Abstract
ARB1 is an essential yeast protein closely related to members of a subclass of the ATP-binding cassette (ABC) superfamily of proteins that are known to interact with ribosomes and function in protein synthesis or ribosome biogenesis. We show that depletion of ARB1 from Saccharomyces cerevisiae cells leads to a deficit in 18S rRNA and 40S subunits that can be attributed to slower cleavage at the A0, A1, and A2 processing sites in 35S pre-rRNA, delayed processing of 20S rRNA to mature 18S rRNA, and a possible defect in nuclear export of pre-40S subunits. Depletion of ARB1 also delays rRNA processing events in the 60S biogenesis pathway. We further demonstrate that ARB1 shuttles from nucleus to cytoplasm, cosediments with 40S, 60S, and 80S/90S ribosomal species, and is physically associated in vivo with TIF6, LSG1, and other proteins implicated previously in different aspects of 60S or 40S biogenesis. Mutations of conserved ARB1 residues expected to function in ATP hydrolysis were lethal. We propose that ARB1 functions as a mechanochemical ATPase to stimulate multiple steps in the 40S and 60S ribosomal biogenesis pathways.
Ribosome synthesis takes place primarily in the nucleus of eukaryotic cells and involves the processing of a large precursor rRNA containing the sequences of 18S rRNA, found in mature 40S subunits, and the 5.8S and 25S rRNAs found in 60S subunits. A large ensemble of trans-acting factors, the U3 snoRNP complex, and ribosomal proteins (mostly of the 40S subunit), interact with the pre-rRNA to form the 90S preribosome, or small subunit processome. Three cleavage reactions that excise the 20S precursor of 18S rRNA from the 35S rRNA precursor (at sites A0, A1, and A2) (see Fig. 2G) take place in the >2-MDa 90S complex (reviewed in references 15, 25, and 70). The pre-40S particle released after cleavage at sites A0-A1-A2 is transported to the cytoplasm with a relatively small number of factors, some of which also reside in the 90S complex, while others appear to join with the pre-40S particle after its separation from the pre-60S particle (62). Cleavage of 20S pre-rRNA to mature 18S rRNA occurs following export of the pre-40S particle to the cytoplasm.
FIG. 2.
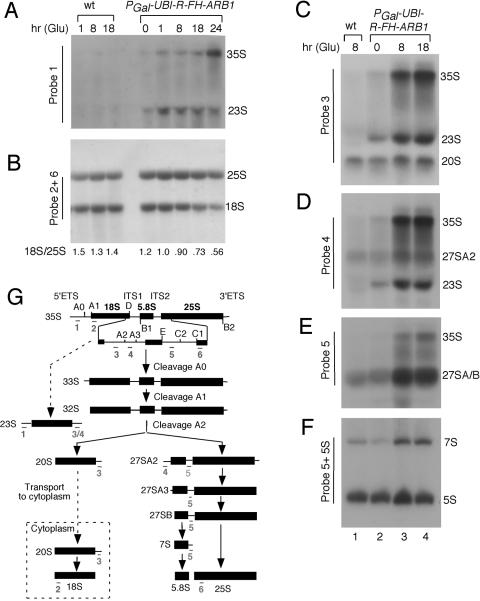
Depletion of ARB1 leads to defects in 35S pre-rRNA processing and a deficit of 18S rRNA. (A to F) Strains BY4741 (WT) and YDH226 (PGAL-UBI-R-FH-ARB1) were cultured as described in the legend of Fig. 1 for the indicated times in SCGlu (Glu) medium, and total RNA was extracted and subjected to Northern analysis using the probes indicated along the side of each blot. (G) Schematic diagram of the pre-rRNA processing pathway. The 35S pre-rRNA contains the sequences for mature 18S, 5.8S, and 25S rRNAs, separated by the two internal transcribed spaces (ITS) and flanked by the external transcribed spaces (ETS). The processing sites are indicated by uppercase letters A to E. The annealing positions of probes 1 to 6 are depicted beneath all of the rRNA species which they detect.
The pre-60S particle contains 27SA pre-rRNA (see Fig. 2G) and a large array of trans-acting factors not found in the 90S particle. It undergoes a series of RNA cleavage reactions that excise 25S and 5.8S from the 27SA pre-rRNA before being exported from the nucleus. Only a small number of factors present in the 90S particle also occur in pre-60S particles, and it is thought that such factors dissociate from the pre-60S particle early in its maturation. Hence, there are few factors found stably associated with both pre-40S and pre-60S particles (52, 70).
A number of ATPases of the AAA family and GTPases interact exclusively with pre-60S particles and may catalyze association/dissociation reactions or molecular rearrangements during 60S maturation. The GTPase Bsm1p, by contrast, is associated with the 90S particle and participates in 18S rRNA processing (70). Putative ATP-dependent RNA helicases have also been implicated in ribosome biogenesis (10, 26, 40, 52, 76), including several involved in A0-A1-A2 cleavage, and have been shown to be physically associated with pre-ribosomal particles.
Most ATPases in the ATP-binding-cassette (ABC) superfamily are membrane transporters that use ATP hydrolysis to transport solute molecules against a concentration gradient (48), and, typically, they contain two ABCs and a transmembrane domain. The ABC harbors a nucleotide binding domain with Walker A and B motifs plus an α-helical domain with the “LSGGQ” signature motif that distinguishes ABC proteins from other ATPases. ABCs bind ATP as a dimer, sandwiching two ATP molecules between the dimerized cassettes. Interactions of the ATP γ-phosphates with the signature motifs in the opposing cassettes clamp together the dimerized ABCs. ATP hydrolysis opens the clamp, and this pivoting movement of the interacting cassettes is thought to perform the mechanical work of opening and closing the solute channel formed by the transmembrane domains (6, 8, 35, 44, 50, 67). A soluble ABC protein could likewise perform work if the ABCs, or tightly associated domains, interact with different molecules or different segments of the same molecule. For example, it was proposed that the ABCs in Rad50 bind and juxtapose the ends of broken DNA molecules destined for repair (32).
The yeast proteins GCN20, YEF3, and RLI1 are soluble ABC proteins in Saccharomyces cerevisiae that interact with ribosomes and have functions connected with protein synthesis, translational control, or ribosome biogenesis. YEF3 is an essential translation elongation factor (69), whereas GCN20 is thought to relay the signal of uncharged tRNA in the ribosomal A site to a protein kinase (GCN2) that down-regulates translation initiation in amino acid-starved cells (46, 47, 73). RLI1 interacts with 40S subunits and translation initiation factors eIF2, eIF3, and eIF5 and stimulates assembly of the 43S preinitiation complex containing these and other essential initiation factors (11). RLI1 also functions in ribosome biogenesis, being required for wild-type (WT) rates of pre-rRNA processing in both the 40S and 60S biogenesis pathways and for nuclear export of both 40S and 60S subunits (37, 76). The mammalian soluble ABC protein, ABC50, interacts with ribosomal subunits and eIF2 and stimulates the formation of eIF2-GTP-Met-tRNAiMet ternary complexes (71). The X-ray crystal structure of an archaeal ortholog of RLI1 revealed that its two ABCs interact to form composite active sites, consistent with the ATP-driven clamp-like motion described for ABC transporters (35).
YER036C is a predicted soluble ABC protein in yeast of unknown function, closely related in sequence to GCN20 and YEF3. YER036C is essential (21) and was reported to interact with IMP4 in a large-scale yeast two-hybrid screen (29) and to copurify with ARX1 in a global analysis of protein complexes in yeast (19). IMP4 and ARX1 are involved in biogenesis of 40S and 60S ribosomal subunits, respectively (42, 52, 74). Accordingly, we set out to determine whether YER036C functions in ribosome biogenesis. We show here that depletion of YER036C in living cells lowers the steady-state level of mature 40S subunits. This phenotype can be attributed to delays in the processing of the 35S and 20S precursors of 18S rRNA and a possible defect in nuclear export of pre-40S particles. Interestingly, rRNA processing reactions in the 60S biogenesis pathway are also delayed by YER036C depletion. We find that YER036C shuttles from nucleus to cytoplasm and is physically associated in vivo with 40S, 60S, and 90S ribosomal species and with various proteins implicated previously in 40S or 60S ribosome biogenesis. Hence, YER036C may function directly in both legs of the ribosome biogenesis pathway. Having established that conserved residues in the ABCs are required for the essential function of YER036C in vivo, we henceforth designate this protein ARB1, for ABC protein involved in ribosome biogenesis. Our findings support the contention that all soluble members of the GCN20 subfamily of eukaryotic ABC ATPases are involved in ribosome biogenesis or protein synthesis.
MATERIALS AND METHODS
Plasmid constructions.
