Abstract
Brome mosaic virus (BMV) RNA replication has been examined in a number of systems, including Saccharomyces cerevisiae. We developed an efficient T-DNA-based gene delivery system using Agrobacterium tumefaciens to transiently express BMV RNAs in Nicotiana benthamiana. The expressed RNAs can systemically infect plants and provide material to extract BMV replicase that can perform template-dependent RNA-dependent RNA synthesis in vitro. We also expressed the four BMV-encoded proteins from nonreplicating RNAs and analyzed their effects on BMV RNA accumulation. The capsid protein that coinfiltrated with constructs expressing RNA1 and RNA2 suppressed minus-strand levels but increased plus-strand RNA accumulation. The replication proteins 1a and 2a could function in trans to replicate and transcribe the BMV RNAs. None of the BMV proteins or RNA could efficiently suppress posttranscriptional silencing. However, 1a expressed in trans will suppress the production of a recombinant green fluorescent protein expressed from the nontranslated portions of BMV RNA1 and RNA2, suggesting that 1a may regulate translation from BMV RNAs. BMV replicase proteins 1a did not affect the accumulation of the BMV RNAs in the absence of RNA replication, unlike the situation reported for S. cerevisiae. This work demonstrates that the Agrobacterium-mediated gene delivery system can be used to study the cis- and trans-acting requirements for BMV RNA replication in plants and that significant differences can exist for BMV RNA replication in different hosts.
Successful viral infection requires specific interactions between the viral genomic RNAs and viral proteins (13). The interactions are likely complex since the virus needs to regulate the amount and timing of RNA synthesis and then escape or counteract cellular defenses (44, 54, 70, 74, 90, 95). We use brome mosaic virus (BMV) as a model system to analyze the interaction of viral proteins and RNAs.
BMV belongs to the alphavirus-like superfamily of plant and animal positive-strand RNA viruses. A number of different systems have been used to analyze the requirements for BMV RNA replication and spread. These include the BMV RNA replicase that can direct RNA synthesis in vitro (1, 88), barley and tobacco protoplasts that can be transfected with BMV RNAs (25, 35, 48), plants that can be used to analyze local and systemic infections (28, 57, 68), and Saccharomyces cerevisiae, which is permissive for BMV RNA replication and transcription (39). These diverse systems have yielded useful insights into the mechanism of viral infection. Furthermore, the results from different systems are generally in agreement, but some notable differences have been observed (31, 65, 86).
The BMV genome is divided into three capped RNAs, designated RNA1, RNA2, and RNA3 (2). The RNAs make four proteins. RNA1 encodes the multifunctional protein 1a, which methylates and caps BMV RNAs (3, 47) and transports BMV RNAs and the replication-associated proteins to the site of RNA replication (18, 24, 77, 87). RNA2 encodes protein 2a, the RNA-dependent RNA polymerase that is the catalytic core of the BMV replicase. RNA3 is a dicistronic RNA that encodes the movement protein (MP) required for spread of the virus from the infected cell and the capsid protein (CP), which encapsidates BMV RNAs.
The viral RNAs provide a number of cis-acting sequences that will direct replication and encapsidation through their interaction with the viral proteins. Sequences for BMV RNA encapsidation are only beginning to be elucidated (20, 22, 23). The sequences required for RNA replication and transcription are somewhat better characterized, and the RNA motifs that bind the replicase in vitro and direct RNA synthesis have been mapped (17, 19, 80, 84). Minus-strand RNA replication requires the nearly identical 3′ noncoding regions of BMV genomic plus-strand RNAs that form a tRNA-like structure in vitro (17, 27) and in vivo (66, 85). A motif named stem-loop C (SLC) within the tRNA-like structure binds the replicase to direct minus-strand RNA initiation (19, 45). Subgenomic RNA synthesis requires a sequence in the central portion of minus-strand RNA3, which contains four key residues within a ca. 20-nucleotide (nt) regulatory sequence (80, 82). Genomic RNA1 and RNA3 synthesis requires ca. 30-nt from the 3′ ends of their respective minus-strand RNAs that can bind the replicase and direct initiation (84). A key element within this sequence is the cB box, which is conserved in other members of the Bromoviridae (82, 83). The cB box is complementary to a previously identified regulatory element, the B box (29, 43, 56, 61). In barley protoplasts, the deletion of the B/cB box resulted in defects in BMV replication and transcription (61, 87). In S. cerevisiae, the B box binds the 1a protein in a process that increases the half-life of the RNA from minutes to hours (24). This interaction is required to translocate the RNA into membranes where the replicase can assemble and replicate the BMV RNAs in a compartment called a spherule (77). BMV replication is also known to take place within membranes in plant cells (72).
In this study, we developed a T-DNA-based Agrobacterium-mediated expression system to examine the effects of BMV proteins on the replication and accumulation of BMV RNAs. Insertion of foreign sequences into the agrobacterial T-DNA has successfully launched a number of plant DNA and RNA viruses, including BMV (5, 21, 32, 33, 49, 51, 62, 69, 73, 89, 91, 92). This system allows the introduction of different combinations of viral cDNAs into a host, thus permitting dissection of the requirements for viral infection. Marillonnet et al. (55) have demonstrated that many copies of the T-DNAs expressing the desired recombinant sequences can be delivered into each cell, thus ensuring that each cell will receive multiple copies of T-DNAs from each culture. We also compared results from agroinfiltrated Nicotiana benthamiana (for brevity, henceforth called “tobacco”) and tobacco protoplasts to those from the more traditionally used barley protoplasts, and we document several common requirements and some significant differences, including a novel regulation of recombinant protein production by the BMV 1a protein.
MATERIALS AND METHODS
Plasmid construction.
To develop an Agrobacterium-mediated gene delivery system to express BMV RNAs for replication and infection in plants, each BMV cDNA (40) was cloned into the T-DNA region of a modified binary vector derived from pCB301 (GenBank accession number AF139061). cDNAs were generated by use of Pfu polymerase and pairs of oligonucleotides (Table 1) that place the appropriate restriction sites, BglII and XmaI (Fig. 1), at the two ends of the cDNA. The PCR fragments were cloned into pGEMT-easy vectors (Promega Inc., Madison, WI) and sequenced in their entirety to verify that no unintended errors were introduced. In its final state, the 5′ end of the cDNA is flanked with a cauliflower mosaic virus (CaMV) 35S promoter with a double enhancer cassette that should initiate transcription at the 5′ nucleotides of the BMV RNAs (Fig. 1). A ribozyme developed from avocado sunblotch viroid and tobacco ringspot virus satellite RNAs (49, 62) was fused to the 3′ end of the viral cDNAs, which generated authentic viral RNA 3′ ends by cis-preferential cleavage of the transcript (Fig. 1). All finished plasmids were sequenced across the cloning junctions to ensure that no unintentional changes were made. The resulting plasmids were named pBR1, pBR2, and pBR3 for the three BMV cDNAs. The plasmids were subsequently mobilized into Agrobacterium tumefaciens strain C58C1 by electroporation.
TABLE 1.
