Salmonella enterica serotypes form a group of pathogens that differ widely in their host range within mammals and birds (Table 1). Members of S. enterica seem to lie along a spectrum in terms of host range. At one end of this spectrum, S. enterica serotype Typhi is perhaps the most highly host-adapted pathogen of this group, causing disease only in humans and higher primates (23). Serotypes that have the ability to cause disease and persist in multiple different species have a broad host range and thus define the other end of this spectrum. Traditionally, S. enterica serotype Typhimurium has been thought of as the prototypical broad-host-range serotype, since it is frequently associated with disease in numerous species, including humans, livestock, domestic fowl, rodents, and birds (Table 1). However, here we present the alternate view that some serotype Typhimurium variants have a very narrow host range, while others are clearly able to infect and persist in multiple species. Therefore, it may be more accurate to describe serotype Typhimurium as a collection of variants that vary significantly in their host range and their degree of host adaptation.
TABLE 1.
Host adaptation of S. enterica serotypes revealed by epidemiological surveys
| Reservoir | Location of survey (time period [yrs] covered) | S. enterica serotypes most frequently associated with disease (% of total isolates) | Reference |
|---|---|---|---|
| Humans | Massachusetts (1940-45) | Typhimurium (31), Typhi (18), Montevideo (10), Newport (8), Oranienburg (7) | 39 |
| Korea (1951) | Paratyphi A (61), Typhi (11), Paratyphi C (10), Enteritidis (8), Paratyphi B (6) | 71 | |
| England and Wales (1958-67) | Typhimurium (64), Paratyphi B (8) | 42 | |
| United States (1987-97) | Typhimurium (23), Enteritidis (21), Heidelberg (8) | 47 | |
| Cattle | Germany (1937) | Dublin (82), Typhimurium (15) | 37a |
| England and Wales (1958-67) | Dublin (71), Typhimurium (27) | 56 | |
| England and Wales (1968-73) | Dublin (79), Typhimurium (17) | 57 | |
| United States (1976) | Typhimurium (54), Dublin (20) | 9 | |
| England and Wales (1982-83) | Typhimurium (63), Dublin (24) | 58 | |
| Swine | United States (1934-41) | Choleraesuis (70), Typhimurium (11) | 16 |
| Germany (1937) | Choleraesuis (50), Typhimurium (27), Dublin (23) | 37a | |
| England and Wales (1958-67) | Choleraesuis (83), Typhimurium (8) | 56 | |
| England and Wales (1968-73) | Choleraesuis (57), Typhimurium (17), Dublin (15) | 57 | |
| United States (1976) | Choleraesuis (52), Typhimurium (21) | 9 | |
| Sheep | England and Wales (1958-67) | Abortusovis (63), Dublin (26), Typhimurium (7) | 57 |
| England and Wales (1968-74) | Dublin (45), Abortusovis (36), Typhimurium (9) | 59 | |
| England and Wales (1975-81) | Typhimurium (26), Dublin (24), Montevideo (11) | 60 | |
| Horses | United States (1934-41) | Abortusequi (68), Typhimurium (32) | 16 |
| England and Wales (1973-79) | Typhimurium (70) | 68 | |
| Rodents | United States (1934-41) | Typhimurium (51), Enteritidis (31) | 16 |
| Germany (1978) | Typhimurium (51), Enteritidis (39) | 33 | |
| Chickens | England and Wales (1933-44) | Gallinarum (92) | 24 |
| Denmark (1934-38) | Typhimurium (96) | 25 | |
| England and Wales (1948-56) | Gallinarum (76), Typhimurium (14), Thompson (8) | 7 | |
| England and Wales (1958-67) | Gallinarum (67), Typhimurium (18) | 56 | |
| England and Wales (1968-73) | Enteritidis (46), Typhimurium (41), Gallinarum (7) | 57 | |
| Turkeys | England and Wales (1933-44) | Typhimurium (46), Gallinarum (27), Enteritidis (27) | 24 |
| England and Wales (1948-56) | Typhimurium (57), Stanleyville (12), Thompson (10) | 7 | |
| Ducks | England and Wales (1933-44) | Typhimurium (71), Enteritidis (21) | 24 |
| Denmark (1934-38) | Typhimurium (58), Enteritidis (42) | 25 | |
| England and Wales (1948-56) | Typhimurium (76), Thompson (9) | 7 | |
| United States (1950-60) | Typhimurium (93) | 51 | |
| Pigeons | England and Wales (1933-44) | Typhimurium (100) | 24 |
| Denmark (1934-38) | Typhimurium (100) | 25 | |
| England and Wales (1948-56) | Typhimurium (100) | 7 | |
| Germany (1956-1958) | Typhimurium (99) | 27 | |
| Massachusetts (1960-64) | Typhimurium (94) | 18 | |
| Finches | England and Wales (1948-56) | Typhimurium (100) | 7 |
This report by Lerche was one of the first to describe host adaptation of Salmonella serotypes.