The plasmids employed are listed in Table 1. Plasmid pDH22 was constructed as follows. The ARB1 gene, including 528 bp upstream of the ATG start codon and 274 bp downstream of the stop codon, was amplified by PCR using the two primers 5′-GGA ATT CCA TTA TAT GCA CAT CTC CTA A-3′ and 5′-CGA TAA GGC AAC GAT GGT CA-3′ and genomic DNA from yeast strain BY4741 as template. The PCR product was cut with EcoRI and ClaI, and the resulting fragment was inserted between the EcoRI and ClaI sites of plasmid pRS316. The DNA sequence of the entire open reading frame (ORF) was verified, and it was shown that this plasmid complements an arb1Δ mutant.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pEMBLyex4 | hc; URA3 leu2-d | 4 |
| pGEX vector 4T-2 | Expression vector for GST fusions | Pharmacia |
| YCplac111 | sc; LEU2 | 22 |
| pRS316 | lc; URA3 | 22 |
| pDH103 | hc; URA3 PGAL-FH-GCN2 in pEMBLyex4 backbone | 12 |
| pGALYERFH | hc; URA3 PGAL-FH-ARB1 in pEMBLyex4 backbone | This study |
| pDH22 | lc; URA3 ARB1 in pRS316 backbone | This study |
| pDH129 | sc; LEU2 FH-ARB1 in YCplac111 backbone | This study |
| pDH144 | sc; LEU2 FH-ARB1-G229D,G230D,G519D in YCplac111 backbone | This study |
| pDH25-1 | sc; LEU2 PGAL-UBI-R-FH-ARB1 in YCplac111 backbone. | This study |
| pTS068 | lc; URA3 NOP1-GFP | 59 |
| pFA6a-GFP(S65T)-His3MX6 | Construct for one-step tagging of chromosomal alleles with GFP coding sequences | 45 |
| pFA6a-13myc-HIS3MX | Construct for one-step tagging of chromosomal alleles with myc13 coding sequences | 45 |
| pRSRPL25-GFP | lc; URA3 RPL25-GFP in pRS316 backbone | 49 |
| pRSRPS2-GFP | lc; URA3 RPL25-GFP in pRS316 backbone | 62 |
| p3397 | sc; LEU2 UBI-Met-NIP9 in YCplac111 backbone | 53 |
| pGEX-ARB1 | Full-length ARB1 ORF in pGEX 4T-2 | This study |
hc, high-copy-number plasmid; lc, low-copy-number plasmid; sc, single-copy plasmid.
To construct plasmid pGalYERFH, a fragment containing the entire ARB1 ORF and the coding sequences for the FLAG-His6 (FH) tag inserted at the 5′ end of the ORF plus 380 bp downstream of the stop codon was PCR amplified using primers 5′-CGA CGC GTC CAC CAG TAT CTG CGT CAA AG-3′ and 5′-CAC TGC AGA CGT AGC TGG TCA TAC GGC-3′ and genomic DNA from BY4741 as a template. The resulting fragment was cut with MluI and Pst1 and used to replace the Mlu1-Pst1 fragment in plasmid pDH103 (12). The entire ARB1 ORF and the sequences encoding the FLAG and His6 peptides appended at the 5′ end of the ORF were verified by DNA sequencing.
Plasmid pDH129 was constructed in several steps. First, ARB1 sequences from −528 to the ATG start codon (+1) were PCR amplified using the primers 5′-GGA ATT CCA TTA TAT GCA CAT CTC CTA A-3′ and 5′-GC GTC GAC TGG CAT ATT TGG TTC TG-3′ and genomic DNA from BY4741 as a template. The PCR product was cut with EcoRI and SalI, and the resulting fragment was inserted between the EcoRI and SalI sites of plasmid YCplac111 (22) to produce the transition plasmid pYER-Pro. Second, the entire ARB1 ORF and coding sequences for the FH tag at the 5′ end of the ORF were PCR amplified using the primers 5′-GG GTC GAC GAC TAC AAG GAC GAC GAT-3′ and 5′-CAC TGC AGA CGT AGC TGG TCA TAC GG-3′ and plasmid pGALYERFH as a template. The PCR product was cut with SalI and PstI and inserted between the SalI and PstI sites of pYER-Pro, resulting in plasmid pDH129. The DNA sequence of the entire FH-tagged ORF was verified, and it was shown that pDH129 can complement an arb1Δ mutant and direct production of FH-tagged ARB1 of the predicted molecular weight in yeast as judged by Western analysis of whole-cell extracts (WCEs) using antibodies against the His6 epitope.
To construct plasmid pDH144, the 1.2-kb BamHI fragment in pDH129 was removed and inserted at the BamHI site of pBLUESCRIPT, resulting in transition plasmid pBSabc. The G229D, G230D, and G519D mutations were introduced into pBSabc by site-directed mutagenesis (11), resulting in transition plasmid pBSabc-m. The sequence of the entire 1.2-kb fragment and the presence of these mutations in pBSabc-m were both verified. Finally, the 1.2-kb BamHI fragment in pDH129 was replaced by the 1.2-kb BamHI fragment from pBSabc-m, producing pDH144.
Plasmid pDH25-1 was constructed in two steps. First, a fragment containing the GAL1 promoter upstream of the ubiquitin coding sequences fused to an Arg codon, and bearing 5′ terminal EcoRI and 3′ terminal SalI sites, was PCR amplified using primers 5′-CG GAA TTC GTA TAC TAA ACT CAC AAA-3′ and 5′-GC GTC GAC GTG CAT ACC ACC TCT TAG CCT-3′ and p3397 (53) as a template. The resulting fragment was cut with EcoRI and SalI and inserted between the EcoRI and SalI sites of YCplac111, producing transition plasmid pUBI-R. Second, the entire ARB1 ORF, containing the coding sequences for the FH tag at the 5′ end, was PCR-amplified using primers 5′-GC GTC GAC GAC TAC AAG GAC GAC GAT-3′ and 5′-AA CTG CAG GCA CGT AGC TGG TCA TAC GGC-3′ and pGALYERFH as a template. The PCR product was cut with SalI and PstI and inserted between the SalI and PstI sites of pUBI-R, resulting in pDH25-1. The entire ARB1 ORF and the coding sequences for the ubiquitin-Arg-FLAG-His6 peptide were verified by sequencing.
To construct plasmid pGEX-ARB1, the entire ARB1 ORF was PCR-amplified using primers 5′-GGA ATT CCC CCACCA GTA TCT GCG TCA-3′ and 5′-GCG TCG ACA ATT ACA AAA CAA CGT TC-3′ and plasmid pDH22 as a template. The resulting fragment was cut with EcoRI and SalI and inserted between the EcoRI and SalI sites of plasmid pGEX vector 4T-2 (Pharmacia), resulting in the plasmid pGEX-ARB1.
Yeast strain constructions.
The yeast strains used in this study are listed in Table 2. To produce strain YDH209, pDH22 was introduced into the ARB1/arb1Δ diploid strain 20168, purchased from Research Genetics. The Ura+ transformants were sporulated and subjected to tetrad analysis. YDH209 was identified as an ascospore clone deleted of chromosomal ARB1 and carrying plasmid pDH22. Strain YDH272 was created by introducing plasmid pDH129 into YDH209. To produce YDH226, plasmid pDH25-1 was introduced into YDH209, and the Ura+ Leu+ transformants were replica plated on synthetic complete medium containing galactose (SCGal) and 5-fluoroorotic acid (5-FOA) to evict pDH22. To make YDH299, plasmid pDH144 was introduced into YDH209. To produce YDH275, pDH129 was introduced into YDH209, and transformants were selected on SC medium containing glucose (SCGlu)-Leu-Ura. The Ura+ Leu+ transformants were replica plated on 5-FOA medium to evict pDH22. To construct YDH338, the coding sequences for green fluorescent protein (GFP) were fused immediately upstream of the stop codon of the chromosomal ARB1 allele by one-step homologous recombination (45). We used the primers 5′-ACC AGA TGG GAC GGA TCC ATT TTG CAA TAT AAG AAC AAA TTG GCC AAG AAC GTT GTT TTG CGG ATC CCC GGG TTA ATT AA-3′ and 5′-TGC TAT TAA AAT ACA TAA AAG GGG AAG TAA AAA CGT GCA CGG CGC AGA TCA ACT TCA CAA TTA GAA TTC GAG CTC GTT TAA AC-3′ and plasmid pFA6a-GFP(S65T)-HIS3MX as a template to PCR amplify the appropriate DNA fragment for transforming strain Y464 to His+, producing YDH338. The presence of ARB1-GFP was verified by Western analysis of WCEs using monoclonal antibodies against GFP. Growth of this strain on yeast extract-peptone-dextrose (YPD) medium was indistinguishable from that of Y464. YDH332 was made by introducing plasmid pRSRPL25-GFP into Y464.