Primers used for plasmid construction
| Plasmid | Primer | Oligonucleotide sequence (5′-3′) | Purpose |
|---|---|---|---|
| pBR1 | 1 | GAGATTCCTGTGGTTGGCATGCACATA | To fuse the 35S promoter to the 5′ end of RNA1 cDNA. Primer 1 adds a BglII site (in italics) at the 5′ end of the 35S promoter, and primer 2 contains the 5′ 29 nt from RNA1 and a SalI site (in italics) at the 3′ end. |
| 2 | GTCGACAAGGGATTGAACCTCGTTCCGTGGTCTACTCCTCTCCAAATGAAATGAACTTCCTTAT | ||
| 3 | ACGTCACTTTGGTTCGGCTTAAGTCGACC | To add a ribozyme to the 3′ end of the RNA1 cDNA. Primer 3 introduces an AflII site (italics) at the 5′ end of the cDNA. Primer 4 adds a 72-nt ribozyme sequence (underlined) and an XmaI site (italics) at the 3′ end of the cDNA. | |
| 4 | CCCGGGCCGTTTCGTCCTCACGGACTCATCAGAAGA CATGTGAATCATGTCTTGACGGCCCTTATTTTTTCT TCCTTGGTCTCTTTTAGAGATTTACAGTGTTTTT | ||
| pBR2 | 5 | GTCGACAAGGGATTGAACCTCGTTCCGTGGTTTACTCCTCTCCAAATGAAATGAACTTCCTTAT | To fuse a 35S promoter to the 5′ end of the RNA2 cDNA. Primer 1 adds a BglII site (in italics) at the 5′ end, and primer 5 contains 29 nt of the RNA2 and a SalI site (italics). |
| 6 | CTAAGGAGCTCCCTGTCAAACGGATCGG | To add a ribozyme to the 3′ end of RNA2 cDNA. Primer 6 adds a SacI site (italics) at the 5′ end, and primer 4 adds a ribozyme sequence and an XmaI site to the 3′ end of the cDNA. | |
| PBR3 | 7 | ATCGAACGAGAATTAGTTGGTATTTTACTCCTCTCCAAATGAAATGAACTTCCTTATATAGAGG | To fuse a 35S promoter sequence to the 5′ end of the RNA3 cDNA. Two PCR products were generated using primers 1 and 7 and primers 8 and 9. The two products were fused using primers 1 and 9, which add a BglII site and an AflIII site at the 5′ and 3′ ends of the cDNA, respectively. |
| 8 | GTAAAATACCAACTAATTCTCGTTCGATTCC | ||
| 9 | ACGTGTGACTTTGGCACCAGGTCTATAT | ||
| 10 | GGCTAAGGTTAAAAGCTTGTTGAATCAG | To add a ribozyme sequence to the 3′ end of the RNA3 cDNA. Primer 10 adds a HindIII site (in italics) at the 5′ end. Primer 4 adds the ribozyme sequence and an XmaI site to the 3′ end of the cDNA. | |
| p1a | 11 | CCATGGGATCAAGTTCTATCGATTTGCTGAAGTTG | To clone the 1a ORF into pCB302. Primer 11 adds an NcoI site (in italics) to the 5′ end of the cDNA, and primer 12 adds an XbaI (in italics) to the 3′ end. |
| 12 | TCTAGATCACTTAACACAATTAAAGATCAAATCC | ||
| p2a | 13 | CCATGGGATCTTCGAAAACCTGGGATGATGATTTC | To clone the 2a ORF into pCB302. Primer 13 adds an NcoI site (in italics) at the 5′ end, and primer 14 adds an XbaI (in italics) site at the 3′ end of the ORF. |
| 14 | TCTAGATCATCTCAGATCAGAGGGCTTAAAAGTTC | ||
| pMP | 15 | CCATGGGATCTAACATAGTTTCTCCCTTCAGTGGT | To clone the MP ORF into pCB302. Primer 15 adds an NcoI site (in italics) at the 5′ end of the cDNA, and primer 16 adds an XbaI site (in italics) to the 3′ end. |
| 16 | TCTAGACTATTTAATTCTAAGCGTAGGACTGGAC | ||
| pCP | 17 | CCATGGGATCGACTTCAGGAACTGGTAAGATG | To clone the CP ORF into the pCB 302 vector. Primer 17 adds an NcoI site (in italics) at the 5′ end, and primer 18 adds an XbaI site (in italics) to the 3′ end. |
| 18 | TCTAGACTACCTATAAACCGGGGTGAAGAAGTCA | ||
| pMPKO | 19 | GTGATACTGTTTTTGTTCCCGCTGTCTAACATAGTT TCTCCCTTCAGTGG | To generate a mutation in the initiation codon of the MP cDNA in within pBR3. The mutation is in bold. |
| 20 | CCACTGAAGGGAGAAACTATGTTAGACAGCGGG AACAAAAACAGTATCAC | ||
| pCPKOI | 21 | GATCTATGTCCTAATTCAGCGTATTAATACTGTCGACTTCAGGAACTGGTAAGATGAC | To generate a mutation in the CP initiation codon in pBR3. The mutation is in bold. |
| 22 | GTCATCTTACCAGTTCCTGAAGTCGACAGTATTAATACGCTGAATTAGGACATAGATC | ||
| pCPKOII | 23 | CGTATTAATACTGTCGACTTCAGGAACTGGTAAGCTGACTCGCGCGCAGCGTCGTGCTGCCGC | To generate mutations in the first and second initiation codons of the CP cDNA in pBR3. The mutation is in bold. |
| 24 | GCGGCAGCACGACGCTGCGCGCGAGTCAGCTTA CCAGTTCCTGAAGTCGACAGTATTAATACG | ||
| pR3−14 U/A | 25 | AAAAAAAAAAAAAAAAAAGATCTAAGTCCTAATTCAGCGTATTAATAATG | To generate a mutation in the −14 nucleotide of the subgenomic promoter in the context of pBR3. The mutation is in bold. |
| 26 | CATTATTAATACGCTGAATTAGGACTTAGATCTTTT TTTTTTTTTTTTT | ||
| pR3-GUA | 27 | GGGGTTCGTGCATGGGCTTGCGTAGCAAGTCTTAGAATGCGGGTAC | To generate a mutation in the core genomic minus-strand promoter in the context of pBR3. The mutation is in bold. |
| 28 | GTACCCGCATTCTAAGACTTGCTACGCAAGCCCATGCACGAACCCC | ||
| pB1ΔBb | 29 | GAGAGGAGTAGACCACGGAACGAGG//TCCCTTGTCGACCACGGTTCTGC | To delete 5 nt from the B-box of pBR1. The location of the deletion is marked by // in the primers. |
| 30 | GCAGAACCGTGGTCGACAAGGGA//CCTCGTTCCGTGGTCTACTCCTCTC | ||
| pR1:GFP | 31 32 | GCCATGGTTTGTTGGTGAAAAACAAAGAACAAG CTCTAGATGCGCTTGTCTCTGTGTGAGACCTCTGC | To replace the 1a ORF with the GFP ORF in between the 5′ and 3′ noncoding regions of RNA1. Two PCR products were generated. The first one was generated using primer 1 (adds a BglII site at the 5′ end) and primer 31 (adds an NcoI site at the 3′ end). The second PCR was generated using primer 32 (adds an XbaI restriction site at the 5′ end) and primer 4 (adds a 72-nt ribozyme and XmaI site at the 3′ end). NcoI and XbaI cut the GFP ORF from the pRTL2-smGFP (43) vector, and the two above-mentioned PCR products were cloned into the pBR1 vector backbone cut with BglII and XmaI restriction endonucleases. |
| pR2:GFP | 33 34 | GGCCATGGCTTGGTGATAGTAGAAAGAACAAG CCTCTAGATCGGTTCTATGATATATGAACCTAAG | To replace the 2a ORF with the GFP ORF in between the 5′ and 3′ noncoding regions of RNA2. Two PCR products were generated. The first one was generated using primer 1 (adds a BglII site at the 5′ end) and primer 33 (adds an NcoI site at the 3′ end). The second PCR was generated using primer 34 (adds an XbaI restriction site at the 5′ end) and primer 4 (adds a 72-nt ribozyme and XmaI site at the 3′ end). NcoI and XbaI cut the GFP ORF from the pRTL2-smGFP (43) vector, and the two above-mentioned PCR products were cloned into the pBR2 vector backbone cut with BglII and XmaI restriction endonucleasees. |
| pR3:MP-GFP | 35 36 37 | GGCCATGGCGGGAACAAAAACAGTATCACTACTG CCTCTAGAGTAAATCCGGTCTAACAAGCTCGGTCC TTATTAATACGCTGAATTAGGACATAGATC | To replace the MP ORF with the GFP ORF in pBR3. Two PCR products were generated. The first one was generated using primer 1 (adds a BglII site at the 5′ end) and primer 35 (adds an NcoI site at the 3′ end). The second PCR was generated using primer 36 (adds an XbaI restriction site at the 5′ end) and primer 37 (adds an AseI site at the 3′ end of the PCR). NcoI and XbaI cut the GFP ORF from the pRTL2-smGFP (43) vector, and the two above-mentioned PCR products were cloned into the pBR3 vector backbone cut with BglII and AseI restriction endonucleases. |
ORF, open reading frame.
FIG. 1.
Schematic representation of BMV cDNA constructs used to clone in the T-DNA region of the binary vector pCB301. The names of the constructs are in bold, and the molecule(s) produced upon transient expression is listed to the right of the constructs. Only the restriction enzyme cleavage sites used to make the constructs are shown. Three constructs used to express the three BMV RNAs are shown first. The boxes labeled with LB and RB denote the left border and right border of the T-DNA sequence, respectively. Large arrows labeled 35S denote the double CaMV 35S promoter elements. The long rectangles represent the protein coding sequences, and the names of the proteins are within the rectangles. The cloverleaf structure represents the 3′ tRNA-like structure of BMV RNAs. The curved arrows represent the cis-cleaving ribozyme sequence. WT, wild type. Construct pCB-302 used to express the BMV proteins from conventional mRNAs is shown at the bottom. All of the transgenes are cloned between the NcoI and XbaI restriction sites.
To express the BMV-encoded proteins in replication-incompetent RNAs, the cDNAs encoding the 1a, 2a, MP, and CP sequences were amplified using appropriate pairs of primers that contain the restriction sites NcoI and XbaI (Table 1) and cloned into the same sites in pCB302 (59, 60), which contained a CaMV 35S promoter and a 5′ nontranslated leader sequence from tobacco etch virus (16) and a 3′ 35S terminator (Fig. 1). An additional codon (GGA) was introduced immediately after the start codon when the NcoI restriction site was introduced into the 5′ ends of all the expressed proteins. This codon severely debilitated 1a's ability to replicate BMV RNAs but had no effects on other proteins constructed in the same manner. Therefore, it was removed from all of the constructs using the Quick Change kit and an appropriate set of primers. The resulting plasmids are designated p1a, p2a, pMP, and pCP.
