae | − | − | − | + | − | − | − | − |
Various gram-negative laboratory strains or pathogens isolated from clinical samples were analyzed by Southern hybridization for the presence of apeE and orf0553 (tRNAArgU), pgtE and nupC (tRNAArgW), sspH2 and msgA (tRNAProL), and vexE and pilV (tRNAPheU).
The number of clinical isolates analyzed is indicated in parentheses. EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; EIEC, enteroinvasive E. coli; EAEC, enteroaggregative E. coli.
These data support the hypothesis that all four tRNA gene-associated elements described in this study are Salmonella-specific insertions. Hybridization data such as those obtained with the nupC probe also indicate that the tRNA-associated elements have a mosaic structure of several independent acquisitions during evolution.
Insertion of tRNA-associated elements.
The characterization of PAI of various pathogens indicated that the insertion points in the chromosome are different in different strains or isolates. We analyzed the vicinities of the four tRNA-associated elements identified in this study. PCR primers were selected for amplicons consisting of the terminal region within the tRNA-associated element, the tRNA gene, and the proximal region outside the tRNA-associated element. The results indicated that the insertion points of the tRNAArgU- and tRNAArgW-associated elements are conserved within the salmonellae (Table 5). Remarkable heterogeneity was observed for the tRNAProL-associated insertion, which may be due to the presence of additional genes in strains of the SARC collection other than S. enterica serovar Typhimurium. The insertion point at tRNAPheU was conserved only between strains SARC 1, SARC 3, and SARC 13. This observation was consistent with different compositions of this region in serovars Typhimurium and Typhi (Fig. 4).
TABLE 5.
tRNA insertion sites of DNA elements within the SARC collection representing S. enterica subspecies I, II, IIIa, IIIb, IV, VI, and VII and S. bongori subspecies V
| Salmonella subspecies (strain) | Presence of PCR productsa
|
|||
|---|---|---|---|---|
| tRNAArgU | tRNAArgW | tRNAProL | tRNAPheU | |
| I (SARC 1) | + | + | + | + |
| I (SARC 2) | + | + | (+) | − |
| II (SARC 3) | + | + | − | + |
| II (SARC 4) | + | + | − | − |
| IIIa (SARC 5) | + | + | (+) | − |
| IIIa (SARC 6) | + | + | − | − |
| IIIb (SARC 7) | + | + | − | − |
| IIIb (SARC 8) | + | + | − | − |
| IV (SARC 9) | + | (+) | (+) | − |
| IV (SARC 10) | + | + | (+) | − |
| V (SARC 11) | + | + | − | − |
| V (SARC 12) | − | + | − | − |
| VI (SARC 13) | + | − | (+) | + |
| VI (SARC 14) | + | + | (+) | − |
| VII (SARC 15) | + | + | − | − |
| VII (SARC 16) | + | + | (+) | − |
(+), PCR products with a size different from that obtained with S. enterica serovar Typhimurium
Role of tRNA-associated elements in Salmonella virulence.
To analyze the contribution to virulence of Salmonella-specific genomic elements identified in this study, deletions were constructed using the one-step inactivation approach. The ykgD-to-apeE region of the tRNAArgU-associated element and the pgt gene cluster of the tRNAArgW-associated element were replaced by a kanamycin resistance cassette as indicated in Fig. 2 and Fig. 3, respectively. Virulence of the mutants was assessed by determination of the CI (1). Four days after intraperitoneal infection of mice with inocula of 104 CFU containing equal amounts of wild-type S. enterica serovar Typhimurium and a mutant strain, mice were sacrificed and the loads of liver and spleen with the wild-type and mutant strains were determined. Control experiments with a mixture of wild-type serovar Typhimurium and a highly attenuated sseC::aph mutant strain resulted in a CI of 0.00071 (±0.00018). Competition experiments with wild-type strain versus the strain with a pgtEABCP deletion resulted in a CI of 1.40 (±0.48), and a CI of 1.62 (±0.38) was obtained for the wild-type strain versus the ykgD-apeE deletion strain. These data indicate that neither the ykgD-apeE gene cluster of the tRNAArgU-associated element nor the pgtEABCP gene cluster of the tRNAArgW-associated element is required for the progression of systemic infections in mice.
DISCUSSION
Acquisition and integration of horizontally acquired genetic information into the chromosome is considered an important factor for the evolution of bacterial species. This process is also important for the evolution of pathogens from their nonpathogenic ancestors. A subset of horizontally acquired genetic elements conferring virulence functions have been defined as PAI.
