Abstract
The calcium-regulated protein phosphatase calcineurin (PP2B) functions as a regulator of gene expression in diverse tissues through the dephosphorylation and activation of a family of transcription factors known as nuclear factor of activated T cells (NFAT). Here we show that NFATc3, in addition to being calcium responsive, is regulated through an indirect recruitment of class II histone deacetylases (HDACs). Specifically, yeast two-hybrid screening with the rel homology domain of NFATc3 identified the chaperone mammalian relative of DnaJ (Mrj) as a specific interacting factor. Mrj and NFATc3 were shown to directly associate with one another in mammalian cells and in vitro. Mrj served as a potent inhibitor of NFAT transcriptional activity within the nucleus through a mechanism involving histone deacetylase recruitment in conjunction with heat shock stimulation. Indeed, Mrj was determined to interact with class II histone deacetylases, each of which translocated to the nucleus following heat shock stimulation. Mrj also decreased NFATc3 occupancy of the tumor necrosis factor-α promoter in cardiomyocytes in an HDAC-dependent manner, and Mrj blocked calcineurin-induced cardiomyocyte hypertrophic growth. Conversely, small-interfering-RNA-mediated reduction of Mrj augmented NFAT transcriptional activity and spontaneously induced cardiac myocyte growth. Collectively, our results define a novel response pathway whereby NFATc3 is negatively regulated by class II histone deacetylases through the DnaJ (heat shock protein-40) superfamily member Mrj.
Regulation of gene expression through inducible transcriptional effector pathways underlie such diverse processes as cellular differentiation, proliferation, senescence, and apoptosis, as well as more acute adaptive responses that modify cellular function. One such inducible response pathway involves the calcium-calmodulin-activated protein phosphatase calcineurin (PP2B) and its downstream transcriptional effector nuclear factor of activated T cells (NFAT). Receptor stimulation that produces a sustained elevation in intracellular calcium concentration, or “store-operated” calcium entry, activates calcineurin. Activated calcineurin directly dephosphorylates NFAT transcription factors within the cytoplasm, permitting their nuclear translocation and participation in transcriptional regulatory complexes (16). Five members of the NFAT family, including NFATc1, NFATc2, NFATc3, NFATc4, and NFAT5, have been identified in mammals, although only NFATc1 through NFATc4 are directly regulated by calcineurin (16). NFAT transcription factors bind DNA as monomers or dimers, although they also typically require interaction with other transcription factors, such as AP-1 or the GATA family of zinc finger-containing proteins (16).
Calcineurin-mediated nuclear import of NFATc1-c4 up-regulates expression of immune response genes in T lymphocytes, including interleukin 2 (IL-2), IL-3, IL-4, IL-5, granulocyte-macrophage colony-stimulating factor, tumor necrosis factor-α (TNF-α), and Fas ligand (5, 16). In other cell types, NFATc1 through NFATc4 have been implicated in regulating developmental patterning and vascular differentiation (13), cardiac hypertrophy (31, 48), skeletal muscle hypertrophy and “fast-slow” fiber transformation (16), chondrogenesis (36, 46), cardiac valve formation (7, 37), osteoclast differentiation (40), embryonic axonal outgrowth (14), and T-cell differentiation (16). The ability of NFAT factors to regulate the differentiation of T lymphocytes, as well as other diverse tissues, has been hypothesized to require chromatin remodeling of downstream target genes (16). Consistent with this hypothesis, NFATc2 has been shown to associate with the histone acetyltransferases (HATs) p300 and cyclic AMP response element binding protein (CREB) binding protein (CBP) in T lymphocytes (12). However, little additional insight into how NFAT factors might regulate cellular differentiation through chromatin remodeling has emerged.
The acetylation of histones represents a fundamental control mechanism whereby specific genetic loci are characterized by a “loose” configuration between DNA and histones or are condensed and relatively inaccessible to transacting factors. In general, the activities of HATs are associated with augmented transcriptional potency, while histone deacetylases (HDACs) render histones hypoacetylated and can repress gene transcription in association with chromatin formation (33). The HDACs are comprised of 11 individual genes segregated as either class I (HDAC1, HDAC2, HDAC3, and HDAC8) or class II (HDAC4, HDAC5, HDAC7, HDAC9, and HDAC10) (8, 51). Class II HDACs are unique, given their ability to shuttle between the nucleus and cytoplasm in a regulated manner and given their ability to directly interact with transcription factors such as myocyte enhancer factor-2 (MEF-2) (27). Indeed, the regulated nuclear import of class II HDACs was shown to profoundly influence skeletal muscle differentiated gene expression through MEF-2 (23, 24). While the interaction between MEF-2 and class II HDACs has been extensively characterized, relatively few additional HDAC-dependent interacting factors have been identified.
Here we performed a yeast two-hybrid screen with NFATc3 as bait, which identified the chaperone protein mammalian relative of DnaJ (Mrj). Mrj is a 242-amino-acid protein within the large family of heat shock protein-40 (HSP40)/DnaJ proteins, although its function is not known (34). Here we show that Mrj serves as a potent inhibitor of NFAT transcriptional activity in the nucleus by directly recruiting class II HDACs. These results implicate class II HDACs as important modulators of NFAT transcriptional responses and provide additional insight into the mechanisms whereby HDACs might have such diverse regulatory effects in vivo. Moreover, these results support the recently proposed model whereby chaperones function as transcriptional disassembly factors as a means of regulating gene expression (11).