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| 20168 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 ARB1/arb1Δ | Research Genetics |
| YDH209 | MATahis3Δ1 leu2Δ0 ura3Δ0 arb1Δ pDH22[ARB1 URA3] | This study |
| YDH226 | MATahis3Δ1 leu2Δ0 ura3Δ0 arb1Δ pDH25-1[PGAL-UBI-R-FH-ARB1 LEU2] | This study |
| YDH272 | MATahis3Δ1 leu2Δ0 ura3Δ0 arb1Δ pDH22[ARB1 URA3] pDH129[FH-ARB1 LEU2] | This study |
| YDH275 | MATahis3Δ1 leu2Δ0 ura3Δ0 arb1Δ pDH129[FH-ARB1 LEU2] | This study |
| YDH299 | MATahis3Δ1 leu2Δ0 ura3Δ0 arb1Δ pDH22[ARB1 URA3] pDH144[FH-ARB1-G229D,G230D G519D LEU2] | This study |
| Y464 | MATahis3 leu2 trp1 ura3 crm1Δ::KANr pDC-crm1[crm1-T539C LEU2] | 51 |
| YDH315 | MATahis3Δ1 leu2Δ0 ura3Δ0 pTS068 [NOP1-GFP URA3] | 11 |
| YDH338 | MATahis3 leu2 trp1 ura3 ARB1-GFP HIS3 crm1Δ::KANr pDC-crm1[crm1-T539C LEU2] | This study |
| YDH332 | MATahis3 leu2 trp1 ura3 crm1Δ::KANr pDC-crm1[crm1-T539C LEU2] pRSRPL25-GFP[RPL25-GFP URA3] | This study |
| YDH1007 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 DED1-myc13 HIS3 | This study |
| YDH1008 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ZUO1-myc13 HIS3 | This study |
| YDH1009 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 CBF51-myc13 HIS3 | This study |
| YDH1010 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 TIF6-myc13 HIS3 | This study |
| YDH1011 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 LSG1-myc13 HIS3 | This study |
| YDH1012 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 SCP160-myc13 HIS3 | This study |
| YDH1013 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ARX-myc13 HIS3 | This study |
| YDH1014 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 IPM4-myc13 HIS3 | This study |
| F1205 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ARX1-TAP::HIS3 | Open Biosystems |
| F1207 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RIO2-TAP::HIS3 | Open Biosystems |
| F1208 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ENP1-TAP::HIS3 | Open Biosystems |
| F1231 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ARB1-TAP::HIS3 | Open Biosystems |
| F1207 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 NSA3-TAP::HIS3 | Open Biosystems |
To construct the eight myc13-tagged strains numbered consecutively from YDH1007 to YDH1014, the coding sequences for myc13 were fused immediately upstream of the stop codons of the chromosomal DED1, ZUO1, CBF5, TIF6, LSG1, SCP160, and ARX1 alleles, respectively, in strain BY4741 by one-step homologous recombination (45), using the eight pairs of primers listed in Table S1 of the supplemental material and plasmid pFA6a-13myc-HIS3MX as a template. Western analysis using monoclonal antibodies against c-myc13 epitope was employed to verify these myc13-tagged strains. Growth of these strains on YPD medium was indistinguishable from that of BY4741.
Biochemical and imaging techniques.
Polysome analysis using extracts from cells treated with cycloheximide was conducted as described previously (72). Northern analysis was conducted as described previously (39) using the following oligonucleotides described in Fig. 2G as probes: probe 1, 5′-GGT CTC TCT GCT GCC GGA; probe 2, 5′-CAT GGC TTA ATC TTT GAG AC; probe 3, 5′-CGG TTT TAA TTG TCC TA; probe 4, 5′-AAT TTC CAG TTA CGA AAA TTC TTG; probe 5, 5′-GGC CAG CAA TTT CAA GTT A; probe 6, 5′-CTC CGC TTA TTG ATA TGC. The sequence of the 5S probe is CGG ACG GGA AAC GGT GCT TTC TGG TAG ATA TGG CCG. Analysis of pre-rRNAs associated with the tandem affinity purification (TAP)-tagged proteins was performed after immunoprecipitation with immunoglobulin G beads as described (10). Pulse-chase labeling analysis of rRNA was carried out as described previously (39). FH-ARB1 protein was purified from strain BY4741 harboring plasmid pGALYERFH as previously described (12), except that 2% galactose was the carbon source and 3-aminotriazole was omitted. Imaging of GFP-tagged fusion proteins in yeast cells was conducted as described previously (68). TAP-tagged ARB1 protein was purified, and the copurifying proteins were analyzed by mass spectrometry essentially as described previously (19, 43). To identify the purified proteins, the TAP-ARB1 complexes were digested with trypsin and analyzed by multidimensional microcapillary liquid chromatography coupled with electrospray ionization-tandem mass spectrometry (43, 58). Only canonical tryptic peptides with SEQUEST cross-correlation scores greater than 2.0 were used for protein identification (14, 43). To estimate the relative abundance of a protein in the TAP-ARB1 and control purifications, a protein abundance factor (PAF) was calculated for each protein identified by the mass spectrometry analysis (57). A protein's PAF value is expressed as the total number of nonredundant spectra that correlate significantly to each ORF normalized to the molecular weight of the cognate protein (104). The PAFs were used to rank and compare proteins by their relative abundance from the TAP-ARB1 and control experiments. Coimmunoprecipitation analysis of myc-tagged proteins was performed as described previously (17).
Antibodies.
To produce antibodies against ARB1, the glutathione transferase (GST)-ARB1 fusion protein encoded by pGEX-ARB1 was expressed in Escherichia coli strain BL21 and purified using glutathione-Sepharose 4B beads according to the vendor's instructions (Pharmacia). The purified GST-ARB1 fusion protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (6% gels) and excised in a gel slice. Rabbits were injected with the GST-ARB1 in gel, and the serum containing polyclonal antibodies against ARB1 was obtained commercially from Covance Research Products (Denver, PA). Antibodies against GCD11 (55) and RPL39 (1) used in this study were described previously. Antibodies against the TAP, myc and His6 tags were purchased from Open Biosystems (item CAB1001) and Santa Cruz Biotechnology, Inc. (items SC40 and SC803), respectively.
RESULTS
Depletion of ARB1 leads to a deficit of 18S rRNA and 40S subunits in vivo.
To examine the requirement for ARB1 in ribosome biogenesis, we constructed a yeast strain (YDH226) in which the sole form of ARB1 is unstable due to attachment of ubiquitin coding sequences, followed by an arginine codon, to the N terminus of the ARB1 ORF. (The coding sequences for FLAG and His6 affinity tags [FH] were also added in tandem for purposes of purification and immunodetection.) The ubiquitin is cotranslationally cleaved, leading to rapid proteosomal degradation of the remaining R-FH-ARB1 moiety by the N-end rule pathway (53). Transcription of this allele, PGAL-UBI-R-FH-ARB1, is regulated by the galactose-inducible GAL1 promoter and, hence, repressed in cells growing with glucose as carbon source.
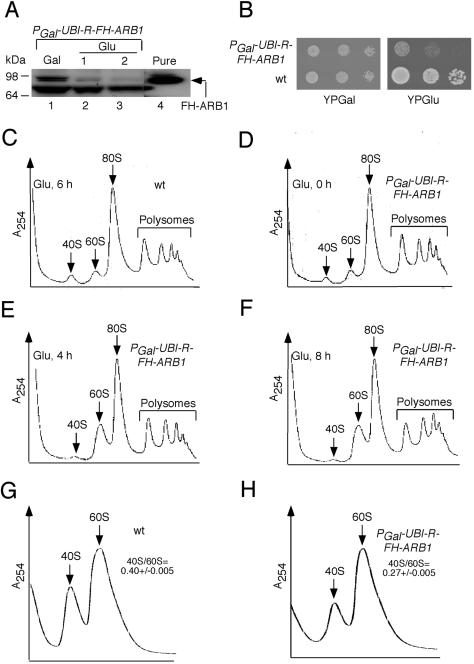
Western analysis of extracts prepared from YDH226 cells using anti-His6 antibodies showed that shifting the culture from galactose to glucose for 2 h greatly reduced R-FH-ARB1 expression (Fig. 1A). (The lower band in the Western blot is an unknown protein that cross-reacts with anti-His6 antibodies, serving as a loading control.) In a separate experiment, we found that the level of R-FH-ARB1 in YDH226 cells grown on galactose was only ∼20% of the level of FH-tagged (but otherwise WT) ARB1 (FH-ARB1) expressed from the native promoter in strain YDH275 (data not shown). Thus, the unstable R-FH-ARB1 protein, even when induced, is probably present at levels below that of native ARB1. Despite the marked depletion of R-FH-ARB1 in glucose medium, YDH226 cells continued to grow for several generations with a doubling time about twofold greater than that of the WT parental strain, BY4741 (data not shown). Moreover, YDH226 can form small colonies from single cells on glucose medium after prolonged incubation (Fig. 1B). Thus, although deletion of ARB1 in BY4741 is lethal (see below), apparently only a very small fraction of the WT level of ARB1 protein can sustain a slow rate of cell growth.