Various approaches have been devised for the identification of pathogen-specific genomic elements, for example, hybridization techniques (7, 9) or mathematical approaches using specific algorithms to detect alien genes (17). Previous observations indicated that a large number of PAI are inserted adjacent to tRNA genes in the chromosomes of various pathogens (reviewed in reference 11). Based on these observations, we devised an approach to identify pathogen-specific genomic insertions at tRNA genes in Salmonella. The genetic organizations of the vicinities of tRNA genes in E. coli and Salmonella spp. were compared. This approach resulted in the identification of 20 loci adjacent to tRNA genes that show a different organization in Salmonella spp. and E. coli strains. In Salmonella spp., several of these species-specific regions also contained genes that are indicative of mobile genetic elements. Insertions, identified by tRNA scanning, comprise several already known elements as well as various new elements. In this study, a subset of these regions was analyzed in detail. Southern blot analyses of the distribution of these loci within the Enterobacteriaceae and other gram-negative pathogens confirmed that these insertions are specific for Salmonella spp. Detailed characterization revealed the presence of several known virulence genes within the tRNA-associated elements: sspH2, encoding a translocated effector protein of the type III secretion system of SPI2 (22) in the tRNAProL-associated element; sopE (12) and the pil (31) and vip (30) gene clusters in the tRNAPheU-associated element; and pgtE in the tRNAArgW-associated element (10). These genes have been previously described as single virulence gene loci. Our analyses indicate that these virulence genes may have been transferred to Salmonella spp. by acquisition of large genetic elements. Comparative analyses also indicate that further species-specific insertions are present at various tRNA genes in an E. coli laboratory strain and a pathogenic strain of E. coli. Remarkably, the cadABC gene cluster located at tRNAPheU in E. coli is not present in this position in Salmonella. The deletion of the cadA region has previously been correlated with increased virulence of Shigella spp. and enteroinvasive E. coli (21). Homologues of cadA have been detected at other locations in the genomes of S. enterica serovars Typhi and Typhimurium (data not shown).
A small insertion at tRNAArgW in serovar Typhi contains a gene for an O-antigen acetylase and is characterized by a low G+C content of 39%. This element is not present in serovar Typhimurium and may represent a recent insertion. We also observed that a different base composition is not always a firm marker for the identification of horizontal acquisitions. Although the tRNA-associated elements described in this study clearly represent horizontal acquisitions, the base composition of these elements is not significantly different from that of the residual genome of Salmonella spp. A similar observation has been made for SPI2, which is composed of two horizontally acquired elements at tRNAValV (13). Only one element, encoding a type III secretion system, shows a significantly lower G+C content, while the G+C content of the second element, encoding metabolic functions, is similar to the average G+C content of the serovar Typhimurium genome. It could be speculated that maintenance of a specific codon usage in a horizontally acquired element is required for the coordinate regulation of these genes and that the divergent base composition is a secondary effect of the biased codon usage.
All of the tRNA-associated elements identified in this study exhibit a remarkable association with phage genes. These genes may represent remnants of phage genomes, since complete phage genomes were not detected in the tRNA-associated elements. The presence of these genes is an indication of the previous acquisition of the genomic elements via phages as vehicles for horizontal gene transfer. Integration of phages at tRNA genes has been observed and may represent a mechanism to extend the host range of a phage, due to the fact that tRNA genes are highly conserved between various bacterial genomes. However, the ancestry and distribution of the tRNA-associated elements remain open to question, as hybridization analyses did not reveal the presence of these elements in other bacterial species.
In summary, all of the new tRNA gene-associated elements identified in this study are specific for Salmonella spp. The distribution of these elements is variable within the salmonellae, ranging from the pilV insertion that is specific for serovar Typhi to the pgtE element present in all subspecies of S. enterica as well as S. bongori. These elements have some characteristics of PAI, but additional work is required to analyze the role of these elements in Salmonella virulence. The divergent distribution of the elements within the subspecies of Salmonella may indicate that these elements contribute to the different pathogenic potentials of the various subspecies, such as host restriction or disease outcome.
Based on genome sequence data, the systematic scanning for tRNA-associated elements of bacterial pathogens represents a useful tool for the rapid identification of species-specific horizontal acquisition and may lead to the identification of new PAI.
Acknowledgments
This work was support by Deutsche Forschungsgemeinschaft grant HE1964/4-2.
We are grateful to Martin Röllinghoff for generous support of our work and to Brad Taylor and Volker Kuhle for critical reading of the manuscript. We thank the Genome Sequencing Center, Washington University (St. Louis, Mo.), and the Sanger Centre (Cambridge, United Kingdom) for communication of DNA sequence data prior to publication.
Editor: B. B. Finlay
REFERENCES
- 1.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of sifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., B. M. Davis, and B. Hochhut. 2001. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9:137-144. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman, E. A., and H. Ochman. 1997. How Salmonella became a pathogen. Trends Microbiol. 5:343-349. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, E. A., M. A. Sturmoski, F. R. Solomon, R. Lin, and H. Ochman. 1993. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc. Natl. Acad. Sci. USA 90:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 12.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella Pathogenicity Island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 14.Hensel, M., J. E. Shea, A. J. Bäumler, C. Gleeson, F. R. Blattner, and D. W. Holden. 1997. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J. Bacteriol. 179:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella Pathogenicity Island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 16.Hou, Y.-M. 1999. Transfer RNAs and pathogenicity islands. Trends Biochem. Sci. 24:295-298. [DOI] [PubMed] [Google Scholar]
- 17.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 18.Liu, S. L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maloy, S. R., V. L. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145-156. [DOI] [PubMed] [Google Scholar]
- 21.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Bäumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 23.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschäpe, H. Rüssmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 25.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 26.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883-891. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, T., S. R. Kim, and T. Komano. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, X. L., C. Morris, and J. Hackett. 1997. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major ‘pathogenicity island' of Salmonella typhi. Gene 202:139-146. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]