MATERIALS AND METHODS
Yeast two-hybrid screen.
The rel homology domain (RHD) (amino acids 405 to 700) of human NFATc3 was PCR amplified and subcloned into a Gal4-DNA binding domain (amino acids 1 to 147)-containing pGBKT7 vector (Clontech). The bait plasmid pGBKT7-NFATc3 was transformed into yeast AH109, along with an adult mouse heart cDNA library fused to the Gal4 activation domain. About 106 cotransformants were grown on plates lacking histidine, tryptophan, and leucine for 5 days to select colonies containing interacting proteins. Fifty positive colonies were picked and replated on selection media with a substrate for the β-galactosidase interaction-specific reporter. To verify and quantify the two-hybrid interaction in yeast, the NFAT bait plasmid and Mrj prey plasmid were cotransformed back into yeast on selection media and a liquid β-galactosidase activity assay was performed with o-nitrophenyl β-d-galactopyranoside as the substrate.
Plasmid constructions.
The 9xNFAT-luc plasmid contains nine copies of NFAT-only site (5′ TACATTGGAAAATTTTATTACAC-3′) from the interleukin-4 promoter. Concatomers of this sequence were fused upstream of a minimal α-MHC promoter (+12 to −164) in pGL3 basic (Promega, Madison, WI). The 10× E-box reporter was generated from the 9xNFAT-luc reporter except that 10 copies of the right E-box from the muscle creatine kinase enhancer (CCAACACCTGCTGC) was substituted for the 9 NFAT sites. The human TNF-α luciferase reporter (−200) was a gift from Anne E. Goldfield (Harvard Medical School, Boston, MA). Muscle creatine kinase-luciferase (MCK-luc) was a gift from Pier L. Puri (Salk Institute, San Diego, CA). HDAC9-Flag was from Richard Rifkind (Memorial Sloan-Kettering, New York, NY). Flag-HDAC7 was from He-Jin Lee (Parkinson's Institute, Sunnyvale, CA). A full-length mouse Mrj cDNA was cloned into pcDNA3-his/Myc vector by PCR amplification from an adult mouse heart cDNA library. Human c-Myc-Mrj, c-Myc-Mrj (amino acids 1 to 146), and c-Myc-Mrj (amino acids 99 to 242) within the pRK expression vector were gifts from Ichiro Izawa and Masaki Inagaki (19). Mrj deletion mutants were generated by PCR and subcloned into pRK containing the c-Myc tag on the N terminus. Glutathione S-transferase (GST)-Mrj and deletion mutants were constructed by subcloning the Mrj cDNA or PCR fragments into the GST-4T-1 vector (Pharmacia). The Mrj H31Q mutant was generated using the QuikChange mutagenesis kit (Stratagene). The plasmids encoding Flag-HDAC4 and mutants were obtained from Xiang-Jiao Yang (McGill University, Montreal, Canada) and Tso-Pang Yao (Duke University, Durham, NC), respectively. HDAC4 (amino acids 117 to 1085) and HDAC5-Flag was from Eric Verdin (University of California—San Francisco). HDAC4 deletion fragments were generated by restriction enzyme digestions or PCR and subcloned into pcDNA3 His 2 vector (Invitrogen). The Hsp40-containing expression vector was a gift from H.Y. Zoghbi (Baylor College of Medicine, Houston, TX). NFATc3 deletion constructs for in vitro transcription-translation reactions were generated by PCR. Sense and antisense strands of Mrj small interfering RNA (siRNA) template oligonucleotides were 5′-GATCCGCCCAGATGATGTCTTCAGGTTCAAGAGA-3′ and CCTGAAGACATCATCTGGGTTTTTTGGAAA-3′ (cloned into pSilencer 2.0-U6 vector [Ambion]). The silent Mrj mutant was generated with the following sense primer: 5′AATCCTGATGATGTCTTCAGGGAA-3′. NFATc3 deletion mutants were generated by PCR and cloned into pcDNA3 and pGBKt7 plasmids.
Adenovirus constructions.
The mouse Mrj cDNA containing a His/c-Myc tag in the C terminus, the Flag-HDAC4 cDNA, and the nuclear Flag-HDAC4-3SA mutant cDNA (15) were subcloned into the adenoviral pShuttle vectors of the Adeno-X expression system (BD Biosciences) according to manufacturer's instruction. AdΔCnA (active calcineurin) and AdGFP-NFATc3 were described previously (9, 18). Adenovirus expressing Mrj siRNA or a random control siRNA were constructed in the Adeno-X expression system (BD Biosciences). The U6 promoter and Mrj siRNA from pSilencer 2.0-U6 were excised at EcoRI and HindIII (partial fill-in) sites and subcloned into the pShuttle vector at EcoRI and SpeI sites (partial fill-in), thereby replacing the cytomegalovirus promoter in the pShuttle vector.
Transient transfection analysis.