Constructs with the BMV protein-coding sequences replaced with the sequence coding for the green fluorescence protein (GFP) were made by replacing the NcoI and XbaI restriction fragments from versions of pBR1, pBR2, and pBR3 with a fragment that can code for GFP (Table 1). The resultant plasmids, named pR1:GFP, pR2:GFP, and pR3MP:GFP, contain all of the nontranslated wild-type BMV sequences.
Analysis of viral replication and infection.
All of the plasmids used for functional analysis were mobilized into Agrobacterium tumefaciens strain C58C1 by electroporation. Agroinfiltration into tobacco leaves was done essentially as described by Llave et al. (53). Briefly, cultures harboring each plasmid were grown at 30°C from single colonies in LB broth containing kanamycin (50 μg/ml), 10 mM MES (morpholine ethanesulfonic acid; pH 5.9), and 50 μM acetosyringone. The cultures were centrifuged at 6,000 rpm for 15 min, and the pellets were suspended in the infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.9, and 150 μM acetosyringone) and incubated at room temperature for a minimum of 3 to 5 h. Bacterial cultures (at an optical density at 600 nm of 0.5) or infiltration medium were mixed in equal proportions prior to infiltration to result in the same bacterial concentrations for all infiltrated samples. Tobacco plant leaves (3 to 4 weeks old) were infiltrated by gently pressing the end of a 3-ml syringe loaded with appropriate culture to the leaf and exerting gentle pressure to flood the interstitial areas within the leaf.
Total RNA was extracted from ∼50 mg of the leaf tissue by macerating the tissue with disposable pestles made to fit into a microcentrifuge tube in the presence of a lysis buffer (0.1 M glycine, pH 9.2, 40 mM EDTA, 100 mM NaCl, 2% sodium dodecyl sulfate [SDS], and 0.05% Bentonite) and then extracted with an equal volume of phenol and chloroform and precipitated with an equal volume of isopropanol. Northern blotting was performed with 4 μg of glyoxylated RNA and strand-specific riboprobes as described by Hema et al. (36). Protein analysis used leaf samples macerated with a pestle in TB buffer (50 mM Tris-acetate, pH 7.4, 10 mM MgCl2, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol). The lysate was centrifuged at 15,000 rpm for 30 min at 4°C, and the supernatant was used for total protein analysis by SDS-polyacrylamide gel electrophoresis (PAGE) and/or subsequently purified by centrifugation over a 10% sucrose cushion prior to SDS-PAGE, as described by Rao et al. (67). Samples for electron microscopy were stained with uranyl acetate as a negative stain, as described by Kao et al. (42). Leaves for purification of the BMV replicase were harvested 2.5 days after infiltration. BMV replicase was prepared from infected barley as described by Sun et al. (88). Replicase activity assays were carried out by the same protocol used by Adkins et al. (1).
RNA silencing assay and visualization of green fluorescence protein expression.
The RNA silencing assay and plasmids of Johansen and Carrington (41) were adapted to detect the reduction of the signal from GFP in Agrobacterium-infiltrated tobacco leaves. The NcoI and XbaI fragment from pRTL2-smGFP and the PstI fragment from pRTL2-dsGFP (41) were excised and subcloned into the pCB-302 vector. The resulting plasmids were designated pGFP and pdsGFP and mobilized into A. tumefaciens strain C58C1. Previously characterized silencing suppressor proteins p19 from tomato bushy stunt virus and 2b from cucumber mosaic virus (CMV) were used as positive controls (12, 34, 81, 93). GFP signals were visualized and photographed using a Zeiss Axioplan 2 microscope using a GFP-optimized fluorescein isothiocyanate (FITC) filter (at 543-nm excitation and 505- to 530-nm emission). Supernatants of plant lysates made with TB buffer were quantified for GFP fluorescence using a PerkinElmer LS55 spectrometer.
Analysis of BMV RNA levels in transfected protoplasts.
Protoplasts were isolated from 6-day-old barley (Hordeum vulgare cv. Apex) as described by Kroner et al. (48). Tobacco protoplasts were generated from tobacco leaves incubated with 0.5% cellulase and 0.25% macerozyme (Calbiochem) for 10 to 12 h in the dark at room temperature as described by Satyanarayana et al. (75). The stability of RNA3 in the presence of different BMV constructs was tested by two methods. First, tobacco protoplasts were isolated from leaves that had been infiltrated 24 h previously with cultures that can express 1a, 2a, or GFP. Protoplasts (1 × 106) were transfected with 0.5 μg of [32P]CMP-labeled RNA made in vitro with the T7 RNA polymerase (Epicenter Biotechnologies, Madison, Wisconsin) as reported by Hema et al. (36). The protoplasts were then washed extensively with a solution containing mannitol to remove the transfection reagents and untransfected RNAs. Total nucleic acids were isolated at hourly intervals for 6 h as described previously. RNA (4 μg per sample) was denatured with glyoxal and run on 1% agarose gels. The gels were dried and the radiolabel quantified using a PhosphorImager and Molecular Dynamics programs. Second, barley protoplasts were transfected with different RNA combinations. At specified times, total nucleic acids were isolated from the protoplasts and analyzed by Northern blotting with a strand-specific riboprobe(s) as reported by Hema et al. (36).
RESULTS
BMV RNA replication in Agrobacterium-infiltrated plants.
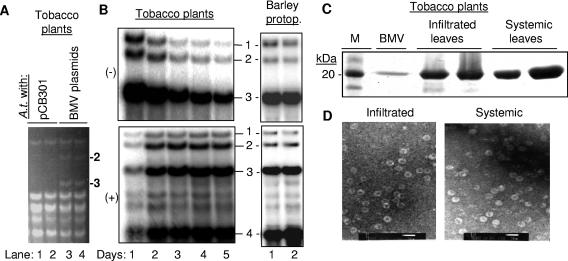
To determine whether A. tumefaciens can launch BMV infection, equal portions of A. tumefaciens cultures harboring plasmids pBR1, pBR2, and pBR3 were combined and infiltrated into tobacco. Every day for 5 days, leaf samples were collected and processed for Northern blot analysis. BMV RNAs could be visualized in ethidium bromide-stained gels of the total RNAs from leaves infiltrated with cultures containing the three BMV plasmids but not from leaves infiltrated with cultures containing the empty cloning vector (Fig. 2A). This result suggests that BMV RNA accumulations in tobacco are at levels approaching that of rRNAs.
FIG. 2.
BMV RNA replication and infection in Tobacco initiated by agroinfiltration. (A) Ethidium bromide-stained agarose gel showing that BMV RNAs launched from T-DNA plasmids replicate to high levels. In this and all other panels, the type of sample from which the material was extracted is indicated above the gel image. The identities of BMV RNA2 and RNA3 are indicated to the right of the gel image. The other bands are RNAs from tobacco. (B) Comparison of minus- and plus-strand BMV RNA accumulation in agroinfiltrated tobacco and transfected barley protoplasts. The sources of the cells used are above the gel image, while the identities of the RNA bands are shown to the right of the gel image. Barley protoplasts (protop.) were harvested at 24 and 48 h after transfection, while the agroinfiltrated plants were collected at 1 to 5 days after infiltration. (C) SDS-PAGE demonstrating that BMV capsid protein is produced in Agrobacterium-infiltrated leaves and in systemic leaves. The infiltrated leaves were harvested 5 days after infiltration, while the systemic leaves were harvested 8 days after infiltration. Two independently infiltrated leaves were examined, while the lane labeled “BMV” shows a preparation of BMV CP purified from infected barley. Lane M, molecular mass standards. (D) Detection of BMV virions by transmission electron microscopy in samples harvested from the agroinfiltrated and noninoculated leaves. The white bar represents 50 nm.
Northern blots were probed for minus- and plus-strand RNAs. Robust amounts of genome length minus-strand BMV RNAs were detected starting at about 20 h after infiltration and decreased gradually over the course of several days. Plus-strand RNA routinely increased with a 1-day delay in comparison to the minus-strand RNAs (Fig. 2B). Since agroinfiltrated plants should transcribe only plus-sense RNA from the T-DNAs, the presence of minus-strand RNA demonstrates that proper RNA processing and RNA replication had occurred. The relative abundance of the minus-strand RNAs was similar to the amount observed in barley protoplasts transfected with BMV transcripts (Fig. 2B). Lastly, subgenomic RNA4 was produced in abundance even though it could be made only from minus-strand RNA3, demonstrating that both BMV RNA replication and transcription occurred. All together, these results show that efficient replication of the BMV RNAs can be launched from Agrobacterium T-DNAs.