FIG. 1.
Depletion of ARB1 reduces abundance of 40S ribosomal subunits. (A) The PGAL-UBI-R-FH-ARB1 strain YDH226 was grown in SCGal-Leu medium to an A600 of ∼1.2 (lane 1), and a portion of the culture was shifted to SCGlu-Leu medium for 1 h (lane 2) or 2 h (lane 3). Twenty micrograms of WCEs (lanes 1 to 3) and 0.1 μg of pure FH-ARB1 (lane 4) were subjected to Western analysis with antibodies against the His6 epitope. (B) Serial dilutions of YDH226 and WT strain BY4741 were spotted on yeast extract-peptone containing galactose (YPGal) and yeast extract-peptone containing glucose (YPGlu). (C to F) Strains BY4741 (C) and YDH226 (D to F) were grown in SCGal medium to an A600 of ∼1.2 and shifted to SCGlu medium for the times indicated in each panel. Cycloheximide was added at 50 μg/ml 5 min before harvesting, and WCEs were prepared in the presence of 10 mM Mg+2 and resolved by velocity sedimentation through 7 to 47% sucrose gradients. Positions of 40S, 60S, 80S, and polysomes are indicated on the A254 tracings. (G and H) BY4741 (WT) and YDH226 were grown in SCGlu-Leu for 18 h as described for panels C to F except that cycloheximide was omitted and WCEs were prepared in the absence of Mg+2 and resolved by velocity sedimentation through 15 to 30% sucrose gradients. The mean ratios of total 40S/60S subunits determined from replicate experiments are indicated with their standard errors.
Although ARB1-depleted cells are capable of dividing, analysis of polysome profiles revealed an obvious increase in free 60S versus 40S subunits by 4 h or 8 h after shifting strain YDH226 from galactose to glucose medium (Fig. 1E and F). By contrast, the ratio of free 60S:40S subunits during growth of this mutant on galactose medium (Fig. 1D, 0 h on glucose) was indistinguishable from that of WT strain BY4741 cultured on glucose medium (Fig. 1C). These results suggest that depletion of ARB1 leads to a specific deficit in 40S subunits. To confirm this conclusion, we measured the total 40S:60S subunit ratio in strain YDH226 after depleting UBI-R-FH-ARB1 for 18 h on glucose medium and resolving the extract in a Mg+2-free buffer where 80S monosomes and polysomes dissociate into free subunits. In several replicate experiments, the 40S:60S ratio was lower by ∼30% in strain YDH226 compared to that measured in the WT strain under the same conditions (0.27 ± .005 versus 0.40 ± .005, respectively) (Fig. 1G and H).
Depletion of ARB1 impedes the first steps in 35S pre-rRNA processing.
We wished to determine whether the deficit in 40S subunits produced by ARB1 depletion is associated with defects in processing of 35S pre-rRNA at the A0-A1-A2 cleavage sites and a reduction in steady-state levels of 18S rRNA (Fig. 2G). Toward this end, we conducted Northern analysis of the mature rRNAs and their precursors after depleting UBI-R-FH-ARB1 in YDH226 for different lengths of time. Depletion of UBI-R-FH-ARB1 for 18 h led to a marked reduction (∼48%) in the ratio of mature 18S:25S rRNA versus the corresponding ratio in the WT, as visualized by hybridization with probes 2 and 6 (Fig. 2B; refer to panel G for locations of probes). This last finding is in reasonable agreement with the 33% reduction in levels of mature 40S subunits on ARB1 depletion shown in Fig. 1G and H. Hybridization with probes 1, 3, 4, and 5, which allows visualization of various precursor rRNAs, revealed accumulation of 35S pre-rRNA on depletion of UBI-R-FH-ARB1, indicating reduced cleavage at the primary processing sites A0-A1-A2 (Fig. 2A, C, D, and E). Consistent with this conclusion, we observed accumulation of the aberrant 23S species (Fig. 2A, C, and D), thought to be produced by cleavage at site A3 prior to cleavage at A0, A1, and A2 (42) (Fig. 2G).
A delay in cleavage at sites A0-A1-A2 would be expected to reduce the steady-state levels of 20S and 27SA2 pre-rRNAs (Fig. 2G). This was not observed in cells depleted of ARB1, however, using probes 3 and 4 designed to measure the levels of these two precursor rRNAs (Fig. 2C and D). Relative to the level of Pol III-transcribed 5S RNA analyzed as an internal control (Fig. 2F), we calculated that the level of 20S pre-rRNA was essentially unchanged while the abundance of 27SA2 pre-rRNA increased about twofold after depleting ARB1 for 8 h in strain YDH226. Similarly, we observed a ∼1.5-fold increase in the ratio of 7S pre-rRNA to 5S RNA upon ARB1 depletion (Fig. 2F). The 7S species is the immediate precursor of 5.8S rRNA (Fig. 2G). To explain these findings it could be proposed that the rates of processing of 20S to 18S and of 27SA2 to 5.8S and 25S rRNAs are reduced in cells depleted of ARB1, thereby compensating for the decreased production of 20S and 27SA2 pre-rRNAs expected from the delay in A0-A1-A2 cleavage.
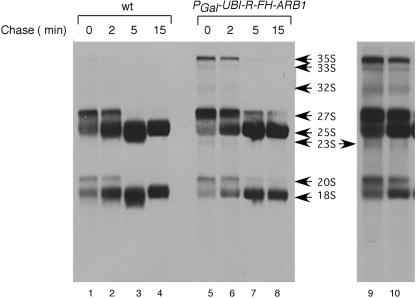
Support for this last interpretation was provided by pulse-chase labeling analysis of pre-rRNA processing. In WT cells, the [methyl-3H]methionine-labeled 35S rRNA precursor was too short-lived to be detected, and the 27S and 20S precursors were rapidly chased to mature 25S and 18S rRNAs (Fig. 3, lanes 1 to 4), all as expected. In contrast, processing of pre-rRNA intermediates was slower in cells depleted of ARB1. We observed marked accumulation of the 35S precursor and a two- to threefold increase in the 20S:18S ratio in the mutant cells at 0 and 2 min of chase, indicating slower cleavage at the A0-A1-A2 sites in 35S pre-rRNA and at site D in 20S pre-rRNA, respectively (Fig. 3, lanes 5 to 8). (With a darker exposure, the presence of the short-lived aberrant 23S RNA can also be seen at 0 min in the mutant cells [Fig. 3, lanes 9 and 10]). Importantly, there was an increased 27S:25S ratio at all four time points in the mutant, confirming that a key processing reaction in the 60S biogenesis pathway occurs more slowly in ARB1-depleted cells (Fig. 3). A 35% reduction in the 18S:25S ratio was evident after 15 min of chase in the mutant versus WT cells (18S:25S ratios of 0.35 versus 0.54, respectively), in accordance with the Northern data in Fig. 2B and the 40S:60S subunit ratios shown in Fig. 1H.
FIG. 3.
Pulse-chase analysis further shows defective pre-rRNA processing in ARB1 depletion cells. Strains YDH209 (WT) and YDH226 (PGAL-UBI-R-FH-ARB1) were grown in SCGal-Met medium to an A600 of 1.2, then shifted to SCGlu-Met medium for 18 h. Cells were labeled with [methyl-3H]methionine for 2 min and subsequently chased with 4 ml of 1 mg/ml unlabeled methionine for the time points indicated. Total RNA was isolated, and samples containing 70,000 dpm were separated on a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and exposed to film. Lanes 9 and 10 depict a longer exposure of lanes 5 and 6.
It is noteworthy that the rate of appearance of mature 18S rRNA between 0 and 5 min of chase was reduced in the ARB1-depleted cells compared to WT (Fig. 3). The reductions in amounts of labeled 18S rRNA present at 0, 2, and 5 min are comparable to those seen after 15 min of chase and also to the steady-state reduction in 18S rRNA measured by Northern analysis in ARB1-depleted cells (Fig. 2B). The nearly identical labeling of 25S rRNA after 15 min of chase in the WT and ARB1-depleted cells insures that equivalent labeling of comparable numbers of cells was achieved for the two strains in this experiment, thus permitting a comparison of their rates of 18S rRNA accumulation. This comparison indicates that the decreased rate of mature 18S rRNA production in ARB1-depleted cells is sufficient to account for the steady-state reduction in 40S subunits.
We presume that the decrease in the steady-state level of 18S rRNA observed in the mutant cells results from degradation of at least a portion of the aberrant 23S RNA generated by premature cleavage at site A3. A relatively greater proportion of the 27S precursors must be converted to mature 25S rRNA, despite slower processing in the 60S pathway, to yield the reduced 18S:25S rRNA ratio seen in the ARB1-depleted cells.