10T1/2 fibroblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. All transfections were performed in six-well plates with Fugene 6 (Roche) for 24 h except for the siRNA experiments (60 h). Cells were transfected with 0.3 μg of the reporter plasmid 9xNFAT-Luc, together with 0.3 μg of expression plasmids. In some control experiments, cells were transfected with MCK-luc or an E-box-dependent reporter together with MyoD and/or Mrj, each at 0.3 μg. Each value presented is the average of triplicate samples and is representative of multiple independent experiments. All transfection experiments were internally normalized by cotransfection with an expression vector encoding β-galactosidase. All data were analyzed for significance with Student's t test.
Rat neonatal cardiomyocyte preparation and transfection.
Cardiomyocyte cultures were prepared as described previously (9). Cultures were infected with adenovirus expressing Mrj and activated calcineurin and/or NFATc3 in 2% M199 medium for 2 h and replaced with serum-free M199 medium for 48 h or 60 h following AdSi-Mrj infection. The cardiomyocytes were stained with mouse α-actinin monoclonal antibody (Sigma), and cell size was measured as described previously (9). One microgram of TNF-α luciferase reporter was cotransfected into one 6-cm plate of cardiomyocytes with 1 μg of expression plasmids encoding NFATc3, activated calcineurin, HDAC-3SA, and Mrj using Tfx20 reagent (Promega, Madison, WI) at a ratio of 2 μl Tfx20 per μg of plasmid in serum-free M199 medium. After 24 h, the cells were lysed and luciferase and β-galactosidase activities were measured.
Nuclear and cytoplasmic extract preparation.
Approximately 4.5 × 106 cardiomyocytes were infected with AdMrj for 24 h and then incubated at 42°C or 37°C for 40 min. Cells were scraped in phosphate-buffered saline (PBS), pelleted, and resuspended in 100 μl buffer A (10 mM Tris-HCl, pH 7.5, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.6% Nonidet P-40, 0.2 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche]) for 5 min at 4°C. After centrifugation, the supernatant was collected as the “cytosolic” fraction, and the nuclearly enriched pellet was incubated in 100 μl buffer C (20 mM Tris-HCl, pH 7.5, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) for 30 min at 4°C. The cytosolic and nuclear protein fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with c-Myc antibody (Sigma) for Mrj, rabbit anti-HDAC2 as a nuclear fraction control (Abcam, Cambridge, MA) and α-tubulin antibody as a cytoplasmic fraction control (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoprecipitation and Western blot analysis.
Rat neonatal cardiomyocytes were infected with adenoviruses encoding Flag-HDAC4 and/or Mrj for 2 h in 2% Dulbecco's modified Eagle's medium. The cultures were grown for 48 h in serum-free M199 medium. 10T1/2 cells were transiently transfected with full-length or mutant Mrj-encoding plasmids, together with pFlag-HDAC4 or pME-NFATc3. Cardiomyocytes and 10T1/2 cells were lysed at 4°C in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40) containing protease inhibitors. Lysates were cleared by centrifugation at 12,000 × g for 10 min. Lysate proteins were precleared with protein A/G-agarose for 1 h and immunoprecipitated overnight at 4°C with mouse anti-Flag antibody (Sigma), rabbit anti-NFATc3 antibody (Santa Cruz), rabbit anti-c-Myc antibody (Santa Cruz), or mouse anti-His-tag antibody (Sigma, St. Louis, MO), followed by incubation with protein A/G-agarose for 2 h. The agarose was washed, and bound proteins were eluted and resolved in SDS-PAGE, followed by Western blotting with mouse anti-Myc (Sigma). Endogenous Mrj detection was performed with rabbit-generated antiserum from Ichiro Izawa and Masaki Inagaki (19).
GST pull-down assays.
All GST fusion proteins were overexpressed in Escherichia coli BL21 cells. Binding assays were performed with labeled proteins synthesized in vitro by using the TNT coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine (Amersham Biosciences). Ten micrograms of immobilized GST fusion protein was incubated for 2 h at room temperature with 10 μl of 35S-labeled proteins in GST binding buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol, 0.1% Nonidet P-40, protease inhibitors, and 1 mg of bovine serum albumin/ml. After four washes in GST binding buffer, beads were boiled in SDS sample buffer to elute bound protein, which was subsequently resolved by SDS-PAGE and analyzed by autoradiography. The GST-Mrj interaction with HDAC4 was performed under more stringent conditions with buffer conditions of 150 mM NaCl, 0.5% NP-40, and 2 mg/ml bovine serum albumin.
Histone deacetylase activity assay.
Cardiomyocytes were infected with adenoviruses encoding Mrj or β-galactosidase (control) for 24 h, and lysates were generated and immunoprecipitated with anti-c-Myc monoclonal antibody (Sigma) or a control hemagglutinin (HA) monoclonal antibody (Santa Cruz), together with protein A/G. HDAC activity was measured using a histone deacetylase assay kit from Upstate Biotechnology. Briefly, biotinylated histone H4 peptide was radiolabeled with p300 coactivator-associated factor and 10,000 cpm [3H]acetyl histone H4 peptide, incubated with or without anti-c-Myc or a control antibody for 15 h at room temperature. Release of [3H]acetate was determined by quantifying radioactivity in the supernatant. The results were averaged from two independent experiments.
TNF-α reverse transcription (RT)-PCR and RNase protection assays.