We next determined whether the agroinfiltrated plasmids could form virions and systemically infect tobacco. No obvious symptoms could be observed in the infiltrated or higher leaves (data not shown). However, both the infiltrated and noninfiltrated upper leaves of plants infiltrated for 8 to 10 days previously had abundant amounts of capsid protein in denaturing PAGE and BMV-like particles of ∼27 nm in diameter in electron micrographs (Fig. 2C and D). All together, these results show that the BMV RNAs transiently expressed from agroinfiltrated plasmids were fully infectious.
BMV replicases from barley and tobacco.
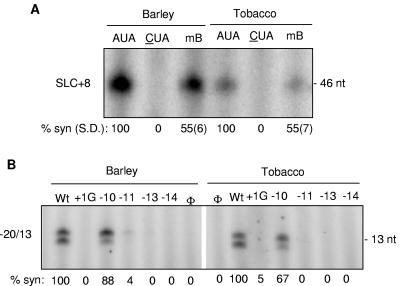
Infiltrated tobacco provides a larger amount of leaf material for RNA replicase purification than virus-infected barley. Using the standard protocol developed for enriching the BMV replicase from barley (1), we attempted to enrich the BMV replicase from tobacco plants infiltrated 2 days previously with cultures harboring pBR1, pBR2, and pBR3. A mutation in the clamp adenine motif in SLC to a cytidine in an RNA named CUA abolished RNA synthesis in vitro and also prevented RNA synthesis by the replicase extracted from agroinfiltrated tobacco leaves in barley protoplasts (17, 45, 46, 85) (Fig. 3A). A mutation in a bulge within SLC named mBulge that reduced but did not abolish RNA synthesis with the replicase from barley (17, 46) decreased RNA synthesis with the replicase from tobacco.
FIG. 3.
Agrobacterium-infiltrated tobacco could be used to enrich a template-specific BMV replicase. (A) Initiation of minus-strand RNA synthesis by the BMV replicase extracted from barley and tobacco. Replicase assays were performed as described in Adkins et al. (1). The image is from a 12% denaturing polyacrylamide gel. Versions of the wild-type and mutant SLC+8 constructs used in each assay are indicated above the lane of the gel. RNAs CUA and mB have mutations in the clamped adenine motif and the bulge sequence, respectively (17, 45). The standard deviations (S.D.) of the results from this and several other experiments (not shown) are shown in parentheses. (B) Initiation of subgenomic RNA synthesis by the BMV replicase from barley and tobacco. The image is from a 20% denaturing polyacrylamide gel. The wild-type (Wt) RNA is named −20/13. The upper of the two bands has a nontemplated nucleotide added to the newly synthesized RNA (79). The numbers above the gel image denote mutations within the subgenomic core promoter that have been previously characterized in Siegel et al. (79). The symbol “Φ” denotes a reaction with no exogenously provided template. % syn, quantification of both bands normalized to the products in the wild-type core.
We further assessed whether the BMV replicase from agroinfiltrated tobacco could recognize the BMV subgenomic core promoter and template (79, 80). A panel of mutations in the BMV subgenomic core promoter and the initiation nucleotide were tested. Of these, the mutation at position −10 relative to the initiation cytidylate was the only one tolerated by the barley BMV replicase for RNA-dependent RNA synthesis in vitro (79). The BMV replicase enriched from tobacco was affected by mutations in the core subgenomic promoter in the same manner as the replicase from barley (Fig. 3B). These results show that the agroinfiltrated tobacco could provide material useful for future biochemical analysis of the BMV replicase. Furthermore, the specific recognition of two of the core promoters was not affected by the factors present in either barley or tobacco.
Cis-acting effects on BMV RNA accumulation in tobacco and barley protoplasts.
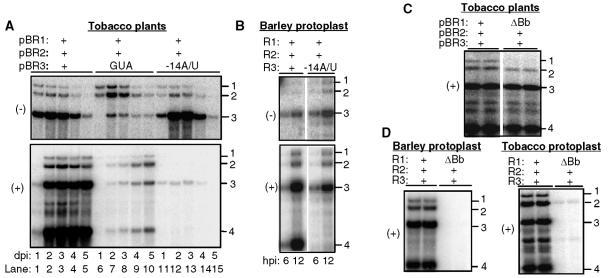
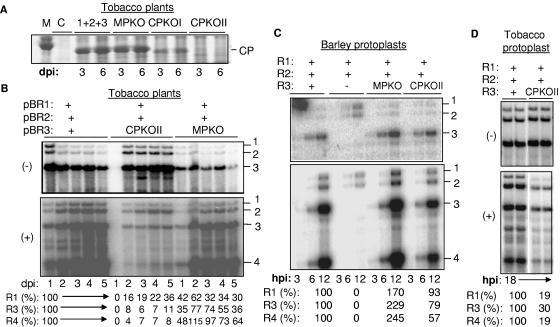
Unlike protoplasts transfected with transcripts, T-DNA-mediated gene expression could produce viral proteins for an extended period without viral RNA replication. Therefore, we compared the effects of mutations in BMV core promoters that were previously characterized in barley protoplasts to those from Agrobacterium-mediated infections. Tobacco infiltrated with cultures containing wild-type pBR1 and pBR2 and pBR3-GUA (with a mutated clamped adenine motif) accumulated low levels of minus-strand RNA3 (Fig. 4, lanes 6 to 10) and decreased genomic plus-strand RNA synthesis. These results from tobacco are consistent with those from barley protoplasts. At 5 days after infection, an increase in plus-strand BMV RNA3 accumulation was observed, possibly due to a suppressor mutation and/or repair of the mutation, as has previously been observed to occur in barley and Chenopodium spp. (64, 76, 58).
FIG. 4.
Effects of cis-acting mutations on plus- and minus-strand BMV RNA accumulation. (A) Northern blot analysis of RNA accumulations in agroinfiltrated tobacco leaves. The infiltrated culture contains a mixture of three Agrobacterium cultures, and the name of the plasmid harboring each culture is shown above the gel image. “GUA” denotes a mutation in pBR3 that affects the minus-strand core promoter (84). “−14A/U” indicates that the culture harbored a plasmid with a mutation in pBR3 that affects the subgenomic core promoter (19). The identities of the RNAs are indicated at the right. (B) Effects of the subgenomic promoter mutation −14A/U in transfected barley protoplasts. A combination of three RNAs (at 0.5 μg each) was transfected into protoplasts as identified above the gel image. Note that the mutation had severe effects only on BMV RNA4 accumulation, unlike the situation in tobacco. (C) Effects of a mutation in the B box of BMV RNA1 on RNA accumulation in agroinfiltrated tobacco leaves. Deletion of the B box is denoted by “ΔBb.” (D) Effects of a mutation in the B box of BMV RNA1 on RNA accumulation in barley and tobacco protoplasts. dpi, days postinfiltration; hpi, hours postinfiltration.
A mutation of the subgenomic core promoter that would prevent replicase binding and subgenomic RNA transcription (19, 85) did not significantly affect Agrobacterium-mediated minus-strand BMV RNA levels but did severely reduce the level of genomic plus-strand and especially subgenomic plus-strand RNAs (Fig. 4A, lanes 11 to 15). Thus, the BMV subgenomic core promoter is also important in Agrobacterium-mediated BMV replication. Notably, BMV RNAs replicated in barley protoplasts had significantly larger amounts of plus-strand RNA3 than their respective tobacco wild types (Fig. 4B). Grdzelishvili et al. (31) have proposed that in S. cerevisiae the subgenomic and genomic RNA3 promoters compete for replicase. However, if agroinfiltration produces higher levels of BMV replicase, then a subgenomic promoter mutation should have a more modest effect on RNA3 levels in tobacco, rather than in barley protoplasts. Instead, we propose that the difference in the effects of the subgenomic promoter mutant is due to the species of the plant and/or a difference between protoplasts and the whole plant.
Next, we examined the accumulation of pBR1 with a 5-nt deletion of the B/cB box, named ΔBb, which should affect plus-strand RNA accumulation. The RNA1 level was severely reduced, confirming the importance of the B/cB box in tobacco plants. However, the other BMV RNAs were not significantly affected (Fig. 4C). In barley protoplasts, RNA1 replication was necessary to sustain the replication of the other RNAs and a mutation in the B/cB box of BMV RNA1 abolished all RNA replication in barley protoplasts (19, 36). Therefore, the result with ΔBb analyzed in agroinfiltrated plant leaves illustrates another difference for BMV RNA replication between agroinfiltrated tobacco plants and barley protoplasts. To ensure that the observations are directly comparable, we examined the effects of ΔBb in transfected barley protoplasts and found that all of the BMV RNAs in barley protoplasts were reduced in number in comparison to wild-type numbers (Fig. 4D). This result documents a second difference in BMV RNA replication in tobacco plants and barley protoplasts.
We hypothesize that the agroinfiltration provides a sufficient amount of 1a from the replication-incompetent mutant RNA1 for the replication of the other RNAs. To test this, N. benthamiana protoplasts were transfected with transcripts of wild-type RNA2, RNA3, and either RNA1 or RNA1 with a deletion of the B/cB box (Fig. 4D). Indeed, ΔBb had similar effects on RNA accumulations in tobacco and barley protoplasts. BMV RNA replication initiated from stable transgenes may thus result in significantly different levels of individual BMV RNAs from what occurs in transient transfections.