ARB1 shuttles from nucleus to cytoplasm.
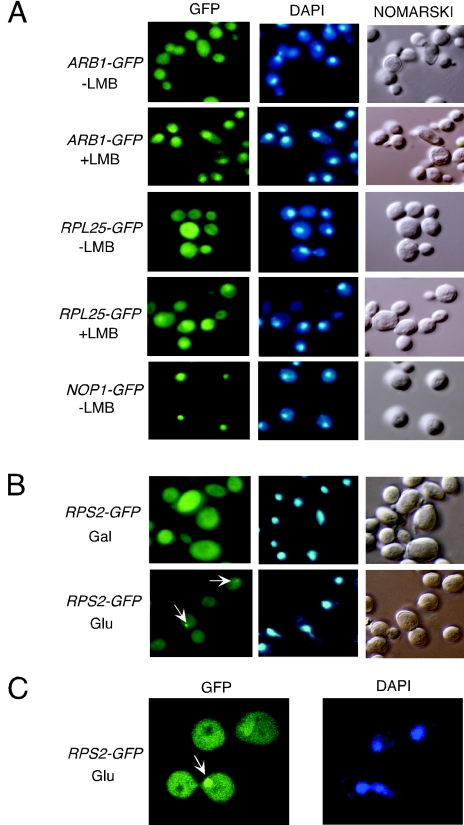
If ARB1 plays a direct role in stimulating the A0-A1-A2 processing steps in the 90S particle and in processing 27S rRNA in the 60S precursor, it should be found in the nucleus (70). Visualization of a functional ARB1-GFP fusion expressed from the normal chromosomal locus in strain YDH338 revealed a nearly uniform distribution of this protein throughout the cells (Fig. 4A, row 1). This strain harbors the crm1-T539C allele, encoding a leptomycin B (LMB)-sensitive form of XPO1, a major β-karyopherin for nuclear export of proteins. Treatment of YDH338 with LMB led to an obvious nuclear accumulation of ARB1-GFP (Fig. 4A, row 2). As expected from previous findings (16, 31), a GFP fusion to the 60S subunit protein RPL25 expressed in the same strain exhibited a similar response to LMB (Fig. 4A, rows 3 and 4), and a NOP1-GFP fusion showed the expected nuclear localization independent of LMB treatment (Fig. 4A, row 5) (70). These results indicate that ARB1 shuttles between nucleus and cytoplasm and becomes trapped in the nucleus when XPO1 function is inhibited by LMB.
FIG. 4.
Evidence that ARB1-GFP shuttles from nucleus to cytoplasm and is required for nuclear export of pre-40S ribosomes. (A) The LMB-sensitive strain crm1-T539C containing chromosomal ARB1-GFP (YDH338) (rows 1 and 2) was grown in YPD to an A600 of ∼0.5. The crm1-T539C strain carrying plasmid-borne RPL25-GFP (YDH332) (rows 3 and 4) and a CRM1 strain carrying NOP1-GFP on a low-copy-number plasmid (YDH315) (row 5) were grown in SCGlu-Ura to an A600 of 1.0 and then diluted in YPD at an A600 of 0.1 and grown to an A600 of ∼0.5. The cells were incubated for 15 min in the presence or absence of LMB, fixed, and incubated with DAPI (4′,6′-diamidino-2-phenylindole) to stain the nuclear DNA. The GFP fusion proteins were visualized by fluorescence microscopy of living cells. Nomarski, phase-contrast imaging of cells. (B) The PGAL-UBI-R-FH-ARB1 strain YDH226 carrying plasmid-borne RPS2-GFP was grown in SCGal-Ura medium to an A600 of ∼1.2, and half of the culture was shifted to SCGlu-Leu medium for ∼18 h. Fluorescence microscopy was conducted as described for panel A. Arrows indicate the fluorescent foci in the nuclei. (C) Enlarged view by confocal laser microscopy of the cells cultured in SCGlu-Leu medium and described in panel B.
Our findings that ARB1 shuttles from nucleus to cytoplasm and that conversion of 20S pre-rRNA to mature 18S rRNA occurred at a lower rate in ARB1-depleted cells could both be explained by proposing that ARB1 stimulates 20S to 18S processing in pre-40S particles, as this reaction occurs in the cytoplasm. Alternatively, ARB1 may participate in nuclear export of pre-40S ribosomes and accompany the latter to the cytoplasm. Consistent with this second possibility, we found that depletion of ARB1 led to nuclear accumulation of a GFP fusion to 40S subunit protein RPS2. In ∼14% of the cells, we observed a bright fluorescent spot restricted to one sector of the nuclear DNA (Fig. 4B and C). This structure was found in another ∼20% of ARB1-depleted cells when we employed a confocal laser microscope to visualize multiple sections through the cell images (see Fig. S1 in the supplemental material). Thus, we estimate that approximately one-third of cells depleted of ARB1 exhibit a concentrated accumulation of pre-40S particles at a discrete location in the nucleus. This structure was never observed for the RPL25-GFP fusion under exactly the same conditions (data not shown), suggesting that export of 60S subunits is relatively unaffected in cells depleted of ARB1. Thus, it is possible that ARB1 is required for an optimal rate of nuclear export of pre-40S particles.
While our results are consistent with a role for ARB1 in nuclear export of 40S subunits, we cannot exclude the possibility that ARB1 depletion leads to accumulation of RPS2-GFP in an aberrant pre-40S particle that is not competent for nuclear export. Moreover, whether ARB1 directly stimulates 20S to 18S conversion in the cytoplasm or promotes this reaction merely by enhancing nuclear export of pre-40S particles cannot be determined from our results.
ARB1 is physically associated with ribosomal particles and proteins involved in ribosome biogenesis in vivo.
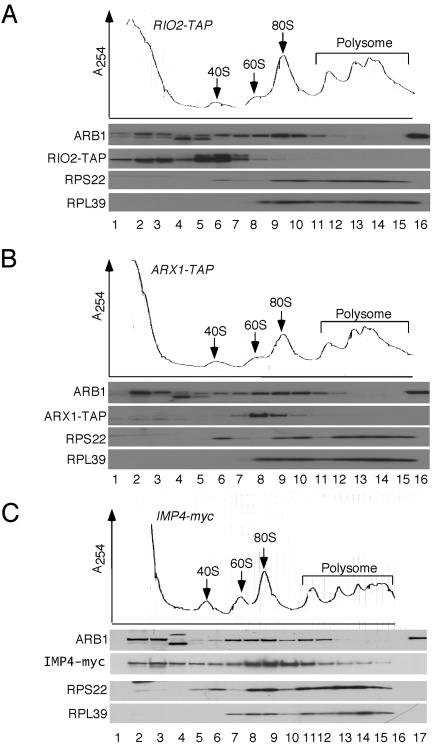
To provide further evidence that ARB1 functions directly in ribosome biogenesis, we asked whether ARB1 physically interacts with pre-ribosomal particles in vivo. In the first approach, we determined the sedimentation profile in sucrose gradients of native ARB1 in cell extracts of strains encoding myc13 or TAP-tagged versions of IMP4, RIO2, and ARX1 proteins reported to associate with 90S, 40S, or 60S preribosomes, respectively (52, 62, 74). We observed that proportions of ARB1 were present in the fractions containing 40S, 60S, and 80S ribosomes, with peak abundance in the 80S to 90S region of the gradient (Fig. 5A to C). The latter corresponds to the peak fractions occupied by the known 90S component IMP4 (Fig. 5C). There was also a pool of ARB1 that sedimented with lower s values visible in the upper fractions of the gradient. By contrast, the majority of RIO2-TAP cosedimented with 40S subunits and virtually all of the ARX1-TAP cosedimented with 60S subunits (Fig. 5A and B), in accordance with previous findings (52, 62). Thus, it appears that ARB1 is more broadly distributed among ribosomal species than either RIO2 or ARX1 and shows a sedimentation profile quite similar to that observed for IMP4. This last conclusion is consistent with the fact that IMP4 was found to interact with ARB1 in the yeast two-hybrid assay (29).
FIG. 5.
ARB1 cosediments with 40S, 60S, and 80S or 90S ribosomal species. Strains F1205 (ARX1-TAP) (B), F1207 (RIO2-TAP) (A), and YDH1014 (IMP4-myc13) (C) were grown at 30°C to an A600 of ∼1.2 in YPD medium and treated with cycloheximide (50 μg/ml) for 5 min before harvesting. WCEs were prepared in buffer containing 1.5 mM Mg+2 and resolved by velocity sedimentation through 7 to 47% sucrose gradients in buffer containing 1.5 mM Mg+2. The gradients were collected with continuous scanning at 254 nm, and fractions were subjected to Western analysis using antibodies against protein A for the TAP-tagged proteins (A and B), myc13 for the myc13 tagged IMP4 protein (C), or the other proteins indicated on the left. Lanes 1 to 15, gradient fractions from top to bottom; lane 16 in panels A and B and lane 17 in panel C contain 100 ng of purified FH-ARB1 protein. Lane 16 in panel C is blank.
