Abstract
Identification of Mycobacterium tuberculosis proteins that can provide immunological protection against tuberculosis is essential for the development of a more effective vaccine. To identify new vaccine targets, we have used immunoaffinity chromatography to isolate class I HLA-A*0201-peptide complexes from M. tuberculosis-infected cells and sequenced the isolated peptides by mass spectrometry. From this material, we have identified three peptides derived from a single M. tuberculosis protein that is encoded by the M. tuberculosis Rv0341 gene. Although no known protein encoded by the Rv0341 gene has been described, it is predicted to give rise to a 479-amino-acid protein with a molecular mass of 43.9 kDa. The three peptides identified are all nested and were found to be antigenic, in that they were capable of inducing peptide-specific, CD8+ T cells from healthy blood donors in vitro and capable of recognizing and lysing M. tuberculosis-infected dendritic cells. This methodology provides a powerful tool for the identification of M. tuberculosis proteins that can be evaluated as potential vaccine candidates.
While it has been estimated that one-third of the world's population is infected with Mycobacterium tuberculosis, the causative agent of tuberculosis, only 10% of the people in this group will develop active disease during their lifetimes (40). The remainder of the infected population harbors M. tuberculosis in a latent form, and reactivation of infection occurs sporadically. The current live attenuated vaccine, bacillus Calmette-Guérin (BCG), has been shown to have variable and limited efficacy against adult pulmonary tuberculosis (20), and the tuberculin skin test, measuring BCG-induced immunity, does not correlate with the partial protection induced by BCG vaccination (8). With the increasing threat that M. tuberculosis presents to the world population due to the worldwide human immunodeficiency virus epidemic and the development of multidrug-resistant strains of M. tuberculosis, the focus of much research is on the development of a new tuberculosis vaccine.
The cellular arm of the immune response has been established as an essential component in the control of tuberculosis infection and the development of a protective immune response against M. tuberculosis with evidence for active roles by both CD4+ and CD8+ T cells (3, 5, 10, 21, 36, 37). Antigen recognition by these T cells requires a complex series of events in which antigen is processed and transported to specific cellular compartments, where it associates with either class I or class II major histocompatibility complex (MHC) molecules. These peptide-MHC complexes are transported to the cells surface, where they are available for recognition by antigen-specific T cells (17, 23, 53). More recently, an MHC-independent pathway for antigen presentation, in which hydrophobic nonpeptides associate with CD1 molecules, has been described (6, 39, 43). While CD1-restricted T cells specific for lipid and glycolipid components of mycobacterial cell walls have been described, a role for these T cells in the M. tuberculosis immune response is still being investigated.
Identification and evaluation of the immune response against defined M. tuberculosis antigens are paramount to the development of a new and more efficacious vaccine. To this end, multiple approaches have been used, with various levels of success, to identify M. tuberculosis proteins and peptides derived from these proteins that may be useful in the formulation of a new vaccine. Early studies described the biochemical fractionation and purification of M. tuberculosis secreted proteins from immunologically active culture filtrates (4, 35). This approach has resulted in the identification of a number of proteins, such as the antigen 85 complex (26), ESAT-6 (early secretary antigen target 6) (45), CFP-10 (culture filtrate protein 10) (7), and Mtb39 (18), which are currently being investigated as potential vaccine candidates. Molecular approaches have also been applied to the identification of immunologically reactive M. tuberculosis proteins. Expression cloning, in which M. tuberculosis-specific CD4+ T cells were used to screen an M. tuberculosis genomic library expressed in Escherichia coli, has led to the identification of a family of highly related M. tuberculosis antigens (2). More recently, the use of computer algorithms designed to predict class I binding peptides has led to the identification of CD8+ T-cell epitopes in known immunologically reactive M. tuberculosis proteins such as the 19-kDa lipoprotein (34), ESAT-6 (35), and Ag85B (22). This approach has also been used to screen M. tuberculosis protein sequences from the GenBank database and has resulted in the identification of new M. tuberculosis target proteins (14).
An alternative approach described here is direct identification of M. tuberculosis peptides that have been processed by the cell and presented on the cell surface in association with class I and class II MHC molecules. These peptides represent naturally processed epitopes presented in a form that is readily recognizable by the immune system. One advantage to this approach is that a list of M. tuberculosis vaccine candidates can be generated without the isolation and characterization of M. tuberculosis-specific T cells. In addition, computer HLA motif algorithms often result in the identification of a large numbers of potential HLA binding peptides with little or no binding affinity, this approach limits the analysis to known HLA-associated peptides.
Using this approach, we have sequenced a number of HLA-A*0201-associated peptides derived from M. tuberculosis-encoded proteins. In this report, we describe the identification of three naturally occurring M. tuberculosis peptides from the Rv0341 gene product. All three peptides were found to be antigenic in that they were capable of inducing the generation of peptide-specific cytotoxic T lymphocytes (CTL) from the peripheral blood of healthy blood donors and were capable of recognizing and lysing M. tuberculosis-infected dendritic cells (DC).
MATERIALS AND METHODS
Cell lines and bacterial cultures.
U937/A2 is a variant of the U937 histiocytic lymphoma cell line transfected with the gene encoding the HLA-A*0201 molecule (52). HLA-A*0201+, TAP-deficient T2 (174xCEM.T2) cells were used as target cells in 51Cr release assays. Cell lines were maintained in RPMI 1640 medium containing 2 mM l-glutamine, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 10 mM HEPES (complete RPMI medium) and supplemented with 5% fetal bovine serum (FBS). M. tuberculosis strain H37Ra was obtained from the American Type Culture Collection (Manassas, Va.) and grown in 7H9 broth (Difco) supplemented with 10% ADC enrichment medium (Difco), 2% glycerol, and 0.005% Tween 20.