Trans-acting proteins and BMV RNA accumulation.
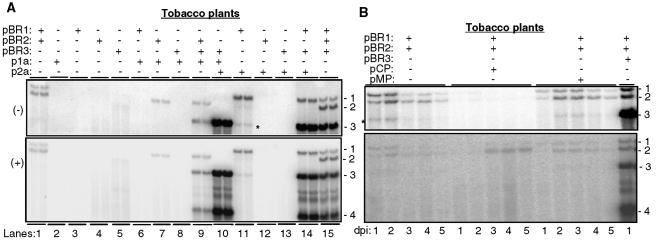
We expressed all four BMV proteins from replication-incompetent RNAs and examined whether and how they could affect BMV RNA accumulation. Tobacco leaves were infiltrated with A. tumefaciens containing various combinations of plasmids producing BMV RNAs and/or BMV proteins. As negative controls, plants receiving pBR1, pBR2, or plasmids expressing the recombinant proteins did not accumulate any BMV RNA (Fig. 5A, lanes 2 to 6). However, pBR2 in combination with p1a resulted in the accumulation of RNA2, and p2a was able to replicate with coinfiltrated pBR1 (Fig. 5A, lanes 7 and 11). Expression of p1a and p2a allowed the replication of RNA3 and the transcription of RNA4 (lanes 9 to 10). Dinant et al. (25) had demonstrated that specific interaction between these proteins expressed in tobacco protoplasts is required for BMV RNA replication. These results demonstrate that 1a and 2a proteins produced from nonreplicating RNAs can form active BMV replicase.
FIG. 5.
Northern blot analyses of the effects of BMV proteins on BMV RNA accumulation. (A) Effects of the 1a and 2a proteins produced independently of BMV RNAs. The plasmids agroinfiltrated into particular tobacco leaves are noted above the gel image. The symbols + and − denote the presence and absence of a plasmid in the Agrobacterium strain used to infiltrate plants, respectively. Every sample was tested in two independently infiltrated leaves and processed for RNA analysis. We did not coexpress a protein with the RNA from which it was derived, since this combination resulted in silencing of the RNA (data not shown), in agreement with the results of Iyer and Hall (38). Also, these results were reproducible in at least three independent experiments that contained different combinations of the expression plasmids. The asterisk denotes what we presume to be a degradation product that comes from RNA1. This RNA is not present in the absence of agroinfiltration. (B) Effects of the movement and coat proteins on BMV RNA accumulation. The symbols used are as explained above. dpi, days postinfiltration. The asterisk denotes what we presume to be a degradation product that comes from RNA1. This RNA is not present in the absence of agroinfiltration.
Plants expressing the CP and the MP were also tested for effects on the accumulation of BMV RNA1 and RNA2 (driven from pBR1 and pBR2) (Fig. 5B). The CP coexpressed with pBR1 and pBR2 in trans significantly reduced the amount of minus-strand RNA1 and RNA2 over the 5 days in which samples were analyzed. It also caused a slight increase in plus-strand RNA2 levels (Fig. 5B). Expression of the MP had relatively minor effects on minus-strand RNA accumulation, although plants infiltrated with pBR1 and pBR2 had more-abundant minus-strand RNAs at days 1 and 2 after infiltration, while plants coinfiltrated with pBR1 and pBR2 with MP caused minus-strand RNAs to be more abundant at days 2 and 3 (Fig. 5B).
The above observations suggest that the CP and MP may regulate the temporal switches from replication to cell-to-cell spread in plants. Therefore, we examined their roles in BMV RNA accumulation further by mutating their initiation codons in the context of pBR3. Changing the first AUG codon of the CP coding sequence to a CUG in construct pCPKOI resulted in the production of a truncated CP (Fig. 6A). Rao and Grantham (68) reported that translation of the BMV CP could also initiate from the second codon. Therefore, we mutated both the first and second AUG codons in the construct named pCPKOII. Plants infiltrated with wild-type pBR1, pBR2, and pCPKOII accumulated BMV RNAs with a 1-day delay in comparison to the positive control. Once the minus-strand RNAs accumulated, however, they did so for several days at two- to threefold higher levels than in the wild-type constructs (Fig. 6B). In contrast, plus-strand RNA accumulation was severely debilitated. RNA1, for example, was present at between 16 and 36% of the wild-type level, while RNA3 and RNA4 were at less than 11% of the amount observed in the wild type (Fig. 6B). These results are complementary to those in Fig. 5B, where CP expression in trans decreased minus-strand RNA levels and increased plus-strand RNA levels, perhaps by binding to plus-strand RNA and preventing minus-strand RNA synthesis by the BMV replicase.
FIG. 6.
Effects of manipulating MP and CP expression in agroinfiltrated tobacco and transfected barley protoplasts. (A) BMV capsid protein expressed in tobacco after agroinfiltration. A slice of a 4 to 12% denaturing protein gel is shown. MPKO and CPKOI indicate that plasmid pBR3 contains a mutation of the translation initiation codon from AUG to CUG. The BMV CP gene has a second initiation codon, which allows expression of a smaller amount of a truncated protein. CPKOII has two mutations that altered the first two initiation codons (codons 1 and 8) to CUGs. The lane marked with “M” denotes a protein molecular mass standard of 20 kDa, and the lane marked with “C” denotes a mock-inoculated control. (B) Northern blot analysis of the RNAs produced by agroinfiltration in tobacco by RNAs with mutations affecting either CP or MP expression from pBR3. dpi, days postinfiltration. The quantifications were performed by adjusting the amount of RNA3 or RNA4 in the wild-type sample each day to 100%. The amount of the comparable RNAs in plants infiltrated with CPKOII or MPKO were normalized to that in the wild-type sample. (C) Effects of the MPKO and CPKOII mutations on BMV RNA accumulation in barley protoplasts. The presence or absence of plasmids encoding wild-type BMV RNAs is denoted with a +. The MPKO or the CPKO substituted for wild-type RNA3 is shown by the names of the mutant construct. hpi, hours postinfiltration. The amount of BMV RNA3 and RNA4 were normalized to the amount present in the protoplasts transfected with wild-type BMV RNAs. (D) Accumulations of the BMV RNAs in tobacco protoplasts transfected with the three wild-type BMV RNAs or wild-type RNA1, RNA2, and CPKOII.
A mutation in the initiation codon of the MP coding sequence in pBR3 (in the construct named pMPKO) reduced minus-strand RNA levels to 30% of those of the positive control. However, there were only modest effects on the accumulation of the plus-strand RNAs, including subgenomic RNA4. The effects of preventing MP translation are thus milder than the effects of preventing CP translation.
We examined the effects of pCPKOII and MPKO on BMV RNA levels in barley protoplasts to allow comparisons to the effects observed in tobacco plants. For the MPKO, no significant effect on minus-strand RNA replication was observed. For the CPKOII, RNA1 and RNA2 levels were not significantly different from those of the wild-type control, while RNA3 and RNA4 were at 79% and 57% of wild-type levels (Fig. 6C). The results with CPKOII can be compared to those from the agroinfiltrated tobacco, where plus-strand RNA3 and RNA4 levels were both at less than 11% of wild-type levels (Fig. 6D).
The difference between the agroinfiltrated tobacco and transfected barley protoplasts seen with the CPKOII could be due to an innate difference between barley and tobacco cells or regulatory input from the entire plant. To distinguish between these two possibilities, tobacco protoplasts were transfected with wild-type RNA3 or CPKOII (along with wild-type RNA1 and RNA2). In tobacco protoplasts, the lack of the capsid protein did not affect minus-strand RNAs but reduced the plus-strand RNA levels. RNA1 was at 19% of wild-type levels, levels similar to those observed in agroinfiltrated plants. RNA3 and RNA4 were reduced to 30 and 19%, respectively, a more severe decrease than was observed in barley protoplasts. These results show that the plant species from which the protoplast was generated may also significantly influence the outcome of BMV RNA replication.
BMV proteins and RNA silencing.
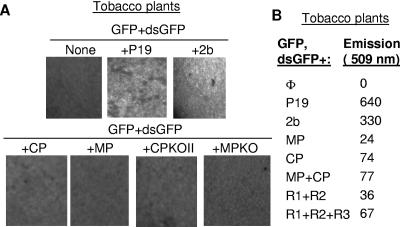
The effects of the CP on BMV RNA accumulation in agroinfiltrated plants could be due to encapsidation of the RNA and/or an effect on posttranscriptional gene silencing (PTGS). While related plant-infecting viruses have been reported to suppress silencing through virus-encoded suppressor proteins (15, 26, 50-52, 63, 71, 94), BMV has not been reported to produce such a protein. To examine possible effects of the CP and other BMV proteins on PTGS, we used an assay developed by Johansen and Carrington (41) in which tobacco transiently expressing a double-stranded RNA containing an inverted repeat of the GFP coding sequence in plasmid dsGFP would induce silencing of a second copy of the GFP expressed from a reporter, pGFP.