METHODS FOR DIFFERENTIATING BETWEEN SEROTYPE TYPHIMURIUM VARIANTS
In order to draw accurate conclusions about the host range of serotype Typhimurium variants, it is necessary to have methods to distinguish isolates from different sources from one another. A number of techniques are presently in use to differentiate serotype Typhimurium isolates for epidemiological analysis. The presence or absence of the O5 antigen is used to differentiate between serotype Typhimurium (which possesses the O5 antigen) and its serological variant Copenhagen (which lacks the O5 antigen) (32). Although serological techniques can be used to differentiate between serotype Typhimurium isolates, differences are revealed at much higher resolution by phage typing.
Phage typing distinguishes serotype Typhimurium variants based on their susceptibility to a set of bacteriophages. The phage typing system developed by Lilleengen in 1948 distinguishes 24 Lilleengen types among serotype Typhimurium isolates (38). A second typing system described by Callow in 1959 distinguishes 17 phage types (10). Between 1974 and 1996, epidemiological national surveillance at the Robert Koch Institute in Germany was performed using a combination of the Callow and Lilleengen typing systems (34). Since 1996, a phage typing system originally described by Felix and Callow (20) and later extended by Anderson et al. (2) that distinguishes more than 300 definitive phage types (DT) (Robert Koch Institut and L. R. Ward, personal communication) has been used for surveillance in Germany. In most cases, the phage type is a stable property that provides an opportunity to follow the spread of a particular serotype Typhimurium clone within different host populations over years or even decades (1). For example, molecular fingerprinting shows that the majority of DT104 isolates from Germany, Austria, the United Kingdom, the United Arab Emirates, the Philippines, and The Netherlands belong to a single clone that has spread pandemically during the 1990s (50). In addition to molecular fingerprinting, serotype Typhimurium phage types can be further differentiated by biotyping, a method based on fermentative characteristics that was introduced by Edwards in 1936 (13). A modified biotyping scheme developed by Duguid and coworkers (12) is presently used to further differentiate between isolates of the same phage type (1).
The sensitivity and stability of phage typing make this a very attractive method for determining the spread of different, yet very closely related, serotype Typhimurium clones over time. Thus, phage typing in combination with other typing methods can be used to determine in which host reservoirs a particular serotype Typhimurium clone is associated with disease.
THE PIGEON-ADAPTED VARIANT OF SEROTYPE TYPHIMURIUM
Based on two lines of evidence, serotype Typhimurium variants that are associated with disease in pigeons may be considered highly host adapted. First, serotype Typhimurium is the only Salmonella serotype frequently associated with serious systemic disease (avian paratyphoid) in pigeons (Table 1) (37) and it causes substantial mortality in this host species (17, 27). Because of its ability to cause fatal systemic disease in pigeons, serotype Typhimurium is similar to other highly host-adapted S. enterica serotypes that are characterized by their ability to cause systemic disease associated with high mortality rates in their respective host reservoirs. These include the poultry-adapted S. enterica serotype Gallinarum (fowl typhoid and pullorum disease caused by biotypes Gallinarum and Pullorum, respectively), the porcine-adapted S. enterica serotype Choleraesuis (pig paratyphoid), the bovine-adapted S. enterica serotype Dublin (bacteremia), and human-adapted serotype Typhi (typhoid fever).
Second, epidemiological evidence indicates that serotype Typhimurium isolates from pigeons are distinct from those cultured from other host species. In 1935, Edwards noted that all serotype Typhimurium isolates from pigeons lack the O5 antigen and thus belong to the serological variant Copenhagen (14, 15), and subsequent investigations have confirmed this finding (Table 2). Since the serological variant Copenhagen is also responsible for sporadic cases of disease in humans and other mammals, there was considerable concern in the first half of the 20th century that pigeons may be a reservoir for salmonellosis in mammalian species. However, subsequent studies have shown that serotype Typhimurium isolates from pigeons differ in their biotype and ribotype from those isolated from humans or other animals, suggesting that pigeons are not a source of infection for other host reservoirs (46, 55, 65, 69). Based on these data, the existence of a serotype Typhimurium “pigeon type” that predominates in pigeons but is rarely transmitted to other host species has been proposed (25, 31, 35, 38).
TABLE 2.