Cardiomyocytes were infected with adenoviruses encoding green fluorescent protein (GFP)-NFATc3, activated calcineurin and Mrj or HDAC4-3SA for 2 h and grown for 48 h in serum-free M199 medium. TNF-α RT-PCR was performed using total RNA isolated from infected cardiomyocytes together with SuperScript III first-strand synthesis system (Invitrogen), and PCR was performed using rat TNF-α primers, which were designed from a region that encompasses an intron to eliminate false signal from genomic DNA, as follows: 5′-ATGGGCTCCCTCTCATCAGTT-3′ (sense) and 5′-CAAAGTAGACCTGCCCGGACT-3′ (sense). RNase protection assay was carried out using 5 μg RNA per reaction using the Riboquant kit (BD Pharmingen). The rat cytokine-1 multiprobe template (BD Pharmingen) was used to prepare 32P-labeled RNA antisense probe that was transcribed using T7 polymerase (BD Pharmingen).
Immunofluorescence and confocal microscopy.
For analysis of Mrj, NFATc3, or HDAC4 intracellular localization, cardiomyocytes were infected with AdMrj, AdNFATc3, or AdHDAC4 for 2 h. After 24 h, cardiomyocytes were incubated with anti-NFATc3 (1:400) (Santa Cruz) and rabbit c-Myc antibody for Mrj (1:500) (Santa Cruz) for 1 h and then stained with secondary antibodies. Myocytes were stained with rabbit anti-HDAC4 antibody (1:200; Santa Cruz) and also mouse anti-His-tag monoclonal antibody for Mrj when needed (1:800; Cell Signaling, Beverly, MA).
Chromatin immunoprecipitation assay.
Cardiomyocytes (4 × 106) on 10-cm plates were infected with AdβGal as a control and AdGFP-NFATc3 and AdΔCnA, with or without AdMrj or AdHDAC4-3SA, for 48 h. Cells were then treated with 1% formaldehyde for 10 min at 37°C. Chromatin immunoprecipitation was performed using an acetyl-histone H3 immunoprecipitation assay kit (Upstate Biotechnology) and NFATc3, HDAC4, and His-tag antibody (Mrj). After immunoprecipitation, the eluted histone-DNA cross-links were reversed by heating at 65°C for 4 h, after which the DNA was purified with QIAGEN PCR quick spin column. TNF-α genomic DNA was PCR amplified from immunoprecipitated and nonimmunoprecipitated chromatin using the primers 5′-TGATGCCTGGGTGTCCCCAAC-3′ (−202) and 5′-TGGTGTCCTCGCTGAGTTCTG-3′ (+28). PCR consisted of 1 cycle at 96°C for 3.5 min and 30 cycles at 96°C for 50 s, 55°C for 45 s, and 72°C for 45 s in the presence of 2.5 μCi of [32P]dCTP. PCR products were resolved by 6% acrylamide gel electrophoresis.
RESULTS
NFAT factors directly interact with Mrj.
Given the prominent role that NFAT transcription factors play as regulators of calcium-inducible gene expression across diverse cell types, here we sought to identify novel interacting factors as a means of further elucidating NFAT-dependent transcriptional regulation. To this end, a yeast two-hybrid assay was performed using the Gal4 DNA binding domain fused to the rel homology domain of NFATc3 (amino acids 405 to 700) in conjunction with a total mouse heart cDNA library fused to the Gal4 transcriptional activation domain. Approximately 106 recombinants were screened, resulting in the identification of the protein Mrj (amino acids 34 to 242 were contained in the rescued prey). Mrj (DnaJB6) is a member of the DnaJ/Hsp40 family of cochaperone proteins that was previously characterized as a relatively ubiquitously expressed factor (4). The first 70 amino acids of Mrj comprise the highly conserved DnaJ domain that defines this family, while the remaining C-terminal amino acids (71 to 242) are conserved only between Mrj and five close relatives but not within Hsp40 (4).
The rescued prey and bait plasmids were retransformed back into yeast, which confirmed the identified interaction assayed with a Gal4-dependent β-galactosidase reporter (Fig. 1A, first lane). Moreover, a series of Gal4-NFATc3 deletion mutants was generated to more carefully assess the interaction domain within NFATc3. The data demonstrate that NFATc3 amino acids 1 to 1075 (full length), 405 to 700, and 641 to 700, comprising the C-terminal rel homology domain, all mediated interaction with Mrj (Fig. 1A). Sequence alignment of the minimal interacting domain in NFATc3 (amino acids 641 to 700) with NFATc1, NFATc2, and NFATc4 showed 60 to 71% pair-wise similarity (see Fig. S1 in the supplemental material).
FIG. 1.