Infection with M. tuberculosis.
U937/A2 cells were expanded in 1-liter spinner bottles in complete RPMI medium with 5% FBS and 300 μg of G418 per ml. U937/A2 cells were pretreated with 20 ng of phorbol myristate acetate (PMA; Calbiochem) per ml for 24 h prior to infection. Aliquots of M. tuberculosis were thawed from −80°C, sonicated for 2 min to dissociate bacterial material, and incubated with 5% human serum for 1 h at 37°C. M. tuberculosis was then added to 109 U937/A2 cells at a multiplicity of infection of 5 in a final volume of 100 ml. After 3 h of incubation, the cells were adjusted to 106/ml and incubated in spinner bottles for an additional 18 to 20 h. The level of infection, determined by auramine staining, was 26 to 32%.
Immunoaffinity purification of HLA-A2 molecules and associated peptides.
H37Ra-infected and uninfected U937/A2 cells were washed twice in phosphate-buffered saline and stored as cell pellets at −80°C. Cell pellets consisting of either 3 × 1010 infected or 5 × 1010 uninfected cells were solubilized, peptide class I complexes were isolated, and peptides were purified as previously described (25). Briefly, the cell pellets were solubilized at 106 cells/ml in 20 mM Tris (pH 8.0)-150 mM NaCl-1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)-5 μg of aprotinin per ml-10 μg of leupeptin per ml-10 μg of pepstatin A per ml-5 nM EDTA-0.2% sodium azide-17.4 μg of phenylmethylsulfonyl fluoride per ml for 1 h at 4°C. The lysates were centrifuged at 100,000 × g, the pellets were discarded, and the supernatant was passed through a 0.22-μm-pore-size filter. The supernatant was then passed over a series of recombinant protein A-Sepharose columns. First, a precolumn of cyanogen bromide-activated Sepharose 4B was used. This was followed by a protein A-Sepharose immunoaffinity column conjugated to anti-HLA-A2.1 monoclonal antibody (MAb) BB7.2 to isolate the HLA-A*0201-peptide complexes. All remaining class I-peptide complexes were isolated on protein A-Sepharose columns conjugated with anti-pan class I MAb W6/32. After a series of wash steps, peptides were eluted from the MAb columns with 10% acetic acid and boiled for 5 min to further dissociate any bound peptide from the class I heavy chain. The peptides were then separated from the copurifying heavy chains and β2-microglobulin by centrifugation on an Ultra-CL membrane with a nominal 5,000-Mr cutoff (Millipore, Bedford, Mass.). The resulting peptide was concentrated in a Speedvac to 50-μl aliquots.
Liquid chromatography-tandem mass spectrometry (LC/MS/MS) on a quadrupole ion trap mass spectrometer.
An aliquot corresponding to approximately 2 × 108 cell equivalents of the peptides from M. tuberculosis-infected U937/A2 cells was loaded onto a reverse-phase C18 microcapillary high-performance liquid chromatography (HPLC) column and gradient (A = 0.1 M acetic acid in H2O, B = 70% acetonitrile-0.1 M acetic acid in H2O, 0 to 60% B in 200 min, 60 to 100% B in 20 min) eluted directly into a Finnigan LCQ DECA quadrupole ion trap mass spectrometer (ThermoFinnigan, San Jose, Calif.). The instrument was operated in a data-dependent fashion, in which acquisition of a full-scan mass spectrum (300 < m/z < 2,000) was followed by collision-activated dissociation analysis of the five most abundant ions. Each cycle (MS scan followed by five MS/MS scans) was performed in approximately 14 s, and cycles were repeated continuously for the 3-h duration of peptide elution. MS/MS spectra were searched by using the SEQUEST algorithm against proteins predicted from the M. tuberculosis genome (The Sanger Centre [http://www.sanger.ac.uk/]) and against the nonredundant protein database maintained at the National Center for Biotechnology Information.
LC/MS on an FTMS.
For each analysis, an aliquot corresponding to approximately 2 × 107 cell equivalents was loaded onto a reverse-phase C18 microcapillary HPLC column and gradient (A = 0.1 M acetic acid in H2O, B = 70% acetonitrile-0.1 M acetic acid in H2O, 0 to 60% B in 40 min, 60 to 100% B in 10 min) eluted directly into a home-built Fourier-transformed mass spectrometer (FTMS) (31). Full-scan mass spectra (350 < m/z < 5,000) were acquired approximately every 1.1 s. LC/MS data from each sample were searched manually for masses corresponding to M. tuberculosis-derived peptides identified in the LC/MS/MS analysis of the M. tuberculosis-infected sample.
Peptide synthesis.
Peptides used for MS analysis were synthesized by standard 9-fluorenylmethoxy carbonyl chemistry with a Gilson AMS422 peptide synthesizer. Peptides used for CTL generation and antigenic analysis (Rv034133-42, GLIDIAPHQI; Rv034133-44, GLIDIAPHQISS; Rv034133-45, GLIDIAPHQISSV) were purchased in bulk from Research Genetics, Huntsville, Ala.
HLA-A*0201 binding assay.