Association of serotype Typhimurium var. Copenhagen with pigeons between 1933 and 1970
| Time period (yrs) of isolation | No. of serotype Typhimurium cultures investigated | % Cultures of variant Copenhagen | Reference |
|---|---|---|---|
| 1933-44 | 5 | 100 | 24 |
| Before 1937 | 35 | 100 | 14 |
| 1934-38 | 79 | 100 | 25 |
| Before 1948 | 14 | 100 | 38 |
| 1955-64 | 25 | 88 | 17 |
| 1960-64 | 16 | 94 | 18 |
| 1968-70 | 45 | 100 | 69 |
| 1969 | 292 | 97 | 55 |
| 1969-70 | 145 | 98 | 70 |
| Total | 656 | 98 |
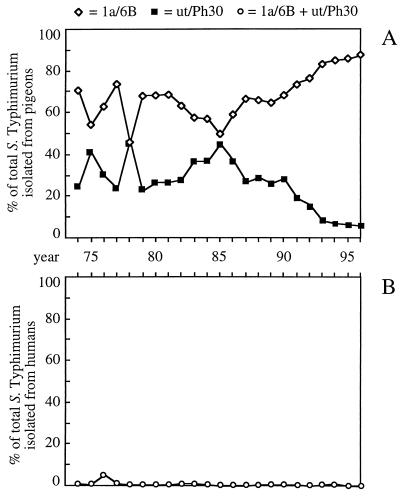
With the advent of phage typing it became increasingly clear that serotype Typhimurium var. Copenhagen isolates from pigeons represent variants with a very narrow host range. Surveillance in western Germany performed between 1969 and 1978 showed that 80% of cases of fatal DT2 infection occur in pigeons (8). Surveillance in eastern Germany (and later in the reunited Germany) performed between 1974 and 1996 showed that the Callow/Lilleengen phage types 1a/6B (which corresponds to DT2 in the Anderson system) and ut/Ph30 have persisted in pigeon populations for decades (Fig. 1A) but were never frequently isolated from humans (Fig. 1B). During this time, Callow/Lilleengen type 1a/6B (DT2) was never frequently isolated from animal sources other than pigeons in Germany (Table 3). Callow/Lilleengen type ut/Ph30 can be differentiated into 14 Anderson phage types; however, retrospective analysis shows that all ut/Ph30 isolates from pigeons correspond to Anderson phage type DT99 (W. Rabsch, Abstr. Epidémiol. Santé Anim., abstr. 12.18, p.31-32, 1997). Since the Anderson system was introduced in 1996 for national surveillance in Wernigerode, Germany, DT99 has rarely been isolated from sources other than pigeons (W. Rabsch and H. Tschäpe, unpublished data). Several studies show that DT2 and DT99 cultures represent the phage types predominantly isolated from pigeons in Europe and North America (1, 4, 8, 35, 43, 70), suggesting a wide geographic distribution of these clones.
FIG. 1.
Association of serotype Typhimurium var. Copenhagen phage types 1a/6B (DT2) and ut/Ph30 (DT99) with disease in pigeons in Germany between 1974 and 1996. Serotype Typhimurium strains were phage typed in the National Reference Laboratory for Salmonella Berlin, Wernigerode Branch, which was a part of the Federal Institute for Health Protection of Consumers and Veterinary Medicine Berlin. (A) Number of annual 1a/6B (open diamonds) and ut/Ph30 (closed squares) isolations from pigeons expressed as a percentage of the total number of serotype Typhimurium cultures from this source. (B) Sum of annual 1a/6B and ut/Ph30 isolations (open circles) from humans in Germany expressed as a percentage of the total number of serotype Typhimurium cultures from this source.
TABLE 3.
Association of serotype Typhimurium phage types 1a/6B (DT2), 4/3 (DT8), and 1aPhi4/6B (DT46) with avian sources in Germany between 1974 and 1996
| Source | Total no. of serotype Typhimurium cultures | % Isolates of indicated phage type
|
||
|---|---|---|---|---|
| 4/3 (DT8) | 1aPhi4/6B (DT46) | 1a/6B (DT2) | ||
| Cattle | 11,558 | 0.6 | 1.4 | 0.5 |
| Pigs | 3,701 | 0.5 | 2.7 | 1.2 |
| Domestic fowl | 3,528 | 5.0 | 2.2 | 2.8 |
| Waterfowl | 2,854 | 18.1 | 36.6 | 1.5 |
| Pigeons | 5,573 | 0.3 | 0.3 | 67.6 |
The available epidemiological evidence supports the idea that disease in pigeons is caused predominantly by serotype Typhimurium variants DT2 and DT99. Phage types DT2 and DT99 are isolated extremely infrequently from species other than pigeons. This observation illustrates that these serotype Typhimurium variants have a narrow host range. Conversely, pigeons are very rarely infected with phage types other than DT2 and DT99, indicating that other serotype Typhimurium isolates are not well adapted to this host species. It is therefore reasonable to conclude that phage types DT2 and DT99 represent a pigeon-adapted variant of serotype Typhimurium.
SEROTYPE TYPHIMURIUM VARIANTS ASSOCIATED WITH OTHER AVIAN SOURCES AND WITH FOOD-BORNE DISEASE IN HUMANS
While pigeon paratyphoid is caused primarily by pigeon-adapted variants of serotype Typhimurium, analysis of isolates from other avian sources reveals that disease is caused by a variety of clones, some of which appear to have a narrow host range while others have a broad host range. Broad-host-range variants that are isolated from avian sources include phage types DT49 and DT104, both of which are frequently isolated from poultry (3, 8, 62). Both phage types are also frequently isolated from livestock and cases of human disease (8, 22, 61, 62). However, several phage types associated with disease in certain avian species seem to have a very narrow host range. Although early evidence for the existence of avian-adapted variants based on biotyping was not compelling, subsequent analysis using phage typing strongly supports this view.