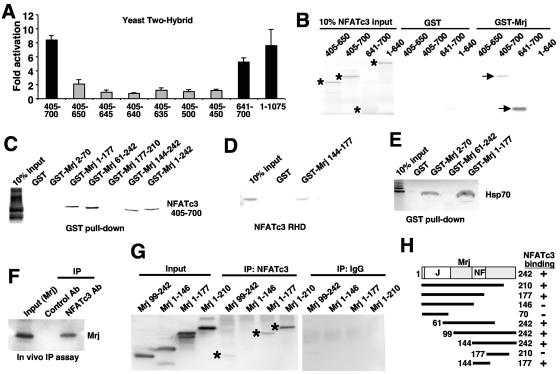
Identification of Mrj as an NFAT interacting factor. (A) A yeast two-hybrid analysis using a liquid β-galactosidase activity assay with the Mrj prey vector and full-length NFATc3 or various NFATc3 deletion mutant domains within the rel homology domain. (B) GST pull-down assay with GST or GST-Mrj and [35S]methionine-labeled NFATc3 deletion mutants. The asterisks show the position of each of the input proteins, while the arrows show the NFATc3 fragments that interact with Mrj. Ten percent of the input is shown in lanes 1 to 4. (C and D) GST pull-down assay with the indicated GST-Mrj deletion mutants in conjunction with [35S]methionine-labeled NFATc3 amino acids 405 to 700. Ten percent of the input is shown in lane 1. (E) A control showing that [35S]methionine-labeled Hsp70 interacts with the N-terminal DnaJ domain of GST-Mrj. (F) Western blot for Mrj after an in vivo immunoprecipitation assay from 10T1/2 fibroblasts that were transfected with expression vectors encoding Mrj and full-length NFATc3. (G) In vivo immunoprecipitation assay was employed in 10T1/2 cells transfected with constructs encoding four different Mrj truncation mutants and full-length NFAT. The asterisks show specific immunoprecipitation. IgG is a nonspecific control antibody. (H) Schematic representation of the different Mrj truncation mutants and their ability to interact with NFATc3.
To further characterize the identified interaction between Mrj and NFATc3, a series of association assays was performed. Radioactively labeled NFATc3 deletion mutants were incubated with GST or GST-Mrj, which confirmed the ability of NFATc3 amino acids 405 to 700 and 641 to 700 to interact with Mrj (Fig. 1B). In addition, the DNA binding RHD from NFATc3 (amino acids 405 to 700) was radioactively labeled and incubated with various GST-Mrj deletion mutants to identify the interacting region within Mrj. The data demonstrate that full-length GST-Mrj was capable of interacting with the NFAT rel homology domain, while GST alone showed no interaction (Fig. 1C). Moreover, deletion mutants encoding Mrj amino acids 1 to 177, 61 to 242, and 144 to 242 all interacted with the NFATc3 RHD, while Mrj amino acids 2 to 70 and 177 to 210 did not interact, suggesting that the interacting domain spans amino acids 144 to 177 (Fig. 1C). Indeed, this region alone was generated as a GST-Mrj fusion and shown to precipitate the NFATc3 rel homology domain (Fig. 1D). As an additional control, the various Mrj-GST deletion mutants were incubated with Hsp70 that was generated by a cotranscription-translation reaction for interaction assessment (Fig. 1E). Hsp70 was previously shown to interact with the DnaJ domain (21), which was also specifically observed here, yet it failed to interact with the remaining C terminus of Mrj (Fig. 1E). In support of these data, an interaction was also observed following immunoprecipitation of NFATc3 from cultured mammalian cells cotransfected with plasmids encoding full-length epitope-tagged Mrj and full-length NFATc3 (Fig. 1F).
To further characterize the NFAT interacting domain within Mrj in vivo, a series of epitope-tagged truncation mutants were generated for immunoprecipitation analysis from cultured cells cotransfected with NFATc3. This assay revealed an association between full-length NFATc3 and amino acids 1 to 210, 1 to 177, and 99 to 242 of Mrj but not amino acids 1 to 146 (Fig. 1G). None of the Mrj truncations showed immunoprecipitation with an unrelated antibody (immunoglobulin G [IgG]), indicating that the observed interaction with Mrj was specific for NFATc3 (Fig. 1G). Collectively, these data support the in vitro results obtained with the various GST-Mrj deletion constructs, demarcating the NFAT interaction domain between amino acids 144 to 177 of Mrj (NF domain) (Fig. 1H).
Mrj functions as an NFAT transcriptional repressor.
To investigate the functional significance underlying the observed interaction between Mrj and NFATc3, transfection assays were performed using promoter-containing reporter plasmids. Transfection of a reporter containing nine multimerized NFAT-only DNA binding elements fused to a basal promoter, and a luciferase cDNA demonstrated transcriptional induction in the presence of cotransfected NFATc3, which was repressed by cotransfection of full-length Mrj but not by a plasmid encoding Hsp40 (Fig. 2A). Cotransfection of Mrj or Hsp40 had no effect on NFATc3 protein expression levels in this experiment (Fig. 2A, right panel). Hsp40 was selected as a control, given that it contains the highly conserved DnaJ domain but is otherwise divergent and lacks homology with the NFAT interacting domain identified in Mrj. It was also of concern that Mrj might simply serve as a generalized inhibitor of gene expression or might be inducing a nonspecific effect on reporter activity. However, using an E-box-dependent reporter that contained 10 E-box sites (substituted from the 9xNFAT-luc construct) or a MCK-luc reporter, cotransfected with MyoD, no significant repression by Mrj was observed (Fig. 2B and C).
FIG. 2.