The binding affinities of M. tuberculosis peptides for the HLA-A*0201 class I molecule were demonstrated by measuring their ability to inhibit the binding of a radiolabeled standard peptide to purified HLA-A*0201. Affinity-purified HLA-A*0201 molecules were incubated at room temperature with the iodinated indicator peptide FLPSDY*FPSV and graded concentrations of the different M. tuberculosis peptides in phosphate-buffered saline, pH 7, containing 0.05% NP-40, 1 μg of β2-microglobulin per ml, 1 mM phenylmethylsulfonyl fluoride, 1.3 mM 1,10-phenanthroline, 73 μM pepstatin A, 8 mM EDTA, and 200 μM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK). After 48 h, class I-peptide complexes were separated from free peptide by gel filtration and the dose of each individual test peptide that reduced the binding of the indicator peptide by 50% (IC50) was calculated.
Generation of DC.
Peripheral blood was obtained from healthy, HLA-A2+ blood donors, and peripheral blood mononuclear cells (PBMC) were isolated by centrifugation over Ficoll-Hypaque. PBMC were resuspended in complete RPMI medium with 10% FBS, and 108 PBMC were plated in T-75 flasks. After a 2-h incubation at 37°C, nonadherent cells were removed and adherent cells were cultured in complete RPMI medium with 10% FBS supplemented with 50 ng of granulocyte-macrophage colony-stimulating factor per ml and 100 ng of interleukin-4 (IL-4; Peprotech, Rocky Hill, N.J.) per ml for 7 days.
Induction of peptide-specific, CD8+ CTL lines.
PBMC isolated from healthy, HLA-A2+ blood donors were used as starting material for the in vitro generation of peptide-specific CTL lines. Donors were HLA typed serologically, and no subtyping was performed. CD8+ T cells were isolated with immunomagnetic beads (Miltenyi Biotec) in accordance with the manufacturer's instructions. DC were irradiated (5,000 rads) and pulsed with 40 μg of peptide per ml and 3 μg of β2-microglobulin per ml for 2 to 4 h. CD8+ T cells were cocultured with DC at a ratio of 20 CD8+ T cells to 1 DC in complete RPMI medium supplemented with 10% FBS and 10 ng of recombinant human IL-7 (Peprotech) per ml in 24-well plates at 2 × 106 CD8+ T cells/well. After 7 days, CTL were harvested and restimulated with DC pulsed with 20 μg of peptide per ml and 3 μg of β2-microglobulin per ml. Recombinant human IL-2 (15 U/ml; Chiron, Emeryville, Calif.) was added to cultures every 3 to 4 days. The peptide concentration used to pulse DC was reduced to 10 μg/ml for subsequent rounds of restimulation. After three or four rounds of restimulation, cultures were tested for cytotoxic activity. A minimum of two CTL lines were generated against each peptide.
Cytotoxicity assays.
Recognition of M. tuberculosis antigens by peptide-specific CTL was measured in a standard 51Cr release assay. T2 cells were labeled with 150 μCi of Na51CrO4 overnight at 37°C before being pulsed with 1 μg of peptide per ml for 2 h at 37°C. Washed target cells (2 × 103) and effector CTL were cocultured at various effector-to-target cell ratios in V-bottom microtiter plates in a final volume of 200 μl of complete RPMI medium supplemented with 5% FCS. The assay plates were centrifuged for 2 min at 200 × g and then incubated at 37°C.
After 4 to 5 h of incubation, 125 μl of supernatant was removed from each well and the amount of 51Cr released from the target cells was measured in a gamma counter (ICN Micromedic, Huntington, Ala.). Maximum release from 51Cr-labeled target cells was determined in the presence of 2% Triton X-100, and spontaneous release was obtained in the presence of medium alone. The percentage of specific 51Cr release was calculated as follows: % 51Cr release = [(Experimental release − Spontaneous release)/(Maximal release − Spontaneous release)] × 100. All assays were repeated a minimum of three times in triplicate.
Infection of DC with M. tuberculosis.
Immature DC were generated from PBMC as described above. On day 6, DC were infected with M. tuberculosis at a multiplicity of infection of 10. Infected DC were labeled with 150 μCi of Na51CrO4 overnight at 37°C and used as target cells 24 h postinfection.
RESULTS
Identification of M. tuberculosis-derived MHC class I-associated peptides.
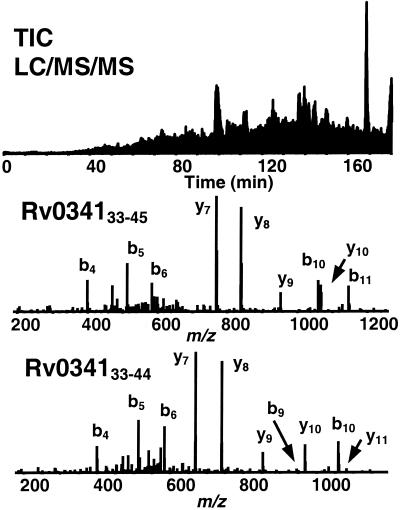
Aliquots of the peptides purified from the BB7.2 and W6/32 antibody columns were analyzed by LC/MS/MS on an LCQ DECA quadrupole ion trap mass spectrometer. Approximately 5,000 MS/MS spectra were acquired during each LC/MS/MS analysis. Use of these spectra to search the Sanger Centre M. tuberculosis predicted protein database yielded three peptides from the same hypothetical protein (Rv034133-42, Rv034133-44, and Rv034133-45) (Fig. 1). After synthesis of the peptides, several femtomoles of each peptide was spiked into an aliquot of the M. tuberculosis-infected sample and the aliquot was analyzed on the LCQ ion trap mass spectrometer. Coelution of the synthetic peptides with the naturally occurring peptides identified in the sample confirmed the identification of the endogenous peptides.
FIG. 1.