The fact that ARB1 is very abundant in the fractions containing 80S monosomes but present at low levels in the large polysome fractions (Fig. 5A to C) likely indicates that ARB1 binds to 90S preribosomes in the nucleus rather than functional 80S ribosomes in the cytoplasm, although we cannot rule out the possibility that ARB1 binds to cytoplasmic 80S couples that are not engaged in translation. While we cannot discern a distinct peak of ARB1 in the 60S fractions, it is noteworthy that ARB1 does not exhibit the low abundance in the 60S region displayed by the 40S ribosomal subunit protein RPS22 (Fig. 5B). Moreover, ARB1 abundance does not sharply decline in the 40S region in the manner observed for the 60S subunit protein RPL39 (Fig. 5B). Thus, it is likely that ARB1 cosediments specifically with 40S, 60S, and 90S preribosomal particles.
We asked next whether ARB1 is associated in vivo with proteins implicated previously in preribosomal processing. Toward this end, we conducted a stringent TAP of functional TAP-tagged ARB1 expressed under the control of the chromosomal ARB1 promoter. A mock purification using an untagged strain was conducted in parallel to control for nonspecific interactions. The proteins copurifying with TAP-ARB1 were identified by multidimensional microcapillary liquid chromatography coupled with electrospray ionization-tandem mass spectrometry (43). We identified 43 proteins with a PAF of 3 or greater for the tagged sample and a PAF of 0 for the untagged control sample (see Table S2 in the supplemental material) (3, 5, 9, 13, 18, 23, 27, 28, 36, 41, 54, 56, 60, 64, 75, 77, 78). The PAF expresses the number of nonredundant peptides identified by mass spectrometry normalized for the molecular weight of the cognate protein. Ten of the 43 proteins that satisfy these criteria are components of mature 60S or 40S ribosomal subunits, consistent with our conclusion that ARB1 directly interacts with ribosomal species. Importantly, 15 proteins are nonribosomal factors shown previously to participate in ribosome biogenesis or to be physically associated with a known ribosome biogenesis factor or preribosomal particle. Five of the remaining proteins are known or predicted translation factors, and the remaining 13 proteins have functions unrelated to ribosomes or have unknown functions.
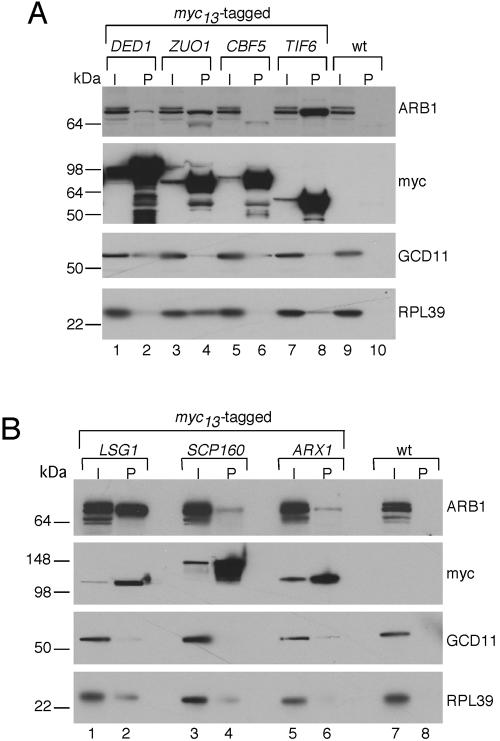
We wished to verify the association of ARB1 with ribosomal processing factors that copurified with TAP-ARB1. Toward this end, we constructed myc13-tagged chromosomal alleles in WT strain BY4741 for 10 of the genes encoding putative ARB1-interacting factors with known or suspected functions in ribosome biogenesis: DED1, ZUO1, TIF6, CBF5, LSG1, ARX1, SOF1, NOP58, UTP7, and SCP160. WCEs from these strains were immunoprecipitated with monoclonal antibodies against the myc epitope, and immune complexes were probed with polyclonal ARB1 antibodies. As shown in Fig. 6A and B, a substantial amount of ARB1 specifically coimmunoprecipitated with myc-tagged TIF6, ZUO1, and LSG1.
FIG. 6.
ARB1 is physically associated with 40S and 60S processing factors. WT BY4741 or its derivatives containing chromosomal DED1-myc13, ZUO1-myc13, CBF5-myc13, or TIF6-myc13 (A) or LSG1-myc13, SCP160-myc13, or ARX1-myc13 (B) were grown to an A600 of ∼1.5 in YPD medium. WCEs were immunoprecipitated with anti-myc antibodies, and the immune complexes were subjected to Western analysis using antibodies against ARB1, myc epitope, GCD11, or 60S subunit protein RPL39, as indicated on the right. I, 1/100 of the input WCE extract; P, pellet fraction. The molecular sizes in kilodaltons are indicated on the left.
TIF6 has been shown to shuttle between nucleus and cytoplasm (65) and is associated with 60S ribosomal species (65, 66) and the pre-60S processing factor NOG1 (61). Depletion of TIF6 impairs conversion of 27SB to 25S rRNA and leads to a strong reduction in 60S subunits; however, it also affects the A0-A1-A2 cleavage of 35S pre-RNA (2). LSG1 is a 60S-associated GTPase required for WT accumulation of 60S subunits (34). It is restricted to the cytoplasm and is needed to recycle the 60S nuclear export factor NMD3 following export to the cytoplasm (30). Thus, ARB1 is tightly associated with two factors implicated previously in various aspects of 40S or 60S biogenesis. (There is no definitive evidence in the literature linking ZUO1 to pre-ribosome processing.) Note that we cannot distinguish between a direct interaction between ARB1 and these proteins and the possibility that ARB1 merely interacts with preribosomal particles containing TIF6 or LSG1.
Although much smaller amounts of ARB1 coimmunoprecipitated with tagged ARX1, DED1, and SCP160, these results may still be significant because no detectable ARB1 was immunoprecipitated from the untagged extract or from extracts containing myc-tagged CBF5, SOF1, NOP58, or UTP7 (Fig. 6A and B and data not shown). In addition, ARB1 was found to interact with ARX1 in a previous genome-wide analysis of protein-protein interactions in yeast (19). ARX1 is associated with pre-60S particles (52), DED1 is associated with both pre-40S and pre-60S particles (62, 76), and SCP160 is associated with DED1 and other pre-40S processing factors (62). These considerations lend additional support to the idea that ARB1 stimulates processing reactions in both the 40S and 60S biogenesis pathways.
Finally, we asked whether pre-rRNAs found in pre-60S or pre-40S subunits could be coimmunoprecipitated with TAP-tagged ARB1 by conducting Northern analysis of RNA extracted from the immune complexes using probes specific for 27S, 7S, and 20S pre-rRNAs. Strains harboring TAP-tagged ENP1 or TAP-NSA3 were analyzed in parallel as controls of proteins known to be associated with 20S (7, 24) and 27S pre-rRNAs (26, 52), respectively. Under conditions where the known interactions of ENP1 and NSA3 with pre-rRNAs were clearly evident, we did not observe significant coimmunoprecipitation of 27S, 7S, or 20S pre-rRNAs with TAP-ARB1 (see Fig. S2 in the supplemental material). However, we note that coimmunoprecipitation of pre-rRNAs has not been reported for any of the three proteins that coimmunoprecipitated at high levels with myc-tagged ARB1, namely TIF6, LSG1, and ZUO1 (Fig. 6). Indeed, this would not be expected for LSG1 considering its cytoplasmic function in recycling NMD3 after nuclear export of nascent 60S subunits (30). We speculate that the association of TAP-ARB1 with immature ribosomal particles containing these pre-rRNAs is too labile to survive the immunoprecipitation regimen, whereas the interactions of TAP-ARB1 with TIF6 and LSG1 can be maintained free of ribosomes.
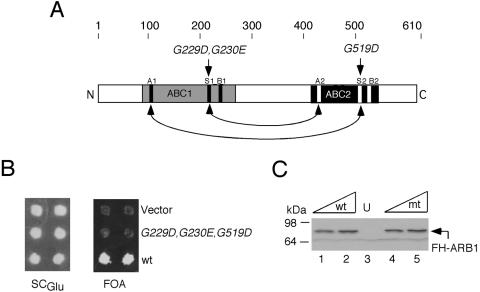
Conserved residues in the ABC cassettes of ARB1 are required for its essential function in vivo.