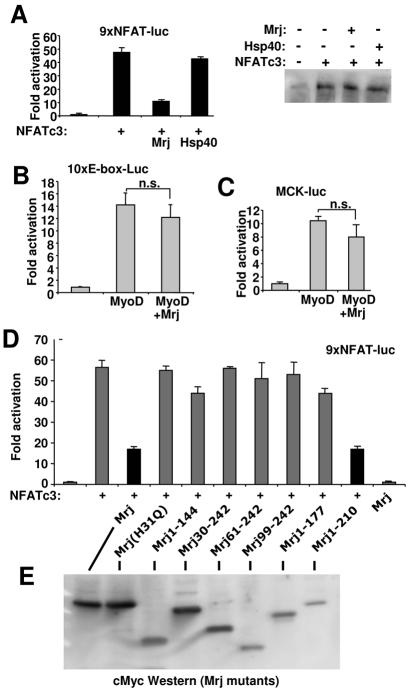
Mrj represses NFAT-dependent transcription. (A) Relative luciferase activity in 10T1/2 cells transfected with the NFAT-dependent reporter and plasmids encoding NFATc3, Hsp40, and Mrj. The data shown were averaged from triplicate transfections (repeated twice). The panel on the right is a Western blot for NFATc3 protein from the cotransfection reactions. (B and C) Relative luciferase activity in 10T1/2 cells transfected with an E-box-dependent luciferase reporter (10xE-box-luc) or a MCK-luciferase reporter and MyoD alone or with Mrj. (D) Relative luciferase activity in 10T1/2 cells transfected with the NFAT-dependent luciferase reporter, NFATc3, and the indicated mutants of Mrj. The data shown were averaged from triplicate transfections (repeated twice). (E) Western blot (c-Myc antibody) from cells transfected with each of the Mrj mutants (all were c-Myc epitope tagged).
The immunoprecipitation-based interaction assays indicated that amino acids 144 to 177 in Mrj are sufficient to interact with NFATc3. It was also of interest to dissect the domains of Mrj involved in transcriptional repression. Accordingly, a series of Mrj deletion and mutation constructs were generated for cotransfection analysis, along with the NFAT-dependent luciferase reporter. Full-length Mrj and a deletion mutant containing amino acids 1 to 210 of Mrj significantly inhibited NFATc3-induced reporter activation, but the deletion of either the DnaJ domain or the C terminus of Mrj blocked repressor function (Fig. 2D). Even partial deletion of the DnaJ domain was sufficient to reverse Mrj-dependent transcriptional repression suggesting a critical role for this domain in mediating the repressor function itself (this domain mediates Hsp70 interaction). Singular mutation of histidine at amino acid 31 in full-length Mrj, which was previously shown to inactivate Hsp40, also failed to function as a transcriptional repressor (28) (Fig. 2D). This mutant form of Mrj fails to localize to the nucleus at baseline or after heat shock (data not shown), suggesting that Mrj functions as a nuclear repressor of NFATc3 transcriptional activity and not as a regulator of NFATc3 shuttling. Western blotting was also performed from cells transfected with each of the deletion constructs to verify effective protein expression, hence dismissing a trivial reason for the lack of repression observed with any single mutant (Fig. 2E).
Mrj-dependent NFAT repression is associated with class II HDAC recruitment.
While Mrj functioned as a NFAT transcriptional repressor within the nucleus, the mechanism underlying this effect was unknown. Given the emerging paradigm that chromatin remodeling serves a critical role in regulating gene expression, we first assessed the ability of histone deacetylases to mediate Mrj-dependent repression. Using an immunoprecipitation-dependent HDAC activity assay, adenovirus-mediated overexpression of Mrj (the Mrj cDNA also contains a c-Myc epitope tag) followed by immunoprecipitation with specific (c-Myc) and nonspecific (HA) antibodies showed recruitment of HDACs but not without Mrj overexpression (control with c-Myc). Specifically, the immunoprecipitates were measured for HDAC enzymatic activity by incubating with p300 coactivator-associated factor-acetylated histone peptide substrate, demonstrating that Mrj specifically recruited HDAC activity (Fig. 3A).
FIG. 3.
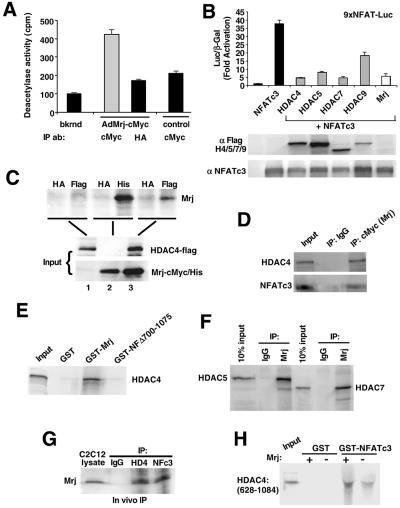
Mrj interacts with class II HDACs. (A) Deacetylase activity assay from immunoprecipitation reactions against Mrj (c-Myc antibody) or a nonspecific antibody (HA) in cardiomyocytes infected with AdMrj or a control adenovirus (AdβGal). (B) Transfection assay showing NFAT-dependent activity (9xNFAT luciferase reporter) in the presence of cotransfected NFATc3 and HDAC4/HDAC5/HDAC7/HDAC9 or Mrj. Western blotting is shown below for each transfection reaction to verify expression of HDAC4/HDAC5/HDAC7/HDAC9 (all are flag tagged) or NFATc3. (C) Coimmunoprecipitation assay followed by Western blotting for Mrj by using a c-Myc antibody (upper panel). Reactions were performed using the inputs shown (lower panels) from cells infected with AdHDAC4 alone, AdMrj alone, or both. The polyhistidine antibody served as a positive control for Mrj precipitation, the HA antibody was a negative control, and the Flag antibody showed a specific interaction between Mrj and HDAC4. (D) Coimmunoprecipitation assay followed by Western blotting for HDAC4 and NFATc3 from cotransfected Cos-7 cells with HDAC4, NFATc3, ΔCnA, and Mrj. Mrj was immunoprecipitated with a c-Myc antibody, which identified HDAC4 and NFATc3. (E) SDS-PAGE of [35S]methionine-labeled HDAC4 precipitated with GST, GST-Mrj, or GST-NFATc3 with amino acids 700 to 1075 (GST-NFΔ700-1075). Thirty percent of the input is shown. (F) Coimmunoprecipitation assay with Mrj antibody (c-Myc tag) or a nonspecific IgG, followed by Western blotting for HDAC5 or HDAC7. Cos-7 cells were first cotransfected with HDAC5 or HDAC7 in combination with Mrj. (G) Immunoprecipitation of endogenous HDAC4 (HD4), NFATc3 (NFc3), or nonspecific IgG from C2C12 protein lysates, followed by Western blotting for endogenous Mrj. (H) SDS-PAGE of [35S]methionine-labeled HDAC4 (amino acids 628 to 1084) in the presence or absence of cotranslated Mrj. These translated products were subjected to pull-down assay with GST or GST-NFATc3 (amino acids 405 to 700).