LC/MS/MS analysis of unfractionated MHC class I peptides isolated from M. tuberculosis-infected human macrophage cell line U937/A2. Shown are the total ion chromatogram of HLA-A*0201 purified peptides (top) and MS/MS spectra of Rv034133-45 (middle) and Rv034133-44 (bottom).
Semiquantitative analysis of differentially expressed M. tuberculosis-derived MHC class I-associated peptides.
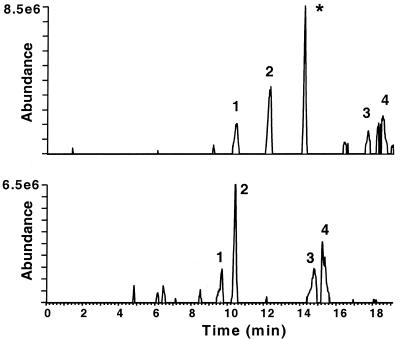
The peptide samples were further analyzed by LC/MS on a home-built FTMS to confirm the differential presentation of the M. tuberculosis-derived peptides. The high duty cycle (>99%), mass accuracy, and sensitivity (low-attomole detection limit) of this instrument provided a more comprehensive analysis of the sample than did LC/MS/MS analysis (31). A search of the FTMS data confirmed the presence of these three peptides in the M. tuberculosis-infected sample and the absence of these peptides in the uninfected sample (based on accurate mass and coeluting peaks). The data for one of the three peptides are shown in Fig. 2. Note that all of the peaks in the infected sample are also present in the uninfected sample, with the exception of the Rv034133-45 peptide. The FTMS data are semiquantitative, allowing estimation of the copy number of each peptide. Based on these data, each of the three peptides was present in the sample at approximately the same level, 50 to 100 copies/cell.
FIG. 2.
Comparative LC/MS analysis of unfractionated MHC class I peptides from uninfected and M. tuberculosis-infected U937/A2 macrophage cells. The similarity of the two samples is evidenced in the selected-ion chromatograms, as each peak in the infected-cell sample is present in the uninfected-cell sample, with the exception of Rv034133-45 (∗).
Determination of peptide-MHC class I binding affinity.
The peptide binding affinity of class I antigens has been shown to be a critical factor in the in vitro induction of CTL with synthetic peptides (50). Before an attempt was made to generate CTL to the newly identified peptides, the binding affinities of three M. tuberculosis-peptides with the HLA-A*0201 class I molecule were determined. Two peptides, Rv034133-42 and Rv034133-45, were characterized as having high binding affinities, exhibiting IC50s of 49 and 40 nM, respectively, while the affinity of Rv034133-44 was somewhat lower (IC50, 173 nM) (data not shown). These binding affinities are similar to those reported for those peptides recognized by CTL derived from patients with cancer and viral disease (50).
Generation of CD8+ CTL lines specific for M. tuberculosis peptides.
To determine whether the identified M. tuberculosis peptides are capable of eliciting CTL responses, PBMC from healthy, HLA-A2+ blood donors were stimulated weekly with autologous DC pulsed with the various peptides. CTL lines generated against all three peptides exhibited a high level of lysis on T2 cells pulsed with the peptide used for in vitro stimulation compared to that of unpulsed targets (Fig. 3). In addition to lytic activity, four peptide-specific CTL lines (two each of the 10- and 13-mer CTL) were tested for the ability to secrete gamma interferon (IFN-γ), as a role for IFN-γ in the CD8+ T-cell response to tuberculosis has been suggested (29, 50). These CTL lines produced marked levels of IFN-γ in response to specific antigen stimulation (data not shown).
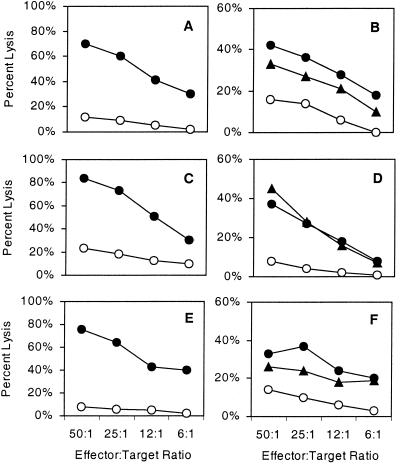
FIG. 3.
Peptide recognition by M. tuberculosis peptide-specific CTL. PBMC from healthy donors were stimulated in vitro with peptide-pulsed autologous DC. CTL lines were generated with peptides Rv034133-42 (a and b), Rv034133-44 (c and d), and Rv034133-45, (e and f). The cytolytic activity of the CD8+ T-cell lines was measured in a 51Cr release assay with either T2 cell (a, c, and e) or DC (b, d, and f) targets pulsed with either no peptide (○) or the peptide used for in vitro stimulation (•) or with cells infected with M. tuberculosis (▾).
The three M. tuberculosis-derived peptides were originally isolated from M. tuberculosis-infected U937/A2, a human histiocytic tumor cell line that had been treated prior to infection with PMA to enhance phagocytosis of the bacilli. To determine whether these peptides are also the result of processing by a naturally occurring antigen-presenting cell, HLA-A2+ DC were infected M. tuberculosis and used as targets in a 51Cr cytotoxicity assay. All three populations of peptide-specific CTL were capable of recognizing both M. tuberculosis-infected DC and DC pulsed with the stimulating peptide (Fig. 3).
Patterns of peptide recognition by the M. tuberculosis peptide CTL lines.