Tobacco agroinfiltrated with the pGFP construct produced fluorescence 1 day after infiltration, and the fluorescence gradually decreased after 3 to 4 days. No detectable GFP fluorescence was observed when dsGFP was coexpressed with GFP, as expected. The presence of silencing suppressors from P19 or 2b rescued GFP fluorescence and allowed the GFP signal in these leaves to be observed for up to 10 days (Fig. 7A and data not shown). These results indicate that the silencing detection assay works in our hands.
FIG. 7.
The BMV proteins do not have potent silencing suppressing activity. (A) Microscopic images of tobacco leaves infiltrated with GFP plus dsGFP, which will induce posttranscriptional silencing, silencing suppressors, and BMV proteins. The upper panel of three images shows the controls where there is silencing and the reversal of silencing by P19 and 2b. The images were taken with a Zeiss Axioplan 2 microscope using an FITC filter (at a 543-nm excitation and a 505- to 530-nm emission). The images were processed with Adobe systems software. (B) Quantitative analysis of GFP expression in plant lysates. The emissions are measured in arbitrary fluorescent units but can be compared to each other.
When various combinations of BMV constructs, including those that express all three BMV RNAs, were coinfiltrated along with dsGFP and pGFP, there was no visible increase in GFP levels. Therefore, none of the BMV proteins or RNAs, either alone or in combination, could effectively suppress posttranscriptional silencing. To examine GFP levels more quantitatively, we made lysates of another series of plants and monitored GFP fluorescence. While the GFP levels in the positive controls were present at between 330 and 640 arbitrary fluorescent units, the presence of the CP constructs was, at best, able to increase GFP expression to only 77 arbitrary fluorescent units in comparison to the bona fide silencing suppressors (Fig. 7B). All combinations that caused a slight increase in GFP activity contained the BMV CP. Together with the effects of the CP on BMV minus- and plus-strand RNAs, our data suggest that the CP can bind BMV RNAs and confer some protection to degradative enzymes. Nonetheless, the BMV CP does not have the silencing suppressor properties in the range of p19 and 2b proteins from tomato bushy stunt virus and CMV, respectively.
BMV replication proteins and GFP expression from BMV RNA.
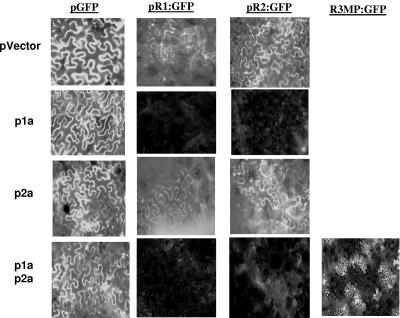
The availability of functional BMV replication proteins expressed in trans of the RNA allows us to examine the requirements for processes other than RNA replication. Viral RNA replication is intimately linked to translation, since both processes use the same template RNA. Furthermore, poliovirus and flavivirus RNA replication and translation are tightly regulated in a temporal manner (4, 7-9, 30, 37, 96). To address how the BMV replication proteins can affect translation, a sequence coding for GFP was used to replace the sequences encoding BMV 1a, 2a, and MP from versions of pBR1, pBR2, and pBR3, respectively. The resulting plasmids are named pR1:GFP, pR2:GFP, and pR3MP:GFP. Since the sequences used to express 1a and 2a in trans are no longer present in these plasmids, RNA silencing should be minimized. To ensure that these constructs are functional for translation, they were introduced into Agrobacterium and infiltrated into tobacco leaves. All three constructs produced easily detectable levels of GFP either by themselves or when they were coinfiltrated with the empty vector used to produce recombinant BMV proteins (Fig. 8). These control experiments corroborate the previous analyses from French and Ahlquist (29) and Annamalai and Rao (5), who had analyzed the expression of foreign proteins in place of the capsid coding sequence.
FIG. 8.
Effects of BMV RNA replication proteins on GFP expression from BMV RNA. The images are from tobacco leaves infiltrated with Agrobacterium cultures transformed with either the plasmid vector or plasmids expressing BMV proteins. The constructs containing GFP are listed above the micrographs. The relevant proteins or the control are identified to the left of the micrographs. All micrographs shown were taken 2 days after infiltration; each image is from an independently infiltrated leaf, and all were taken at a magnification of ×20 using the FITC filter.
When pR1:GFP and pR2:GFP were visualized for GFP expression in leaves coinfiltrated with p1a or both p1a and p2a, GFP signals were reduced to background levels (Fig. 8). However, tobacco infiltrated to express the 2a protein had higher levels of GFP fluorescence, suggesting that 1a is responsible for suppressing GFP expression. Protein 1a did not reduce GFP expression from pGFP expressed independently of the BMV nontranslated sequences (Fig. 8), indicating that the reduction that we observed requires specific interaction between 1a and a cis-acting sequence from BMV RNA1 and RNA2. Furthermore, 1a and 2a did not prevent GFP expression from pR3MP:GFP, although the pattern of expression was restricted to individual cells (Fig. 8). These results demonstrate that, in addition to its role in directing the replication of BMV RNAs, 1a could differentially regulate expression from the three BMV RNAs. The Agrobacterium system could thus be used to examine the relationship between viral RNA replication and protein production.
BMV replication proteins and RNA stability.
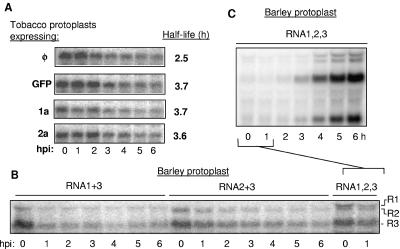
One observation that we have consistently made with agroinfiltrated tobacco leaves is that the expression of 1a or RNA1 does not obviously affect the stability of other BMV RNAs (e.g., compare Fig. 5, lanes 5 and 8). These results are in contrast to the dramatic 1a-induced stabilization of BMV RNA3 and RNA2 when they were coexpressed in S. cerevisiae (18, 87). Based on our analysis of the differences in cis and trans requirements in agroinfiltrated plants and transfected barley protoplasts, clear differences can be due to the host system used. Therefore, we examined the in vivo stability of RNA3 in the presence of tobacco cells expressing either 1a, 2a, GFP, or no recombinant protein. Protoplasts were made from plants that had been infiltrated 2 days before. These protoplasts obviously express GFP and 1a protein, as detected by fluorescence and Western blot analysis using an antibody specific to 1a (data not shown). Equal numbers of cells were transfected with radiolabeled full-length RNA3, and aliquots were then extracted for total RNA and analyzed for the full-length radiolabeled RNA3 over a 6-hour period. With all three samples, the half-life of BMV RNA3 was approximately 2 to 3 h and did not depend on 1a expression (Fig. 9A). A similar experiment with BMV RNA2 yielded the same result (data not shown). These results suggest that the rapid degradation of BMV RNAs observed in yeast is specific to yeast.
FIG. 9.
BMV proteins and the stabilities of BMV RNAs. (A) Effects of BMV replication proteins on transfected BMV RNA3. The gel images are all from one experiment examining the stability of radiolabeled BMV RNA3 transfected into tobacco protoplasts that express GFP, 1a, or 2a. Total RNAs were harvested at 0 to 6 h posttransfection and then electrophoresed in an agarose gel. The half-lives of the RNAs were determined by deriving the time in which half of the initial amount of RNA was present. (B) The effects of BMV RNAs transfected into barley protoplasts. The transcripts transfected into protoplasts are identified above and to the right of the gel image. The times (in hours) when the RNAs were harvested after inoculation of the protoplasts are noted below the gel image. (C) Accumulations of the BMV RNAs in barley protoplasts transfected with all three BMV RNAs. This gel image is identical to the last two lanes of the gel in panel C. However, it was exposed for a shorter period of time to allow visualization of the replication products. hpi, hours postinfiltration.
Next, we determined whether the BMV RNA replication proteins would affect the stability of the BMV RNAs in barley protoplasts as well as in tobacco. Barley protoplasts were transfected with various combinations of BMV RNAs, and the input and replicated RNAs were analyzed over time. Transfection of RNA1 and RNA3 did not result in an obvious stabilization of either RNA over a 6-h period (Fig. 9B). Transfection of RNA2 and RNA3 also did not result in an obvious stabilization of either RNA (Fig. 9B). Only when all three BMV RNAs were present did we observe an increase in the abundance of the plus-strand BMV RNAs (Fig. 9C). This increase could be attributed to BMV RNA replication, rather than an increase in RNA stability. Even in this sample, we note that prior to the replication of BMV plus-strand RNAs, which takes place 4 to 6 h after transfection (36), there were no obvious differences in the stabilities of the RNAs that could be attributed to the presence of RNA1 (Fig. 9C).