In the 1960s, Morgenroth and Duguid described a non-type 1, fimbriate, non-inositol-fermenting, and non-rhamnose-fermenting biotype of serotype Typhimurium that was designated FIRN (45). A survey of serotype Typhimurium cultures isolated from 57 different countries between 1920 and 1975 reveals that FIRN variants are primarily associated with an avian reservoir (1). In this study, of 124 FIRN cultures from animal sources, 78% were from birds and 22% were from mammals. Of the remaining 470 non-FIRN cultures from animal sources, only 23% were from birds (1). However, FIRN variants of serotype Typhimurium are a heterogeneous group with regard to phage type. FIRN strains belong to 27 different phage types (with DT13, DT14, DT40, and DT80 being the most common), and most of these also contain non-FIRN isolates (1). Thus, FIRN strains are not likely to represent a clonal group and this biotype is not an adequate criterion to distinguish between avian and mammalian isolates.
Phage typing has subsequently provided more compelling evidence for the association of particular serotype Typhimurium isolates with particular avian hosts. In wild birds, surveillance in England and Germany showed that DT40 isolates of the FIRN biotype are the serotype Typhimurium variants most commonly associated with disease (8, 49; Rabsch, Abstr. Epidémiol. Santé Anim.). Phage type DT40 has been reported to be responsible for annual spring incidents of wild bird (mainly songbirds and finches) mortality in England (40). In Germany, most fatal cases of disease in animals associated with DT40 occur in wild or zoo-kept birds, and this phage type appears to cause only rare isolated cases of disease in other species (8, 35). Phage type DT40 was responsible for an epizootic of salmonellosis in feeder birds (mainly northern flocking songbirds) in northeastern America in the winter of 1997-1998, and this phage type was noted to be very rarely isolated from other species (11, 51). These data suggest that phage type DT40 represents an avian-adapted serotype Typhimurium variant with narrow host range and wide geographic distribution.
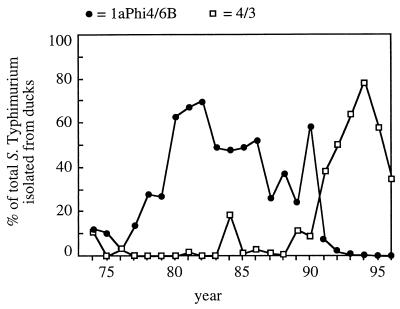
Serotype Typhimurium cultures from ducks are a source of human infection but appear to differ from those isolated from livestock or other avian reservoirs. This was first noted by Hohn and Herrmann in 1937 using biotyping (28, 29) and was subsequently confirmed by phage typing (Table 3). Surveillance in Germany performed between 1974 and 1996 revealed that the majority of duck cultures belong to two Callow/Lilleengen phage types, 1aPhi4/6B and 4/3 (Fig. 2). During this time period, 1aPhi4/6B and 4/3 were associated with a small number of human serotype Typhimurium infections traced back to the consumption of raw or undercooked duck eggs but were rarely isolated from other sources (Table 3). Phage types 1aPhi4/6B and 4/3 correspond to DT46 and DT8 in the Anderson typing system, respectively, and both are characterized by a positive reaction with typing phage F4 of Callow (10) that is rarely observed for other groups. However, clones persisting in populations of ducks seem to be less conserved temporally than those isolated from pigeons and wild birds. For instance, the preponderance of 1aPhi4/6B (DT46) in the 1980s and of 4/3 (DT8) in the 1990s suggests that the persistence of serotype Typhimurium in populations of ducks is characterized by successive epidemics caused by distinct bacterial clones (Fig. 2).
FIG. 2.
Association of serotype Typhimurium phage types 1aPhi4/6B (DT46) and 4/3 (DT8) with disease in ducks in Germany between 1974 and 1996. Serotype Typhimurium strains were phage typed in the National Reference Laboratory for Salmonella Berlin, Wernigerode Branch, which was a part of the Federal Institute for Health Protection of Consumers and Veterinary Medicine Berlin. The number of annual 1aPhi4/6B (closed circles) and 4/3 (open squares) isolations from ducks is expressed as a percentage of the total number of serotype Typhimurium cultures from this source.
In conclusion, a subset of serotype Typhimurium clones associated with disease in ducks (i.e., DT8 and DT46) and wild birds (DT40) appear to have a narrow host range and may be considered host adapted. These host-adapted variants coexist in nonpigeon avian reservoirs with serotype Typhimurium phage types (i.e., DT49 and DT104) that appear to represent true broad-host-range variants.
SEROTYPE TYPHIMURIUM STRAINS ASSOCIATED WITH DISEASE IN LIVESTOCK AND WITH FOOD-BORNE DISEASE IN HUMANS
Serotype Typhimurium isolates cultured from livestock are a diverse group of organisms (as indicated by the presence of >80 phage types) that are frequently transmitted to humans via food products. Phage types of serotype Typhimurium associated with disease in livestock generally are associated with a broader host range. These isolates can be differentiated from those phage types associated with disease in pigeons. However, as pointed out above, a distinction between isolates from mammalian sources and those cultured from avian sources other than pigeons is only possible for a subset of clones.