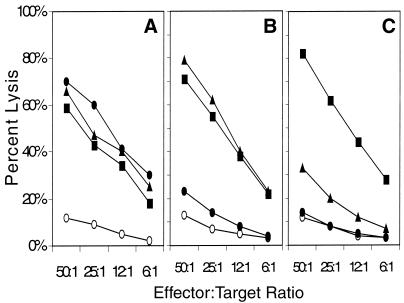
Because the three peptides represent a nested set of peptides, all sharing the same 10 amino-terminal residues, we were interested in examining the pattern of reactivity of the three peptide-induced CD8+ CTL lines. As shown in Fig. 4, the patterns of cross-reactivity of the three CTL with the various peptides were different. CTL generated against the 10-mer (Rv034133-42) exhibited high levels of cross-reactivity on the longer 12 (Rv034133-44)- and 13 (Rv034133-45)-amino-acid peptides. Lytic activity by CTL generated against the 12-amino-acid peptide was only observed on targets pulsed with the 12-mer peptide and the longer 13-mer peptide but not the shorter 10-mer peptide. Finally, CTL generated against the 13-mer peptide strongly recognized target cells pulsed with the 13-mer peptide, weakly recognized targets pulsed with the 12-mer peptide, and showed no recognition of targets pulsed with the 10-mer peptide. The data suggest that the three peptides induce CTL responses that recognize both shared and unique CTL epitopes.
FIG. 4.
Patterns of peptide recognition exhibited by M. tuberculosis peptide-generated CTL. The patterns of recognition by the CD8+ T-cell lines raised against M. tuberculosis peptides Rv034133-42 (a), Rv034133-44 (b), and Rv034133-45 (c) were measured in a 51Cr release assay. T2 target cells were either unpulsed (○) or pulsed with peptide Rv034133-42 (•), peptide Rv034133-44 (▾), or peptide Rv034133-45 (▪).
DISCUSSION
The study presented here focused on the identification of class I-associated T-cell epitopes recognized by CD8+ CTL. Several studies using animal models have indicated a role for CD8+ T cells in protective immunity to M. tuberculosis (19, 21, 28, 46, 51). Effector mechanisms suggested for CD8+ T cells include secretion of IFN-γ and tumor necrosis factor alpha (48), direct lysis of infected macrophages, and antimicrobial activity effected by the release of granulysin from cytotoxic granules (47). While the role of CD8+ T cells in human recovery from tuberculosis has yet to be established, M. tuberculosis-specific CD8+ T cells have been isolated from tuberculosis-infected individuals in a number of studies (11, 29, 30).
The epitopes presented by class I MHC molecules are generated by the intracellular processing of proteins and their subsequent binding and transport to the cell surface. Recent reports have suggested that within M. tuberculosis-infected macrophages, vacuoles containing tubercle bacilli become permeable to macromolecules, enabling bacterial proteins to enter the cytoplasm (32, 41, 49). These proteins are then available for processing by the proteasome, transport by TAP to the lumen of the endoplasmic reticulum, and association with class I molecules. Although this represents just one of the many mechanisms by which peptide presentation can occur, only a few studies have been able to confirm the conduit between a phagosome and the surrounding cytoplasm. There is also evidence for an alternative processing pathway for MHC class I-presented M. tuberculosis antigens that is insensitive to brefeldin A (11). In addition, DC can also take up apoptotic infected macrophages and re-present MHC class I-bound antigens to T cells (1). Thus, by sampling the spectrum of peptides associated with class I molecules isolated from the infected cell and comparing it to the peptides present on uninfected cells, it should be possible to identify M. tuberculosis-derived peptides that are available for presentation to the immune system.
Using MS to sequence M. tuberculosis-derived peptides eluted from immunoaffinity-purified HLA-A*0201 molecules, we have identified three peptides encoded by a single gene, Rv0341. The Rv0341 gene is encoded by an open reading frame within the H37Rv genome and is predicted to encode a 479-amino-acid protein with a molecular mass of 43.9 kDa (The Sanger Centre) and is referred to here as Mtb43.9. No product of the Rv0341 gene has been identified; however, the amino acid sequence it is predicted to encode has a high degree of homology to a glycine-rich cell wall structural protein from Arabidopsis thaliana (The Sanger Centre).
The three peptides described in this study represent a nested set of peptides, all beginning at the same amino acid but with lengths of 10, 12, and 13 residues. All three of these peptides elicited CD8+ CTL responses in vitro from peripheral blood obtained from healthy donors. Although the peptides were originally isolated from an M. tuberculosis-infected histiocytic tumor cell line activated with PMA to enhance phagocytosis, these CTL were capable of recognizing and lysing M. tuberculosis-infected DC, indicating that these peptides are also presented during the normal course of infection of a naturally occurring antigen-presenting cell.
The predominant lengths of class I binding peptides have been shown to be 8 to 10 amino acids. This is due, in part, to the fact that the peptide binding cleft in the class I molecule is blocked at both ends by bulky amino acid side chains, such as Tyr 84 and Trp 167 in the HLA-A*0201 molecule (42). While peptides of this length may predominate, many studies have shown that longer peptides can indeed bind to class I molecules and that the affinity of these extended peptides for class I antigens can be comparable to that of shorter peptides (13). It has been suggested, based on crystallographic modeling of HLA-Aw68 and associated peptides, that longer species could be accommodated by additional kinking of the central region of the peptide backbone while the peptide maintains hydrogen bonding interactions between the amino and carboxyl termini with the ends of the binding groove (24). This type of structure is more likely to occur with class I molecules, where the distance between anchor residues is greater, such as that between P2 and P9 for HLA-A0201 and -A6801 rather than that between P5 and P9, as is the case with H-2Kb and Db molecules.