DISCUSSION
We have developed an Agrobacterium-mediated T-DNA gene delivery system to analyze the requirements for BMV RNA replication, gene expression, and systemic spread in tobacco plants. BMV launched from agroinfiltrated T-DNAs could successfully infect local and systemic leaves and produce virions and RNA in large quantities. The agroinfiltrated leaves could also be used to enrich viral replicase that will accept exogenously provided templates. Furthermore, we could separate the cis and trans requirements for BMV RNA replication, since replication proteins produced from nonreplicating messenger RNAs are functional for RNA replication. Interestingly, the production of the BMV RNA replication or capsid proteins affected not only the requirements associated with replication, but also the regulation of protein expression. The agroinfiltration system has the added advantage of introducing the viral sequences into virtually every mesophyll cell of the plant and could allow robust transient expression of specific viral proteins. Importantly, this system can be used to examine the contributions of host responses to viral infections. In addition, this system could facilitate future biochemical analyses of BMV infection and be of use in recombinant technology.
BMV and RNA silencing.
The expression of all four BMV proteins in replication-incompetent constructs allows us to analyze their interactions with BMV RNAs. While the closely related CMV is known to produce a potent suppressor for posttranscriptional silencing (34, 50), we did not identify a convincing silencing suppressor activity for any of the four BMV proteins expressed either alone or in combination. Minor effects of CP on the silencing signal could be observed with quantitative analysis of the GFP signal (Fig. 7B). However, this could be attributed to nonspecific binding by the BMV capsid protein to RNA. BMV RNAs are susceptible to PTGS (38). Annamalai and Rao (5) have shown that BMV CP can stabilize BMV RNAs by encapsidation. Therefore, BMV must use one or more alternative mechanisms to escape the detrimental effects of PTGS in comparison to viruses with active silencing suppressors.
BMV replication proteins and gene expression.
We have observed that the BMV 1a protein had the novel activity of shutting down GFP expression when GFP replaced the 1a or 2a coding sequences. This repressive effect was specific to GFP expressed from BMV cis-acting sequences (Fig. 8), suggesting that a specific interaction with the 1a protein is required. A likely candidate for the cis-acting sequence responsible for the repression is the B box located in RNA1 and RNA2. Intriguingly, both B boxes in RNA1 and RNA2 are upstream of the translation initiation codon in these RNAs. This suggests that 1a binding to the RNAs could prevent access to the translation factors. GFP expression from RNA3 is not as severely affected by the presence of 1a. In RNA3, the B box is within the intercistronic sequence, downstream of the MP coding sequence, perhaps providing an explanation for 1a having a less dramatic effect on translation from the MP open reading frame.
Diez et al. (24) had identified a cellular factor, Lsm1P, that is required to decrease translation and perhaps shuttle the BMV RNA into membrane-associated replication complexes. Together with our results, 1a protein is likely responsible for the temporal switch between viral RNA replication and translation. Perhaps when 1a concentration increases in the newly infected cell, 1a shuts down translation to allow RNA replication to commence. Confirmation of this model awaits additional experimentation.
BMV replication in different systems.
To validate the use of the Agrobacterium-mediated gene delivery system, we compared the replication requirements for BMV RNA-infiltrated tobacco and -transfected barley protoplasts. The results can also be compared to the in vitro results from the enriched BMV RNA replicase. For the cis-acting signals, mutations that affected BMV RNA-dependent RNA synthesis in vitro generally correlated well with the results from barley protoplasts (84, 85). As is to be expected, the in vitro replicase assay did not identify some mutations that have detrimental effects in the infected cell, but any residue that affected RNA synthesis in vitro had negative effects in the cell (84). When the results from the BMV replicase and transfected barley are compared to those from tobacco infiltrated with Agrobacterium, mutations in the core promoters for genomic minus-, genomic plus-, and subgenomic plus-strand RNAs all had detrimental effects in tobacco. However, the requirements for RNA accumulation in transfected barley protoplasts and in plants infiltrated with recombinant Agrobacterium are quite different. For example, RNA1 replication is required for the replication and repair of the other BMV RNAs in barley protoplasts (36, 61). With the agroinfiltrated plants, we could produce 1a in trans or from a replication-defective RNA1, and the replication of the other BMV RNAs, including subgenomic RNA4, seemed to be unaffected. Annamalai and Rao (5) made similar observations. We also observed that a mutation in the subgenomic core promoter had severe effects on RNA3 levels in tobacco infiltrated with Agrobacterium but not in transfected barley protoplasts (Fig. 4A and B).
Several factors may contribute to the observed differences in the accumulations of BMV RNAs in different host species, such as (i) the defense responses in whole plants, (ii) high-level expression of the BMV proteins from the recombinant T-DNAs, (iii) a difference between monocot and dicot plants, and (iv) a combination of several effects, including potential ones from the manipulation of the plants or plant cells. With the B box deletions, it is clear that transcription from the strong CaMV 35S promoter in agroinfiltrated tobacco would provide sufficient amounts of the 1a protein even though the mutated RNA1 is incapable of replication (Fig. 4C). The effects of other mutations, however, cannot simply be explained by the expression level of the replicase. For example, even though BMV replicase could be produced in higher levels in agroinfiltrated tobacco, a mutation in the core subgenomic promoter lowered RNA3 levels in comparison to the effects of the same mutation in barley protoplasts (Fig. 4A). In transfected tobacco protoplasts expressing the CPKOII, the effect on plus-strand BMV RNA accumulation was more similar to the levels seen in tobacco plants than to those in barley protoplasts (Fig. 6C and D). These results suggest that the capsid has a greater role in maintaining the stability of the BMV RNAs in tobacco than in barley protoplasts. Binding of the capsid to BMV plus-strand RNA may down-regulate minus-strand BMV RNA accumulation, perhaps by preventing the BMV replicase from gaining access to the RNA (Fig. 4B). All of our results indicate that the difference in BMV RNA levels observed in different host systems is due to a complex array of influences from the host system and/or the expression system used.
Our observations and anecdotal ones reported in the literature indicate that results and conclusions for BMV RNA replication in different systems need to be interpreted cautiously. For example, tobacco plants expressing 1a, 2a, or GFP by agroinfiltration all had similar stabilities for BMV RNA2 and RNA3. We also did not observe obvious stabilizing effects of the BMV RNAs by their encoded proteins in barley protoplasts (Fig. 9). It is logical that an RNA virus such as BMV that replicates at high levels (Fig. 2A) would select for elements in its RNA to render its genome resistant to cellular RNA degradation. Such stabilizing elements may not be functional in the yeast system, where 1a has a dramatic effect in stabilizing BMV (18, 24, 87). This conclusion does not affect the validity of a model where BMV RNA replication takes place in a factory protected by membranes (77). In fact, it is quite logical for viral RNA replication and transcription, and perhaps the sequestration of the viral RNAs from the cellular defense systems, to be located in factories, and there is ample precedent for this being the case in the cell and with other viruses (5, 6, 10, 11). Also, these factories may exclude translation factors, perhaps accounting for the role of 1a in inhibiting translation (Fig. 8). However, these results do point to the need for additional characterization of BMV infection in whole plants. In support of there being differences between hosts, the Ahlquist group already has observed that the requirements for BMV RNA3 and subgenomic RNA4 synthesis are different in barley protoplasts and in S. cerevisiae (31).
Different host systems are increasingly used to analyze the requirements for viral infection. We expect that the differences we documented with BMV in tobacco and in barley will reflect requirements with other viruses. In fact, the BMV capsid protein has been reported to evolve rapidly in adaptation to plant host species (78); mutations that increased the replication of the hepatitis C virus subgenomic replicon are detrimental in chimpanzees, and the virus rapidly reverted to the original sequence (14).
Acknowledgments
We thank the Texas A&M University Cereal Killers for helpful discussions; James Carrington, V. Dolja, and W. Dawson for plasmids and reagents to perform RNA-silencing assays; and A. L. N. Rao for helpful discussions.
The Kao lab is supported by the NSF grant MCB0332259. The Dragnea lab acknowledges funding by NSF grant BES0322767.