To address whether the predicted ATPase activity of ARB1 is required for its essential function in vivo, we mutagenized three of the conserved Gly residues in the LSGGQ signature motifs of its two ABCs (G229D, G230E, and G519D) (Fig. 7A). Mutations in the corresponding residues of certain ABC transporter proteins were shown to inactivate transporter function in vivo and ATPase activity in vitro (38, 63). The FH-ARB1 allele containing these mutations was unable to support growth of an arb1Δ strain following eviction of WT plasmid-borne FH-ARB1 (Fig. 7B). Western analysis of viable strains harboring the mutant or WT FH-ARB1 alleles together with untagged ARB1 showed that the mutations did not affect the steady-state level of ARB1 (Fig. 7C). Hence, conserved residues involved in ATP binding and/or hydrolysis in other ABC transporters are critical for the essential function of ARB1 in vivo.
FIG. 7.
Signature sequences in the ABCs of ARB1 are essential in vivo. (A) The primary structure of ARB1 is depicted schematically with amino acid positions and the locations of mutations made in conserved residues (in italics) shown at the top, regions of sequence similarity to ATP-binding domains in other ABC proteins are shaded, and locations of the Walker A and B motifs and signature sequences (S) are indicated with black (ABC1) or white (ABC2) rectangles. The predicted interactions that would stabilize formation of a dimer between ABC1 and ABC2, sandwiching two molecules of ATP, are indicted with arrows. (B) Patches of an arb1Δ strain containing ARB1 URA3 plasmid pDH22 and LEU2 plasmids YCplac111 (empty vector; row 1), pDH144 (containing FH-ARB1-G229D-G230E-G519D; row 2), or pDH129 (containing FH-ARB1; row 3), were replica plated to SCGlu-Leu-Ura (left panel) and medium containing 5-FOA (right panel). (C) WCEs of the strains described in panel B were grown in SCGlu-Leu-Ura to an A600 of ∼1.2, and 10 or 20 μg of WCE from cells containing pDH129 (lanes 1 and 2), pDH144 (lanes 4 and 5), or empty vector (lane 3) in addition to pDH22 were subjected to Western analysis with anti-His6 antibodies. mt, mutant.
DISCUSSION
In this report we provide evidence that the essential protein YER036C/ARB1 in the GCN20 subfamily of ABC proteins is involved in the biogenesis of both 40S and 60S ribosomal subunits. Depletion of ARB1 in vivo led to a 30% to 40% reduction in the steady-state level of mature 18S rRNA and 40S subunits relative to 25S rRNA and 60S subunits. This deficit in 40S subunits can be attributed at least partly to a reduced rate of cleavage at the A0-A1-A2 sites in 35S pre-rRNA that produces the 20S precursor of 18S rRNA. Thus, depletion of ARB1 led to steady-state accumulation of 35S pre-rRNA and the aberrant 23S transcript that is generated by inappropriate cleavage at site A3 prior to A0-A1-A2 cleavage. We confirmed by pulse-chase analysis that the processing of 35S pre-RNA at the A0-A1-A2 sites is delayed in ARB1-depleted cells, and we also detected a diminished rate of 20S to 18S conversion in the same cells. We presume that both of these processing defects contribute to the overall ∼40% reduction in the synthesis of mature 18S seen in ARB1-depleted cells, but it is difficult to assess their relative contributions to the defect in 40S biogenesis.
Apart from these defects in the rRNA processing reactions that produce 18S rRNA, we observed accumulation of the RPS2-GFP fusion protein in nuclear foci (one per cell) in roughly one-third of the ARB1-depleted cells, suggesting a partial defect in nuclear export of pre-40S particles. It is unclear whether ARB1 functions directly to stimulate 40S export or whether the apparent delay in export is an indirect consequence of the accumulation of aberrant pre-40S complexes in ARB1-depleted cells that are incompetent for nuclear export. As 20S to 18S rRNA processing clearly continues, albeit at reduced rates, in ARB1-depleted cells (Fig. 3), the proposed defect in nuclear export of pre-40S particles would amount to a delay rather than a tight block of pre-40S export from the nucleus. However, such a delay in nuclear export could be responsible for the reduced rate of 20S to 18S rRNA processing detected in the pulse-chase analysis shown in Fig. 3. ARB1 could also enhance 20S to 18S conversion more directly by associating with pre-40S particles in the cytoplasm, as ARB1 itself is exported to the cytoplasm (Fig. 4). In any event, it seems clear that ARB1 stimulates multiple steps of the 40S biogenesis pathway.
The A0-A1-A2 processing events occur in the 90S pre-ribosomal particle. The fact that the majority of ARB1 in cell extracts cosediments through sucrose gradients with IMP4 at a position corresponding to ∼90S particles (Fig. 5C) is consistent with the possibility that ARB1 directly stimulates these cleavage reactions in the 90S preribosome. We also obtained evidence consistent with the possibility that ARB1 functions directly in pre-40S particles to stimulate nuclear export or to enhance 20S to 18S processing in the cytoplasm. Thus, we showed that ARB1 shuttles from nucleus to cytoplasm, and we observed that a fraction of ARB1 in cell extracts cosediments with 40S subunits. In addition, we found that ARB1 is weakly associated with DED1 and SCP160, both of which copurify with processing factors present in pre-40S particles (62) and with 20S pre-rRNA (for DED1) (76). If ARB1 is associated with 90S pre-ribosomes, as suggested above, it could remain bound to the pre-40S particle after cleavage of 35S pre-rRNA at the A0-A1-A2 sites and subsequently accompany the pre-40S to the cytoplasm. Alternatively, ARB1 could interact transiently with 90S and 40S preribosomal particles at various points along the processing pathway. The latter possibility is more consistent with the fact that ARB1 was not identified previously as a stable constituent of any preribosomal particles.
It is intriguing that depletion of ARB1 also produced a delay in processing of 27S pre-rRNA to 25S rRNA, leading to steady-state accumulation of 27S precursors. Accumulation of the 7S precursor to 5.8S rRNA was also evident in cells lacking ARB1. However, we did not observe a defect in nuclear export of GFP-RPL25 nor reduced steady-state levels of either 25S rRNA or 60S subunits in ARB1-depleted cells. Thus, while ARB1 is required for a WT rate of 60S biogenesis, it seems that all of the 27S pre-rRNA is eventually converted to 25S rRNA so that a normal steady-state level of 60S subunits is achieved in ARB1-depleted cells.
Several observations support the possibility that ARB1 also functions directly in pre-60S particles to stimulate processing of 27S and 7S pre-rRNAs. First, we found that a fraction of ARB1 cosediments with 60S ribosomal particles. Second, ARB1 is tightly associated with TIF6 and LSG1 and weakly associated with ARX1, all of which interact with pre-60S subunits and stimulate different aspects of 60S biogenesis. It is interesting that LSG1 appears to interact with pre-60S subunits only in the cytoplasm (30), whereas ARX1 resides in nuclear pre-60S particles at a stage just prior to nuclear export (52). Although TIF6 shuttles to the cytoplasm (65), its function is also required further upstream in the 60S biogenesis pathway for efficient conversion of 27SB to 25S rRNA. Based on its physical association with these three factors, it is possible that ARB1 interacts with pre-60S subunits in both the nucleus and cytoplasm and, similar to TIF6 (65), may accompany pre-60S subunits during export from the nucleus.
Most ribosome biogenesis factors present in the 90S preribosome are not stable constituents of 60S or 40S preribosomal particles, and many factors join the pre-40S or pre-60S particles after cleavage of 35S pre-rRNA in the 90S particle. Thus, the majority of biogenesis factors seem to function exclusively in pre-40S or pre-60S particles (24). ARB1 belongs to a small group of factors that stimulate steps in both the pre-40S and pre-60S pathways, as well as the initial cleavage of 35S rRNA in the 90S particle. Interestingly, DED1, which interacts with ARB1, is associated with both pre-40S and pre-60S particles. It is also intriguing that RLI1, another soluble ABC protein, is a shuttling factor that interacts with pre-40S and pre-60S particles, enhances pre-rRNA processing reactions in both pathways, and stimulates export of both pre-40S and pre-60S particles (37, 76).
A number of GTPases and AAA-type ATPases have been implicated in 60S processing, but only the GTPase BMS1 (20) and the ABC ATPase RLI1 (37, 76) have been identified in the 40S processing pathway (20, 70). Our genetic analysis indicates that the ATPase activity of ARB1 is required for its essential function in ribosome biogenesis. By analogy with other ABC proteins, and RLI1 in particular (35), it can be predicted that binding of ATP will tightly clamp together the ABCs in ARB1, whereas ATP hydrolysis will lead to partial dissociation of the two cassettes. The opening and closing of this clamp could produce conformational changes in segments of the preribosomes or in ribosome-associated factors that stimulate rRNA processing or subunit assembly reactions. Given that ribosomal subunit export is energy dependent (33), ATP hydrolysis by ARB1 might also perform work connected with the nuclear export of 40S preribosomes.