Identification of multiple nested peptides from a single protein associated with the same class I antigen has been described previously. One example is the nested peptides derived from the IP-30 signal sequence and eluted from the HLA-A201 class I molecule. These peptides had lengths of 9 amino acids (IP-3027-35), 10 amino acids (IP-3026-35), 11 amino acids (IP-3026-36), and 12 amino acids (IP-3026-37) (27). It is, however, the first time, to our knowledge, that the individual nested peptides elicited different patterns of reactivity, as demonstrated by CTL recognition. Here, the pattern of Rv0341 peptide recognition indicated that CTL induced by one peptide efficiently recognized targets pulsed with that peptide and longer homologues but not shorter homologues.
In the binding of extended peptides to the HLA-A0201 class I molecule, the anchor residue at position 2 was found to be more critical that the carboxy-terminal anchor residue at position 9 (13, 27). The data from those studies suggested that binding of extended peptides to the class I molecule may occur in various conformations. How the associations of the nested Rv0341 peptides with the class I molecules differ from each other is not clear; however, presentation of these peptides by the class I molecule induces the expansion of T-cell populations with distinct and overlapping specificities. The ability of nested peptides of differing lengths that are bound to the same class I molecule in different conformations may be advantageous to the immune system, eliciting distinct but overlapping T-cell specificities, as observed with the Rv0341 peptides.
We have successfully used the approach of sequencing of immunoaffinity-purified peptides to identify T-cell epitopes in cancer tumor antigens (16, 25, 44). In these experiments, tumor-specific CTL had been generated and were used to identify the peptide defining the T-cell epitopes. Here we demonstrate that, in the absence of antigen-specific CTL, a search of the genomic database of an infectious pathogen with peptide sequences can identify immunologically relevant targets that represent candidate T-cell epitopes for vaccine development. The M. tuberculosis bacillus has a 4.4-Mb genome that has been shown to encode 3,924 open reading frames (15). The number of M. tuberculosis proteins that have been identified as having any immunological role in the M. tuberculosis immune response represents a very small percentage of the total number of M. tuberculosis-encoded proteins.
In addition to the three peptides described in this report, we have also sequenced two additional HLA-A2-associated M. tuberculosis peptides. The first was derived from M. tuberculosis heat shock protein hsp65, a protein previously identified as a target for CD4+ T cells (38) and found to induce protection in murine models when administered as a DNA vaccine (9). Recently, this peptide and others derived from hsp65 have been shown to associate with the HLA-A0201 molecule and induce CTL responses in HLA-A0201/Kb transgenic mice (12). The second peptide was derived from galactofuranosyl transferase (33), an M. tuberculosis protein not previously shown to be recognized by the immune system. Use of this methodology to sequence additional class I-associated peptides, as well as class II-associated peptides, should make it possible to construct a database of M. tuberculosis proteins, processed by the infected cell, that represent potential vaccine candidates.
Acknowledgments
This work was supported in part by grants NIHR37 AI20963 (V.H.E.), GM37537 (D.F.H.), and K08-AI015801 (D.H.C.).
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Albert, M., B. Sauter, and N. Bhardwaj. 1998. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature 392:86-89. [DOI] [PubMed] [Google Scholar]
- 2.Alderson, M. R., T. Bennet, C. H. Day, L. Zhu, D. Molesh, Y. A. W. Skeiky, R. Coler, D. M. Lewinsohn, S. G. Reed, and D. C. Dillon. 2000. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using CD4+ T cells. J. Exp. Med. 191:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, P., A. B. Andersen, A. L. Sorensen, and S. Nagai. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J. Immunol. 154:3359-3372. [PubMed] [Google Scholar]
- 4.Anderson, P. 1994. The T cell response to secreted antigens of Mycobacterium tuberculosis. Immunobiology 91:537-547. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, P. F., J. S. Abrams, S. Lu, P. A. Sieling, T. H. Rea, and R. L. Modlin. 1993. Cytokine production at the site of disease in human tuberculosis. Infect. Immun. 61:197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckman, E. M., S. A. Porcelli, C. T. Moroita, S. M. Behar, S. T. Furlong, and M. B. Brenner. 1994. Recognition of a lipid antigen by CD1-restricted αβ+ T cells. Nature 372:691-694. [DOI] [PubMed] [Google Scholar]
- 7.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195-3203. [DOI] [PubMed] [Google Scholar]