An interesting lesson to be learned from epidemiological surveillance of livestock, particularly cattle, in England and Germany is that the persistence of serotype Typhimurium in this reservoir is characterized by a series of small epidemics, each caused by a distinct bacterial clone defined by phage typing. After dominating for a period of time in their bovine reservoir, each epidemic serotype Typhimurium strain is replaced by a new clone, as indicated by the dominance of a new phage type (reviewed in reference 53). For instance, after its appearance in Britain in 1974, DT204 spread epidemically among cattle herds and became the predominant serotype Typhimurium clone associated with bovine infections in 1977 (64). DT204 was replaced by DT204c, a strain first isolated in England during an outbreak in a calf-rearing complex in 1979, as the phage type most commonly associated with disease in cattle (63). DT204c became the predominant phage type causing salmonellosis in calves by the end of 1980 and caused an epidemic in Britain that lasted until 1991 (67). DT204c coexisted in its bovine animal reservoir with other serotype Typhimurium phage types, most importantly DT49 (62). In 1992 DT204c was replaced as the most prevalent cattle isolate by DT104, a phage type which is presently dominant in Britain and that was first isolated in 1984 (61). A similar rise and fall of serotype Typhimurium clones defined by phage typing has also been observed in cattle populations in Germany (30, 36). Although there is a succession of epidemic phage types, these variants appear to have a broad host range. For example, in addition to circulating within cattle populations, DT49 was also commonly isolated from poultry and was the phage type most frequently associated with human disease in England and Wales during the years 1985 through 1988 (62). In Germany, multidrug-resistant DT204 was frequently isolated from pigs and poultry in addition to its association with disease in cattle. Similarly, DT104 spread from cattle to other food animals, including swine and poultry (3). In conclusion, serotype Typhimurium variants circulating in livestock appear to have a true broad host range.
CONCLUSIONS
Serotype Typhimurium is frequently associated with disease in many different mammalian and avian host species (Table 1). Serotype Typhimurium cultures can be differentiated into numerous distinct variants by phage typing and other epidemiological typing methods. Some of these variants are frequently associated with disease in a single host reservoir but are rarely cultured from other sources, indicative of a narrow host range. The most striking example of this apparent adaptation to one particular host reservoir is in serotype Typhimurium phage types DT2 and DT99 cultured from pigeons. Serotype Typhimurium pigeon isolates are almost exclusively associated with disease in this reservoir (and not in other birds or mammals) since surveillance began some seven decades ago. Furthermore, DT2 and DT99 seem to be the only serotype Typhimurium variants to infect pigeons, as truly broad-host-range serotype Typhimurium variants, such as phage types DT49 and DT104, are very rarely isolated from this source. Apparently, host-adapted variants of serotype Typhimurium also infect ducks (DT8 and DT46) and wild birds (DT40), but these hosts are also susceptible to infection with true broad-host-range serotype Typhimurium variants.
Serotype Typhimurium isolates from different species are more closely genetically related to each other than they are to other Salmonella serotypes (5, 6). This close genetic relatedness of serotype Typhimurium variants suggests that relatively few genetic changes may account for their apparent adaptations to different host reservoirs. One possible mechanism by which such variants may have arisen is phage-mediated transfer of a small number of host-specific virulence factors, although there is no direct evidence for this hypothesis at this point. Phage-mediated transfer of virulence factors may not be uncommon in serotype Typhimurium since putative virulence genes have been identified in the genomes of several of its prophages, including nanH and sodCIII in FELS-1, gogB in Gifsy-1, sodCI and sseI (srfH) in Gifsy-2, sspH1 in Gifsy-3, and sopE1 in SopEΦ (19, 21, 26, 44, 66). Exchange of these phages between serotype Typhimurium isolates may result in a reassortment of virulence factors, thereby likely generating new virulence traits (19, 21, 44, 66). This process may have contributed to the emergence of host-adapted serotype Typhimurium variants that can be distinguished by phage typing because they carry distinct repertoires of prophages.
The identification of serotype Typhimurium variants with a narrow host range is significant since it provides a new opportunity to study host adaptation. Past attempts to identify genes involved in host adaptation by introducing genes from a broad-host-range serotype (e.g., serotype Typhimurium) into a serotype with a narrow host range (e.g., serotype Typhi or serotype Gallinarum) in order to confer mouse virulence have not been successful (54; L. Pascopella, S. Falkow, and P. L. C. Small, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., abstr. B-109, p. 173, 1996). Genome analysis suggests that it may indeed prove experimentally challenging to construct a fully virulent (in mice) serotype Typhi strain in the laboratory by transferring serotype Typhimurium DNA because this approach is complicated by the large amount of serotype-specific DNA present in these bacterial genomes. For instance, the serotype Typhi genome contains 601 genes (on 82 genetic islands) that are absent from serotype Typhimurium while serotype Typhimurium contains 479 genes (on 80 genetic islands) that are unique relative to serotype Typhi (41, 48). These considerations suggest that comparison of pathogens which are more closely related than serotype Typhimurium and serotype Typhi, yet still differ markedly with regard to host range, is likely to be a more amenable approach for identifying genes involved in host adaptation. The finding that serotype Typhimurium isolates are closely related genetically but differ with regard to host range thus opens a new opportunity to apply this strategy.