Transfection experiments with a series of plasmids encoding class II HDACs revealed a prominent role in mediating NFATc3 repression (Fig. 3B). For example, class II HDAC4/HDAC5/HDAC7/HDAC9 each repressed NFAT-dependent reporter activity, although the effectiveness of repression may vary slightly (Fig. 3B). As an important control, Western blotting showed similar NFATc3 overexpression levels in each of the cotransfection reactions and showed that each of the individual HDACs was expressed at detectable levels (Fig. 3B). Since each of the class II HDACs demonstrated repression, we examined the ability of Mrj to specifically interact with at least one family member, HDAC4, using adenoviral overexpression followed by immunopreciptation. Cells were infected with AdMrj (c-Myc and His epitope) and/or AdHDAC4 (Flag epitope), followed thereafter with a specific (Flag or His) or nonspecific antibody (HA) in the immunoprecipitation procedure (Fig. 3C). The results demonstrate that HDAC4 was associated with Mrj when both were overexpressed and HDAC4 was first precipitated (Fig. 3C, lane 6, top). These results were extended and confirmed by performing a similar experiment, except that Mrj was immunoprecipitated (c-Myc antibody) from cells cotransfected with Mrj, HDAC4, NFATc3, and activated calcineurin, identifying both HDAC4 and NFATc3 as Mrj interacting factors (Fig. 3D, lane 3). To further confirm the identified interaction, HDAC4 was radioactively labeled in a cotranscription-translation reaction followed by incubation with a GST-Mrj fusion protein showing a specific interaction (Fig. 3E). As a control, HDAC4 did not interact with GST alone or the C terminus of NFATc3 (NFΔ700-1075) (Fig. 3E).
The cotransfection experiment discussed above in Fig. 3B suggests that each of the class II HDACs can mediate NFATc3 transcriptional repression. To extend this result, we assayed for the ability of HDAC5 and HDAC7 to immunoprecipitate with Mrj from cotransfected Cos-7 cells. Mrj immunoprecitiation with a c-Myc antibody, but not with a nonspecific IgG, demonstrated an association with HDAC5 and HDAC7 (Flag antibody), similar to that observed for HDAC4 (Fig. 3F). Thus, class II HDACs appear to uniformly interact with Mrj.
Each of the assays presented above utilized overexpression of NFATc3, Mrj, or a class II HDAC for purposes of investigating a potential interaction and mapping the relevant domains. To confirm interaction among endogenous levels of each protein, C2C12 myoblasts were employed since they express detectable levels of each protein, unlike cardiomyocytes (levels of NFATc3 and HDAC4 are too low). The data demonstrate that immunoprecipitation of either HDAC4 or NFATc3 identified Mrj, while a nonspecific IgG did not isolate Mrj (Fig. 3G). To further characterize this complex, we also observed that Mrj enhanced the ability of GST-NFATc3 to interact with the C terminus of HDAC4 (Fig. 3H) (see below for mapping of HDAC4).
The interacting domains present within Mrj and HDAC4 were each separately mapped by transfection and immunoprecipitation or by using GST pull-down assays. Full-length HDAC4 associated with Mrj deletion constructs encoding amino acids 1 to 210, 1 to 177, and 99 to 242 but not amino acids 1 to 146, suggesting that the HDAC4 interacting domain in Mrj consists of amino acids 146 to 177, which essentially overlaps with the NFAT interacting domain (Fig. 4A). The interaction between HDAC4 and Mrj was also mapped using a series of GST-Mrj deletion mutants in conjunction with radioactively labeled HDAC4 encompassing amino acids 628 to 1084 (from a cotranscription-translation reaction). The data suggest that Mrj amino acids 144 to 177 are required for HDAC4 binding (Fig. 4B and C).
FIG. 4.