- 8.Bloom, B. R., and P. E. M. Fine. 1994. The BCG experience: implications for future vaccines against tuberculosis, p. 531-558. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 9.Bonato, V. L. D., V. M. F. Lima, R. E. Tascon, D. B. Lowrie, and C. L. Silva. 1998. Identification and characterization of protective T cells in hsp65 DNA-vaccinated and Mycobacterium tuberculosis-infected mice. Infect. Immun. 66:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boom, W. H., R. S. Wallis, and K. A. Chervenak. 1991. Human Mycobacterium tuberculosis-reactive CD4+ T-cell clones: heterogeneity in antigen recognition, cytokine production, and cytotoxicity for mononuclear phagocytes. Infect. Immun. 59:2737-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canaday, D. H., C. Ziebold, E. H. Noss, K. A. Chervenak, C. V. Harding, and W. H. Boom. 1999. Activation of human CD8+ alpha beta TCR+ cells by Mycobacterium tuberculosis via an alternate class I MHC antigen-processing pathway. J. Immunol. 162:372-379. [PubMed] [Google Scholar]
- 12.Charo, J., A. Geluk, M. Sundback, B. Mmirzai, A. D. Diehl, K. J. Malmberg, A. Achour, S. Huriguchi, K. E. Meijgaarden, J. W. Drijfhout, N. Beekman, P. P. Veelen, F. Ossendorp, T. H. Ottenhoff, and R. Kiessling. 2001. The identification of a common pathogen-specific HLA class I A*0201-restricted cytotoxic T cell epitope encoded within the heat shock protein 65. Eur. J. Immunol. 31:3602-3611. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Y. E., J. Sidney, S. Southwood, A. L. Cox, K. Sakaguchi, R. A. Henderson, E. Appella, D. F. Hunt, A. Sette, and V. H. Engelhard. 1994. Naturally processed peptides longer than nine amino acid residues bind to the class I MHC molecule HLA-A2.1 with high affinity and in different conformations. J. Immunol. 152:2874-2881. [PubMed] [Google Scholar]
- 14.Cho, S., V. Mehra, S. Thoma-Uszynski, S. Stenger, N. Sebina, R. J. Mazzaccaro, J. L. Flynn, P. F. Barnnes, S. Southwood, E. Celis, B. R. Bloom, R. L. Modlin, and A. Sette. 2000. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc. Natl. Acad. Sci. USA 97:12210-12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, L. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:357-544. [DOI] [PubMed] [Google Scholar]
- 16.Cox, A. L., J. Skipper, Y. Chen, R. A. Henderson, T. L. Darrow, J. Shabanowitz, V. H. Engelhard, D. F. Hunt, D. F., and C. L. Slingluff. 1994. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science 264:716-719. [DOI] [PubMed] [Google Scholar]
- 17.Cresswell, P. 1994. Assembly, transport and function of MHC class II molecules. Annu. Rev. Immunol. 12:259-293. [DOI] [PubMed] [Google Scholar]
- 18.Dillon, D. C., M. R. Alderson, C. H. Day, D. M. Lewinsohn, R. Coler, T. Bement, A. Campos-Neto, Y. A. W. Skeiky, I. M. Orme, A. Roberts, S. Steen, W. Dalemans, R. Badaro, and S. G. Reed. 1999. Molecular characterization and human T-cell responses to a member of a novel Mycobacterium tuberculosis mtb39 gene family. Infect. Immun. 67:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Souza, T. J., J. E. Pearl, P. Marietta, M. Noel, A. A. Frank, R. Appelberg, I. M. Orme, and A. M. Cooper. 2001. CD8- and CD95/95L-dependent mechanisms of resistance in mice with chronic pulmonary tuberculosis. Am. J. Respir. Cell Mol. Biol. 24:203-209. [DOI] [PubMed] [Google Scholar]
- 20.Fine, P. E. 1995. Variation in protection by BCG: implications of and for heterologous immunity. Lancet 346:1339-1345. [DOI] [PubMed] [Google Scholar]
- 21.Flynn, J. L., M. M. Goldstein, K. J. Triebold, B. Koller, and B. R. Bloom. 1992. Major histocompatibility complex class I-restricted T cells are required for resistance to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA 89:12013-12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geluk, A., K. E. vanMeijgaarden, K. L. M. C. Franken, J. W. Drijfhout, S. D. Souza, A. Necker, K. Huygen and T. H. M. Ottenhoff. 2000. Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLA-A*0201-restricted CD8+ T cells in HLA-transgenic mice and humans. J. Immunol. 165:6463-6471. [DOI] [PubMed] [Google Scholar]
- 23.Germain, R. N., and D. H. Margulies. 1993. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 11:403-450. [DOI] [PubMed] [Google Scholar]
- 24.Guo, H. C, T. S. Jardetzsky, T. P. J. Garrett, W. S. Lane, J. L. Strominger, and D. C. Wiley. 1992. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360:364-366. [DOI] [PubMed] [Google Scholar]
- 25.Hogan, K. T., D. P. Eisinger, S. B. Cupp, K. L. Lekstrom, D. D. Deacon, J. Shabanowitz, D. F. Hunt, V. H. Engelhard, C. L. Slingluff, Jr., and M. M. Ross. 1998. The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res. 58:5144-5150. [PubMed] [Google Scholar]
- 26.Horwitz, M. A., B. W. Lee, B. J. Dillon, and G. Harth. 1995. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt, D. F., R. A. Henderson, J. Shabanowitz, K. Sakaguchi, H. Michel, N. Sevilir, A. Cox, E. Appella, and V. H. Engelhard. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255:1261-1263. [DOI] [PubMed] [Google Scholar]
- 28.Ladel, C. H., S. Daugelat, and S. H. Kaufmann. 1995. Immune response to Mycobacterium bovis bacille Calmette Guerin infection in major histocompatibility complex class I- and II-deficient knock-out mice: contribution of CD4 and CD8 T cells to acquired resistance. Eur. J. Immunol. 25:377-384. [DOI] [PubMed] [Google Scholar]
- 29.Lalvani, A., R. Brookes, R. J. Wilkenson, A. S. Malin, A. A. Panthan, P. Anderson, H. Dockrell, G. Pasvol, and A. V. Hill. 1998. Human cytolytic and interferon gamma-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewinsohn, D. M., M. R. Alderson, A. L. Briden, S. R. Riddell, S. G. Reed, and K. H. Grabstein. 1998. Characterization of human CD8+ T cells reactive with Mycobacterium tuberculosis-infected antigen-presenting cells. J. Exp. Med. 187:1633-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, S. E., J. Shabanowitz, D. F. Hunt, and J. A. Marto. 2000. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 72:4266-4274. [DOI] [PubMed] [Google Scholar]