Acknowledgments
Work in A.B.'s laboratory is supported by the Texas Advanced Research (Technology) Program under grant number 000089-0051-1999 and by Public Health Service grants #AI40124 and #AI44170. This work was supported by a grant from the Federal Ministry of Education and Research, Germany (project # 01KI9901).
Editor: D. A. Portnoy
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. De Saxe, D. C. Old, R. Barker, and J. P. Duguid. 1978. Correlation of phage type, biotype and source in strains of Salmonella typhimurium. J. Hyg. (London) 81:203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1997. Reports of national reference laboratories on Salmonella typhimurium DT104. Newsletter, vol. 3, no. 3. Community Reference Laboratory for Salmonella, Bilthoven, The Netherlands.
- 4.Baggesen, D. L., M. N. Skov, D. J. Brown, and M. Bisgaard. 1997. Separation of Salmonella typhimurium DT2 and DT135: molecular characterization of isolates of avian origin. Eur. J. Epidemiol. 13:347-352. [DOI] [PubMed] [Google Scholar]
- 5.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, M. W. Frerichs, K. Wachsmuth, K. Ferris, A. C. McWorter, J. G. Wells, A. Cravioto, and R. K. Selander. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. [DOI] [PubMed] [Google Scholar]
- 7.Blaxland, J. D., J. W. Sojka, and A. M. Smither. 1958. Avian salmonellosis in England and Wales 1948-56 with comment on its prevention and control. Vet. Rec. 70:374-382. [Google Scholar]
- 8.Brandis, H., J. Posch, G. Oberhoffer, L. Andries, and U. Lehmacher. 1980. Contributions to the epidemiology of Salmonella typhimurium, analyzed according to the results of phage typing in the period between 1969-1978. Offentl. Gesundheitswes. 42(Suppl. 2):75-128. (Author's translation.) [PubMed] [Google Scholar]
- 9.Bryan, F. L., M. J. Fanelli, and H. Riemann. 1979. Salmonella infections, p. 73-130. In H. Riemann and F. L. Bryan (ed.), Food-borne infections and intoxications, 2nd ed. Academic Press, New York, N.Y.
- 10.Callow, B. R. 1959. A new phage-typing scheme for Salmonella typhimurium. J. Hyg. 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daoust, P. Y., D. G. Busby, L. Ferns, J. Goltz, S. McBurney, C. Poppe, and H. Whitney. 2000. Salmonellosis in songbirds in the Canadian Atlantic provinces during winter-summer 1997-98. Can. Vet. J. 41:54-59. [PMC free article] [PubMed] [Google Scholar]
- 12.Duguid, J. P., E. S. Anderson, G. A. Alfredsson, R. Barker, and D. C. Old. 1975. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J. Med. Microbiol. 8:149-166. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, P. R. 1936. Fermentative varieties of Salmonella aertrycke. J. Infect. Dis. 58:225-229. [Google Scholar]
- 14.Edwards, P. R. 1938. Further studies on IV-variants of Salmonella typhi-murium (aertrycke) with special reference to cultures from pigeons. J. Bacteriol. 35:123-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, P. R. 1935. A serological variant of Salmonella aertrycke from pigeons. J. Bacteriol. 30:465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, P. R., and D. W. Bruner. 1943. The occurrence and distribution of Salmonella types in the United States. J. Infect. Dis. 72:58-67. [DOI] [PubMed] [Google Scholar]
- 17.Faddoul, G. P., and G. W. Fellows. 1965. Clinical manifestations of paratyphoid infection in pigeons. Avian Dis. 9:377-381. [PubMed] [Google Scholar]
- 18.Faddoul, G. P., and G. W. Fellows. 1965. A five-year survey of the incidence of salmonellae in avian species. Avian Dis. 10:297-304. [Google Scholar]
- 19.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felix, A., and B. R. Callow. 1943. Typing of paratyphoid B bacilli by means of Vi bacteriophage. Br. Med. J. 2:127-130. [DOI] [PMC free article] [PubMed]
- 21.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 22.Fone, D. L., and R. M. Barker. 1994. Associations between human and farm animal infections with Salmonella typhimurium DT104 in Herefordshire. Commun. Dis. Rep. CDR Rev. 4:R136-R140. [PubMed] [Google Scholar]
- 23.Geoffrey, E., S. Gaines, M. Landy, W. D. Tigertt, H. Sprintz, R.-J. Trapani, A. D. Mandel, and A. S. Benenson. 1960. Studies on infection and immunity in experimental typhoid fever: typhoid fever in chimpanzees orally infected with Salmonella typhosa. J. Exp. Med. 112:143-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, R. F., and A. Buxton. 1946. A survey of avian salmonellosis in Great Britain. Vet. J. 102:187-206. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, A. C. 1942. Die beim Hausgeflügel in Dänemark festgestellten Salmonellatypen. Zentbl. Bakteriol. Orig. 149:222-235. [Google Scholar]