REFERENCES
- 1.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. Kao. 1998. Mechanistic analysis of RNA synthesis by an RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:271-276. [DOI] [PubMed] [Google Scholar]
- 3.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96-111. [DOI] [PubMed] [Google Scholar]
- 6.Asurmendi, S., R. H. Berg, J. C. Kao, and R. N. Beachy. 2004. Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 101:1415-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 15:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 11.Bienz, K., D. Egger, D., T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigneti, G., O. Voinnet, L. Wan-Xiang, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Buck, K. W. 1996. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canizares, M. C., K. M. Taylor, and G. P. Lomonossoff. 2004. Surface-exposed C-terminal amino acids of the small coat protein of Cowpea mosaic virus are required for suppression of silencing. J. Gen. Virol. 85:3431-3435. [DOI] [PubMed] [Google Scholar]
- 16.Carrington, J. C., and D. D. Freed. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64:1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 18.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi, S. K., M. Hema, K. Gopinath, J. Santos, and C. Kao. 2004. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 78:13420-13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi, Y. G., T. W. Dreher, and A. L. N. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Commandeur, U., and R. Martin. 1993. Investigations into the molecular biology of potato leaf roll luteovirus by means of agroinfection. Phytopathology 83:1426. [Google Scholar]
- 22.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of brome mosaic virus D-RNA in barley. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 23.Damayanti, T. A., S. Tsukaguchi, K. Mise, and T. Okuno. 2003. Cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 77:9979-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinant, S., M. Janda, P. A. Kroner, and P. Ahlquist. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 67:7181-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong, X., R. van Wezel, J. Stanley, and Y. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201:31-40. [DOI] [PubMed] [Google Scholar]
- 28.Dreher, T. W., A. L. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425-438. [DOI] [PubMed] [Google Scholar]
- 29.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grdzelishvili, V. Z., H. Garcia-Ruiz, T. Watanabe, and P. Ahlquist. 2005. Mutual interference between genomic RNA replication and subgenomic mRNA transcription in brome mosaic virus. J. Virol. 79:1438-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimsley, N., T. Hohn, J. W. Davies, and B. Hohn. 1987. Agrobacterium-mediated delivery of infectious maize streak virus into maize plants. Nature 325:177-179. [Google Scholar]
- 33.Grimsley, N. T., B. Hohn, T. Hohn, and R. Walden. 1986. Agroinfection as an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. USA 83:3282-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hema, M., and C. C. Kao. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hema, M., K. Gopinath, and C. Kao. 2005. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J. Virol. 79:1417-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isken, O., C. W. Grassmann, H. Yu, and S. E. Behrens. 2004. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer, L. M., and T. C. Hall. 2000. Virus recovery is induced in brome mosaic virus p2 transgenic plants showing synchronous complementation and RNA-2-specific silencing. Mol. Plant-Microbe Interact. 13:247-258. [DOI] [PubMed] [Google Scholar]
- 39.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 40.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 41.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot: induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kao, C., S. Green, B. Stein, and S. Golden. 2005. Diel regulation of a photosynthetic cyanobacterium by a contractile DNA phage. Appl. Environ. Microbiol. 71:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao, C. C. 2002. Lessons learned from the core RNA promoters of Brome mosaic virus and Cucumber mosaic virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 44.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 45.Kim, C. H., and C. Kao. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through an exposed adenine. RNA 7:1476-1485. [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, C.-H., C. C. Kao, and I. Tinoco. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7:415-423. [DOI] [PubMed] [Google Scholar]
- 47.Kong, F., K. Sivakumaran, and C. Kao. 1997. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200-210. [DOI] [PubMed] [Google Scholar]
- 48.Kroner, P. A., B. M. Young, and P. Ahlquist. 1990. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J. Virol. 64:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leiser, R. M., V. Ziegler-Graff, A. Reutenauer, E. Herrbach, O. Lemaire, H. Guilley, K. Richards, and G. Jonard. 1992. Agroinfection as an alternative to insects for infecting plants with beet western yellows luteovirus. Proc. Natl. Acad. Sci. USA 89:9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 51.Liu, L., and G. Lomonossoff. 2002. Agroinfection as a rapid method for propagating Cowpea mosaic virus-based constructs. J. Virol. Methods 105:343-348. [DOI] [PubMed] [Google Scholar]
- 52.Liu, L., J. Grainger, M. C. Canizares, S. M. Angell, and G. P. Lomonossoff. 2004. Cowpea mosaic virus RNA-1 acts as an amplicon whose effects can be counteracted by a RNA-2-encoded suppressor of silencing. Virology 323:37-48. [DOI] [PubMed] [Google Scholar]
- 53.Llave, C., K. D. Kasschau, and J. C. Carrington. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu, R., A. M. Martin-Hernandez, J. R. Peart, I. Malcuit, and D. C. Baulcombe. 2003. Virus-induced gene silencing in plants. Methods 30:296-303. [DOI] [PubMed] [Google Scholar]
- 55.Marillonnet, S. A., M. Giritch, R. Gils, R. Kandzia, V. Klimyuk, and Y. Gleba. 2004. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 101:6852-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marsh, L. E., and T. C. Hall. 1987. Evidence implicating a tRNA heritage for the promoters of positive-strand RNA synthesis in brome mosaic and related viruses. Cold Spring Harbor Symp. Quant. Biol. 52:331-341. [DOI] [PubMed] [Google Scholar]
- 57.Mise, K., R. F. Allison, M. Janda, and P. Ahlquist. 1995. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 67:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsthoorn, R. C. L., A. Bruyere, A. Dzianott, and J. J. Bujarski. 2002. RNA recombination in brome mosaic virus: effects of strand-specific stem-loop inserts. J. Virol. 76:12654-12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng, C. W., and V. V. Dolja. 2000. Leader proteinase of the beet yellows closterovirus: mutation analysis of the function in genome amplification. J. Virol. 74:9766-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng, C. W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pogue, G. P., and T. C. Hall. 1992. The requirements for 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol. 66:674-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prokhnevsky, A. I., V. V. Peremyslov, A. J. Napuli, and V. V. Dolja. 2002. Interaction between long-distance transport factor and Hsp70-related movement protein of beet yellows virus. J. Virol. 76:11003-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rao, A. L., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rao, A. L., B. P. Sullivan, and T. C. Hall. 1996. Use of Chenopodium hybridum facilitates isolation of brome mosaic virus RNA recombinants. J. Gen. Virol. 71:1403-1407. [DOI] [PubMed] [Google Scholar]
- 66.Rao, A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao, A. L. N., R. Duggal, F. C. Lahser, and T. C. Hall. 1994. Analysis of RNA replication in plant viruses. Methods Mol. Genet. 4:216-236. [Google Scholar]
- 68.Rao, A. L. N., and G. L. Grantham. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus. Virology 211:42-52. [DOI] [PubMed] [Google Scholar]
- 69.Ratcliff, F., A. M. Martin-Hernandez, and D. C. Baulcombe. 2001. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25:237-245. [DOI] [PubMed] [Google Scholar]
- 70.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reed, J. C., K. D. Kasschau, A. I. Prokhnevsky, K. Gopinath, G. P. Pogue, J. C. Carrington, and V. V. Dolja. 2003. Suppressor of RNA silencing encoded by Beet yellows virus. Virology 306:203-209. [DOI] [PubMed] [Google Scholar]
- 72.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rigden, J. E., I. B. Dry, L. R. Krake, and M. A. Rezaian. 1996. Plant virus DNA replication processes in Agrobacterium: insight into the origins of geminiviruses? Proc. Natl. Acad. Sci. USA 93:10280-10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 75.Satyanarayana, T., S. Gowda, V. P. Boyko, M. R. Albiach-Martí, M. Mawassi, J. Navas-Castillo, Karasev, V. V. Dolja, M. E. Hilf, and D. J. Lewandowski. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. USA 96:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmitz, I., and A. L. Rao. 1996. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology 226:281-293. [DOI] [PubMed] [Google Scholar]
- 77.Schwartz, M., J. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. USA 101:11263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.She, Y. M., S. Haber, D. L. Seifers, A. Loboda, I. Chernushevich, H. Perreault, W. Ens, and K. G. Standing. 2001. Determination of the complete amino acid sequence for the coat protein of brome mosaic virus by time-of-flight mass spectrometry. Evidence for mutations associated with change of propagation host. J. Biol. Chem. 276:20039-20047. [DOI] [PubMed] [Google Scholar]
- 79.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siegel, R. W., L. Bellon, L. Beigelman, and C. C. Kao. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 95:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyán. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sivakumaran, K., and C. C. Kao. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 73:6415-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sivakumaran, K., C. H. Kim, R. Tayon, and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sivakumaran, K., S. K. Choi, M. Hema, and C. Kao. 2004. Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 78:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sivakumaran, K., M. Hema, and C. C. Kao. 2003. Brome mosaic virus RNA syntheses in vitro and in barley protoplasts. J. Virol. 77:5703-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smirnyagina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations ion the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198:427-436. [DOI] [PubMed] [Google Scholar]
- 87.Sullivan, M., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun, J., S. Adkins, G. Faurote, and C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the brome mosaic virus RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 89.Turpen, T. H., A. M. Turpen, N. Weinzettl, M. H. Kumagai, and W. O. Dawson. 1993. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA from tobacco mosaic virus. J. Virol. Methods 42:227-240. [DOI] [PubMed] [Google Scholar]
- 90.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 91.Vlot, A. C., A Menard, and J. F. Bol. 2002. Role of the alfalfa mosaic virus methyltransferase-like domain in negative RNA synthesis. J. Virol. 76:11321-11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vlot, A. C., L. Neeleman. H. J. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Voinnet, O., V. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Voinnet, O., S. Rivas, P. Mestre, and D. C. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 95.Wang, M. B., and M. Metzlaff. 2005. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 96.Yu, H., O. Isken, C. W. Grassmann, and S. E. Behrens. 2000. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J. Virol. 74:5825-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]