- 32.Mazzaccaro, R. J., M. Gedde, E. R. Jensen, H. M. van Santen, H. L. Ploegh, K. L. Rock, and B. R. Bloom. 1996. Major histocompatibility class I presentation of soluble antigen facilitated by Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA 93:11786-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikusova, K., T. Yagi, R. Stern, M. R. McNeil, G. S. Besar, D. C. Crick, and P. J. Brennan. 2000. Biosynthesis of the galactan component of the mycobacterial cell wall. J. Biol. Chem. 275:33890-33897. [DOI] [PubMed] [Google Scholar]
- 34.Mohagheghpour, N., D. Gammon, L. M. Kawamura, A. van Vollenhoven, C. J. Benike, and E. G. Engelman. 1998. CTL response to Mycobacterium tuberculosis: identification of an immunogenic epitope in the 19-kDa lipoprotein. J. Immunol. 161:2400-2406. [PubMed] [Google Scholar]
- 35.Nagi, S., H. G. Wiker, M. Harboe, and M. Kinomoto. 1991. Isolation and partial characterization of major proteins in the culture fluid of Mycobacterium tuberculosis. Infect. Immun. 59:372-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orme, I. M. 1987. The kinetics of emergence and loss of mediator T lymphocytes acquired in response to infection with Mycobacterium tuberculosis. J. Immunol. 138:293-298. [PubMed] [Google Scholar]
- 37.Orme, I. M., A. D. Roberts, J. P. Griffin, and J. S. Abrams. 1993. Cytokine secretion by CD4 T lymphocytes acquired in response to Mycobacterium tuberculosis infection. J. Immunol. 151:518-525. [PubMed] [Google Scholar]
- 38.Ottenhoff, T. H., B. K. Ab, J. D. Van Embden, J. E. R. Thole, and R. Kiessling. 1988. The recombinant 65 kD heat shock protein of Mycobacterium bovis bacillus Calmette-Guerin/M. tuberculosis is a target molecule for CD4+ cytotoxic T lymphocytes that lyse human monocytes. J. Exp. Med. 168:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porcelli, S., C. T. Moroita, and M. B. Brenner. 1992. CD1b restricts the response of CD4−8− T lymphocytes to a microbial antigen. Nature 360:593-597. [DOI] [PubMed] [Google Scholar]
- 40.Raviglione, M. C., D. E. Snider, Jr., and A. Kochi. 1995. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA 273:220-226. [PubMed] [Google Scholar]
- 41.Rock, K. L., and A. L. Goldberg. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739-779. [DOI] [PubMed] [Google Scholar]
- 42.Saper, M. A., P. J. Bjorkman, and D. C. Wiley. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J. Mol. Biol. 219:277-319. [DOI] [PubMed] [Google Scholar]
- 43.Sieling, P. A., D. Chatteriee, S. A. Porcelli, T. I. Prigozy, R. J. Mazzaccaro, T. Soriano, B. R. Bloom, M. B. Brenner, M. Kronenberg, P. J. Brennen, et al. 1965. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science 269:227-230. [DOI] [PubMed] [Google Scholar]
- 44.Slingluff, C. L., Jr., A. L. Cox, R. A. Henderson, D. F. Hunt, and V. H. Engelhard. 1993. Recognition of human melanoma cells by HLA-A2.1-restricted cytotoxic T lymphocytes is mediated by at least six shared peptide epitopes. J. Immunol. 150:2955-2963. [PubMed] [Google Scholar]
- 45.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sousa, A. O., R. J. Mazzaccaro, R. G. Russell, F. K. Lee, O. C. Turner, S. Hong, L. V. Kaer, and B. R. Bloom. 1999. Relative contributions of distinct MHC class I-dependent cell populations in protection to tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 97:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenger, S., D. A. Hanson, R. Teitelbaum, P. Dewan, K. R. Niazi, C. J. Froelich, T. Ganz, S. Thoma-Uszynski, A. Melian, C. Bogdan, S. A. Porcelli, B. R. Bloom, A. M. Krensky, and R. Modlin. 1988. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282:121-125. [DOI] [PubMed] [Google Scholar]
- 48.Tascon, R. E., E. Stavropoulos, K. V. Lukacs, and M. J. Colston. 1988. Protection against Mycobacterium tuberculosis infection by CD8+ T cells requires the production of gamma interferon. Infect. Immun. 66:830-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teitelbaum, R., M. Cammer, M. L. Maitland, N. E. Freitag, J. Coldeelis, and B. R. Bloom. 1999. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc. Natl. Acad. Sci. USA 96:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai, V., S. Southwood, J. Sidney, K. Sakaguchi, Y. Kawakami, E. Appella, A. Sette, and E. Celis. 1997. Identification of subdominant CTL epitopes of the GP100 melanoma-associated tumor antigen by primary in vitro immunization with peptide-pulsed dendritic cells. J. Immunol. 158:1796-1802. [PubMed] [Google Scholar]
- 51.van Pinxterenm, L. A. H., J. P. Cassidy, B. H. Smedegaard, E. M. Agger, and P. Anderson. 2000. Control of latent Mycobacterium tuberculosis infection is dependent on CD8 T cells. Eur. J. Immunol. 30:3689-3698. [DOI] [PubMed] [Google Scholar]
- 52.Wourela, M., S. Jalkanen, J. Kirveskari, P. Laitio, and K. Granfors. 1997. Yersinia enterocolitica serotype O:3 alters the expression of serologic HLA-B27 epitopes on human monocytes. Infect. Immun. 65:2060-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yewdell, J. W., and J. R. Bennink. 1992. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule restricted T lymphocytes. Adv. Immunol. 52:1-123. [DOI] [PubMed] [Google Scholar]