All of the previously characterized soluble ABC proteins in the GCN20 subfamily were shown to interact with ribosomes and to function in translation initiation (RLI1 and ABC50), translation elongation (YEF3), translational control (GCN20), or ribosome biogenesis (RLI1). Our findings allow us to extend this generalization to ARB1. Moreover, preliminary results indicate that the last uncharacterized member of this subfamily in yeast, NEW1, is also involved in ribosome biogenesis (J. Dong and A. G. Hinnebusch, unpublished observations). It will be interesting to identify the molecular features of the members of this subfamily of ABC proteins that dictate their interactions with ribosomes and identify the nature of the mechanical work they carry out to promote ribosome biogenesis or protein synthesis.
Supplementary Material
Acknowledgments
We thank Leos Valasek, Sander Granneman, Susan Baserga, and Sungpil Yoon for suggestions and advice; Boots Quimby for technical assistance with confocal microscopy; Maurice Swanson for RPL39 antibodies; Jan van't Riet for RPS22 antibodies; Thorsten Schafer and Herbert Tschochner for the RPL25-GFP and RPS2-GFP plasmids; Yoshiko Kikuchi for the NOP1-GFP plasmid; and Kate Abruzzi and Michael Rosbash for the crm1-T539C strain.
J.L.J. is supported by NIH grants GM64779 and HL68744. A.J.L. is supported by NIH grants GM64779, HL68744, ES11993, and CA098131.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, J. T., M. R. Paddy, and M. S. Swanson. 1993. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:6102-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, C. R., J. A. McCann, and H. L. Chiang. 2000. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J. Cell Biol. 150:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesareni, G., and J. A. H. Murray. 1987. Plasmid vectors carrying the replication origin of filamentous single-stranded phages, p. 135-154. In J. K. Setlow and A. Hollaender (ed.), Genetic engineering: principals and methods, vol. 9. Plenum Press, New York, N.Y. [Google Scholar]
- 5.Chakraburtty, K. 1992. Elongation factor 3: a unique fungal protein, p. 114-142. In P. B. Fernandes (ed.), New approaches for antifungal drugs. Birkhauser, Basel, Switzerland.
- 6.Chen, J., G. Lu, J. Lin, A. L. Davidson, and F. A. Quiocho. 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell 12:651-661. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., J. Bucaria, D. A. Band, A. Sutton, and R. Sternglanz. 2003. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31:690-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, A. L. 2002. Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J. Bacteriol. 184:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Cruz, J., I. Iost, D. Kressler, and P. Linder. 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dez, C., C. Froment, J. Noaillac-Depeyre, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2004. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 24:6324-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, J., R. Lai, K. Nielsen, C. A. Fekete, H. Qiu, and A. G. Hinnebusch. 2004. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 279:42157-42168. [DOI] [PubMed] [Google Scholar]
- 12.Dong, J., H. Qiu, M. Garcia-Barrio, J. Anderson, and A. G. Hinnebusch. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6:269-279. [DOI] [PubMed] [Google Scholar]
- 13.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eng, J. K., A. L. McCormack, and J. R. Yates Spaceiiiqq. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 15.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 16.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Barrio, M., J. Dong, S. Ufano, and A. G. Hinnebusch. 2000. Association of GCN1/GCN20 regulatory complex with the conserved N-terminal domain of eIF2α kinase GCN2 is required for GCN2 activation in vivo. EMBO J. 19:1887-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier, T., T. Berges, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 20.Gelperin, D., L. Horton, J. Beckman, J. Hensold, and S. K. Lemmon. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7:1268-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 22.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 23.Graack, H. R., and B. Wittmann-Liebold. 1998. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J. 329:433-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 25.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 26.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 27.Haselbeck, R. J., and L. McAlister-Henn. 1993. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 268:12116-12122. [PubMed] [Google Scholar]
- 28.Hayano, T., M. Yanagida, Y. Yamauchi, T. Shinkawa, T. Isobe, and N. Takahashi. 2003. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 278:34309-34319. [DOI] [PubMed] [Google Scholar]
- 29.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 30.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27:580-585. [DOI] [PubMed] [Google Scholar]
- 34.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karcher, A., K. Buttner, B. Martens, R. P. Jansen, and K. P. Hopfner. 2005. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure 13:649-659. [DOI] [PubMed] [Google Scholar]
- 36.Kettner, C., G. Obermeyer, and A. Bertl. 2003. Inhibition of the yeast V-type ATPase by cytosolic ADP. FEBS Lett. 535:119-124. [DOI] [PubMed] [Google Scholar]
- 37.Kispal, G., K. Sipos, H. Lange, Z. Fekete, T. Bedekovics, T. Janaky, J. Bassler, D. J. Aguilar Netz, J. Balk, C. Rotte, and R. Lill. 2005. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24:589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koronakis, E., C. Hughes, I. Milisav, and V. Koronakis. 1995. Protein exporter function and in vitro ATPase activity are correlated in ABC-domain mutants of HlyB. Mol. Microbiol. 16:87-96. [DOI] [PubMed] [Google Scholar]
- 39.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:7283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 42.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 44.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 45.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 46.Marton, M. J., D. Crouch, and A. G. Hinnebusch. 1993. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 13:3541-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marton, M. J., C. R. Vasquez de Aldana, H. Qiu, K. Charkraburtty, and A. G. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of the eIF2α kinase GCN2. Mol. Cell. Biol. 17:4474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKeegan, K. S., M. I. Borges-Walmsley, and A. R. Walmsley. 2003. The structure and function of drug pumps: an update. Trends Microbiol. 11:21-29. [DOI] [PubMed] [Google Scholar]
- 49.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 50.Moody, J. E., L. Millen, D. Binns, J. F. Hunt, and P. J. Thomas. 2002. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277:21111-21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18: 3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park, E.-C., D. Finley, and J. W. Szostak. 1992. A strategy for the generation of conditional mutations by protein destabilization. Proc. Natl. Acad. Sci. USA 89:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul, M. F., S. Ackerman, J. Yue, G. Arselin, J. Velours, A. Tzagolof, and S. Ackermann. 1994. Cloning of the yeast ATP3 gene coding for the gamma-subunit of F1 and characterization of atp3 mutants. J. Biol. Chem. 269:26158-26164. [PubMed] [Google Scholar]
- 55.Phan, L., X. Zhang, K. Asano, J. Anderson, H. P. Vornlocher, J. R. Greenberg, J. Qin, and A. G. Hinnebusch. 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 18:4935-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Planta, R. J., and W. H. Mager. 1998. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14:471-477. [DOI] [PubMed] [Google Scholar]
- 57.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki, T., E. A. Toh, and Y. Kikuchi. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20:7971-7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato, H., and I. Miyakawa. 2004. A 22 kDa protein specific for yeast mitochondrial nucleoids is an unidentified putative ribosomal protein encoded in open reading frame YGL068W. Protoplasma 223:175-182. [DOI] [PubMed] [Google Scholar]
- 61.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schafer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmees, G., A. Stein, S. Hunke, H. Landmesser, and E. Schneider. 1999. Functional consequences of mutations in the conserved “signature sequence” of the ATP-binding-cassette protein MalK. Eur. J. Biochem. 266:420-430. [DOI] [PubMed] [Google Scholar]
- 64.Scott, B. L., J. S. Van Komen, H. Irshad, S. Liu, K. A. Wilson, and J. A. McNew. 2004. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J. Cell Biol. 167:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 66.Si, K., and U. Maitra. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith, P. C., N. Karpowich, L. Millen, J. E. Moody, J. Rosen, P. J. Thomas, and J. F. Hunt. 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol Cell 11: 3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Triana-Alonso, F. J., K. Chakraburtty, and K. H. Nierhaus. 1995. The elongation factor unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270:20473-20478. [DOI] [PubMed] [Google Scholar]
- 70.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 71.Tyzack, J. K., X. Wang, G. J. Belsham, and C. G. Proud. 2000. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275:34131-34139. [DOI] [PubMed] [Google Scholar]
- 72.Valásek, L., L. Phan, L. W. Schoenfeld, V. Valásková, and A. G. Hinnebusch. 2001. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 20:891-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vazquez de Aldana, C. R., M. J. Marton, and A. G. Hinnebusch. 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 14:3184-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wehner, K. A., and S. J. Baserga. 2002. The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]
- 75.Wenz, P., S. Schwank, U. Hoja, and H. J. Schuller. 2001. A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product. Nucleic Acids Res. 29:4625-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yarunin, A., V. G. Panse, E. Petfalski, C. Dez, D. Tollervey, and E. Hurt. 2005. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24:580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zelenaya-Troitskaya, O., P. S. Perlman, and R. A. Butow. 1995. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 14:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zoll, W. L., L. E. Horton, A. A. Komar, J. O. Hensold, and W. C. Merrick. 2002. Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 277:37079-37087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.