- 26.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centrisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauser, K. W. 1959. Die Salmonellosis der Tauben. Berl. Muench. Tieraerztl. Wochenschr. 72:126-129. [Google Scholar]
- 28.Hohn, J., and W. Herrmann. 1939. Die epidemiologische Bedeutung der Typen des Breslau-Bacteriums. Z. Hyg. Infektkrankh. 122:227-237. [Google Scholar]
- 29.Hohn, J., and W. Herrmann. 1937. Die Typen des Breslaubakteriums. Z. Hyg. Infektkrankh. 119:369-382. [Google Scholar]
- 30.Jacob, W. K., H. Kuhn, H. Kurschner, and W. Rabsch. 1993. The epidemiological analysis of Salmonella typhimurium infections in cattle--results of lysotyping and biochemotyping in the region of East Thuringia from 1974 to 1991. Berl. Muench. Tieraerztl. Wochenschr. 106:265-269. [PubMed] [Google Scholar]
- 31.Jansen, J. 1940. Die epidemiologische und epizootische Bedeutung der Typen des Breslau-Bacteriums. Z. Hyg. Infektkrankh. 122:412-419. [Google Scholar]
- 32.Kauffmann, F. 1934. Über die serologische and kulturelle Varianten der Paratyphus D- und Mäusetyphus-Bacillen. Z. Hyg. 116:368-384. [Google Scholar]
- 33.Krabisch, P., and P. Dorn. 1980. Epidemiologic significance of live vectors in the transmission of salmonella infections in broiler flocks. Berl. Muench. Tieraerztl. Wochenschr. 93:232-235. [PubMed] [Google Scholar]
- 34.Kühn, H., R. Falta, and H. Rische. 1973. Salmonella typhimurium, p. 101-140. In H. Riche (ed.), Infektionskrankheiten und ihre Erreger, vol. 14. Gustav Fischer Verlag, Jena, Germany. [Google Scholar]
- 35.Kuhn, H., W. Rabsch, and H. Tschape. 1989. Diagnosis of salmonellosis. Z. Gesamte Hyg. 35:681-683. [PubMed] [Google Scholar]
- 36.Kuhn, H., W. Rabsch, H. Tschape, and E. Tietze. 1982. Characterization and epidemiology of a Salmonella typhimurium epidemic strain. Z. Aerztl. Fortbild. (Jena) 76:607-610. [PubMed] [Google Scholar]
- 37.Lerche, M. 1937. Die beim Tier vorkommenden Erkrankungen der Bakterien der Paratyphus-Enteritidis-Gruppe und ihre Epidemiologie. Sammelbericht III. Bericht über die 17. Tagung der Deutschen Vereinigung für Mikrobiologie, vom 20, bis 22. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Abt. I Orig. 140:39-52. [Google Scholar]
- 38.Lilleengen, K. 1948. Typing of Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:2-125. [Google Scholar]
- 39.MacCready, R. A., J. P. Reardon, and I. Saphra. 1957. Salmonellosis in Massachusetts. N. Engl. J. Med. 256:1121-1128. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald, J. W., and D. D. Brown. 1974. Salmonella infection in wild birds in Britain. Vet. Rec. 94:321-322. [DOI] [PubMed] [Google Scholar]
- 41.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 42.McCoy, J. H. 1975. Trends in salmonella food poisoning in England and Wales 1941-72. J. Hyg. (London) 74:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menzies, F. D., S. D. Neill, E. A. Goodall, and S. G. McIlroy. 1994. Avian salmonella infections in Northern Ireland 1979-1991. Prev. Vet. Med. 19:119-128. [Google Scholar]
- 44.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 45.Morgenroth, A., and J. P. Duguid. 1968. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha-strains of Salmonella typhimurium: an essay in bacterial archaeology. Genet. Res. 11:151-169. [DOI] [PubMed] [Google Scholar]
- 46.Nastasi, A., C. Mammina, and M. R. Villafrate. 1993. Epidemiology of Salmonella typhimurium: ribosomal DNA analysis of strains from human and animal sources. Epidemiol. Infect. 110:553-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen, S. J., R. Bishop, F. W. Brenner, T. H. Roels, N. Bean, R. V. Tauxe, and L. Slutsker. 2001. The changing epidemiology of salmonella: trends in serotypes isolated from humans in the United States, 1987-1997. J. Infect. Dis. 183:753-761. [DOI] [PubMed] [Google Scholar]
- 48.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 49.Pennycott, T. W., H. M. Ross, I. M. McLaren, A. Park, G. F. Hopkins, and G. Foster. 1998. Causes of death of wild birds of the family Fringillidae in Britain. Vet. Rec. 143:155-158. [DOI] [PubMed] [Google Scholar]
- 50.Prager, R., A. Liesegang, W. Rabsch, B. Gericke, W. Thiel, W. Voigt, R. Helmuth, L. Ward, and H. Tschape. 1999. Clonal relationship of Salmonella enterica serovar typhimurium phage type DT104 in Germany and Austria. Zentbl. Bakteriol. 289:399-414. [DOI] [PubMed] [Google Scholar]
- 51.Prescott, J. F., C. Poppe, J. Goltz, and G. D. Campbell. 1998. Salmonella typhimurium phage type 40 in feeder birds. Vet. Rec. 142:732.. [PubMed] [Google Scholar]
- 52.Price, J. I., E. Dougherty III, and D. W. Bruner. 1962. Salmonella infections in white pekin duck. A short summary of the years 1950-60. Avian Dis. 6:145-147. [Google Scholar]
- 53.Rabsch, W., H. Tschape, and A. J. Baumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 54.Roudier, C., M. Krause, J. Fierer, and D. G. Guiney. 1990. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonella serotypes in mice. Infect. Immun. 58:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scholtens, R. T., and G. Caroli. 1971. Role of pigeons in the spread of salmonellosis: incidence of different types of Salmonella typhi-murium var. copenhagen in pigeons, man, and other animals. Antonie Van Leeuwenhoek 37:473-476. [DOI] [PubMed] [Google Scholar]
- 56.Sojka, W. J., and H. I. Field. 1970. Salmonellosis in England and Wales 1958-1967. Vet. Bull. 40:515-531. [Google Scholar]
- 57.Sojka, W. J., and C. Wray. 1975. Incidence of Salmonella infection in animals in England and Wales, 1968-73. Vet. Rec. 96:280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sojka, W. J., C. Wray, and I. McLaren. 1986. A survey of drug resistance in salmonellae isolated from animals in England and Wales in 1982 and 1983. Br. Vet. J. 142:371-380. [DOI] [PubMed] [Google Scholar]
- 59.Sojka, W. J., C. Wray, J. Shreeve, and A. J. Benson. 1977. Incidence of salmonella infection in animals in England and Wales 1968-1974. J. Hyg. (London) 78:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sojka, W. J., C. Wray, J. E. Shreeve, and J. C. Bell. 1983. The incidence of salmonella infection in sheep in England and Wales, 1975 to 1981. Br. Vet. J. 139:386-392. [DOI] [PubMed] [Google Scholar]
- 61.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1994. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577.. [DOI] [PubMed] [Google Scholar]
- 62.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1990. Plasmid profile typing can be used to subdivide phage-type 49 of Salmonella typhimurium in outbreak investigations. Epidemiol. Infect. 104:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Threlfall, E. J., B. Rowe, J. L. Ferguson, and L. R. Ward. 1985. Increasing incidence of resistance to gentamicin and related aminoglycosides in Salmonella typhimurium phage type 204c in England, Wales and Scotland. Vet. Rec. 117:355-357. [DOI] [PubMed] [Google Scholar]
- 64.Threlfall, E. J., L. R. Ward, and B. Rowe. 1978. Epidemic spread of a chloramphenicol-resistant strain of Salmonella typhimurium phage type 204 in bovine animals in Britain. Vet. Rec. 103:438-440. [DOI] [PubMed] [Google Scholar]
- 65.van Oye, E., and J. Borghijs. 1979. Do pigeons play a part in human infections with Salmonella typhimurium var. copenhagen. Dtsch. Tierarztl. Wochenschr. 89:306-307. (Author's translation.) [PubMed] [Google Scholar]
- 66.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 67.Wray, C., I. M. McLaren, and Y. E. Jones. 1998. The epidemiology of Salmonella typhimurium in cattle: plasmid profile analysis of definitive phage type (DT) 204c. J. Med. Microbiol. 47:483-487. [DOI] [PubMed] [Google Scholar]
- 68.Wray, C., W. J. Sojka, and J. C. Bell. 1981. Salmonella infection in horses in England and Wales, 1973 to 1979. Vet. Rec. 109:398-401. [DOI] [PubMed] [Google Scholar]
- 69.Wuthe, H. H. 1971. Studies on types of Salmonella typhimurium in domestic pigeons in Schleswig-Holstein and their possible epizootological significance. Berl. Muench. Tieraerztl. Wochenschr. 84:290-292. [PubMed] [Google Scholar]
- 70.Wuthe, H. H., H. Brandis, L. Andries, and S. Wuthe. 1975. Phage typing and biotyping of Salmonella typhi-murium. Zentbl. Bakteriol Orig. 230:172-185. (Author's translation.) [PubMed] [Google Scholar]
- 71.Zimmerman, L. E., M. Cooper, and C. D. Graber. 1952. Bacteriologic studies on an outbreak of salmonellosis in Korea. Am. J. Hyg. 56:252-264. [DOI] [PubMed] [Google Scholar]