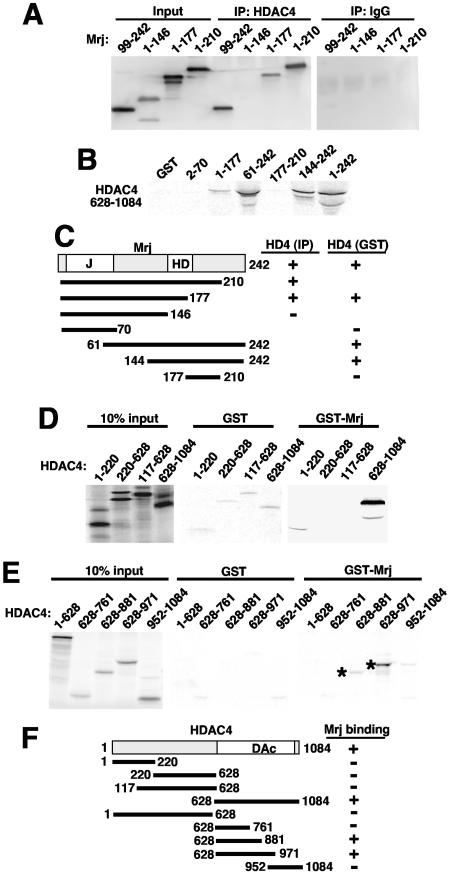
Mrj specifically interacts with HDAC4. (A) Western blot for Mrj after an in vivo immunoprecipitation assay (against HDAC4 or a nonspecific IgG) from 10T1/2 cells transfected with expression vectors encoding truncation mutants of Mrj and full-length HDAC4. (B) GST pull-down assay with the indicated Mrj mutants and [35S]methionine-labeled HDAC4 amino acids 628 to 1085. (C) A schematic representation of the different Mrj truncation mutants and their interaction with HDAC4. (D) SDS-PAGE of [35S]methionine-labeled HDAC4 truncation mutants precipitated with either GST or GST-Mrj. (E) SDS-PAGE of [35S]methionine-labeled HDAC4 deletion fragments subjected to pull-down with GST or GST-Mrj. The asterisks show the specific interactions. (F) A schematic representation of the different HDAC4 truncation mutants and their abilities to interact with Mrj.
A full-length Mrj-GST fusion protein was also generated in bacteria and incubated with a series of radioactively labeled HDAC4 deletion mutants. This assay showed a specific interaction between amino acids 628 to 1084 of HDAC4 and GST-Mrj but not the N-terminal amino acids 1 to 628 or GST alone (Fig. 4D). The C-terminal amino acids 628 to 1084 of HDAC4 encompass the deacetylase catalytic domain, which is conserved in class II HDACs. This interaction was more finely mapped using a series of HDAC4 deletion mutants generated as radiolabeled in vitro transcription-translation products and incubated with GST or GST-Mrj. Interactions were observed with HDAC4 amino acids 628 to 881 and 628 to 971 but not 628 to 761 or 952 to 1084 (Fig. 4E). Taken together, the results suggest that the Mrj interacting domain in HDAC4 encompasses amino acids 761 to 881 (Fig. 4F). Indeed, this region of sequence in HDAC4 has approximately 80% identity with HDAC5, HDAC7, and HDAC9 but only 20% identity with the class I HDAC1 (see Fig. S1 in the supplemental material). Collectively, these results demonstrate that Mrj can physically interact with HDAC4, further supporting the mechanism whereby Mrj mediates NFATc3 transcriptional repression through HDAC recruitment. Indeed, cotransfection of plasmids encoding Mrj and HDAC4 together promoted a greater than additive repression of NFATc3-induced transcriptional activity in 10T1/2 fibroblasts (Fig. 5A). Cotransfection of Mrj or HDAC4 did not alter expression of NFATc3 in this experiment (Fig. 5A, lower panel). That HDAC4 cotransfection represses better or to the same extent as Mrj alone might suggest that class II HDACs are potentially limiting in these cells or that HDAC4 overexpression also reciprocally recruits some endogenous Mrj.
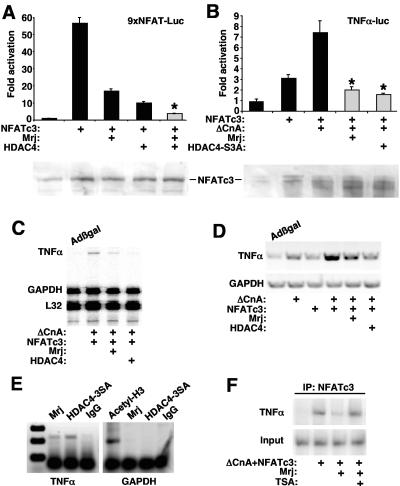
FIG. 5.
Mrj represses NFAT-dependent transcription through class II HDAC recruitment. (A) Relative luciferase activity from 10T1/2 cells transfected with the NFAT-dependent luciferase reporter, NFATc3, Mrj, and HDAC4. The asterisk signifies a P of <0.05 versus NFATc3 plus HDAC4. The lower panel shows a Western blot for NFATc3 protein from the various cotransfection conditions. (B) Relative luciferase activity from cardiomyocytes transfected with a TNF-α promoter-luciferase fusion reporter together with expression plasmids encoding NFATc3, Mrj, activated calcineurin, and HDAC4-3SA (constitutively nuclear). The asterisk signifies a P of <0.05 versus NFATc3 plus ΔCnA. The lower panel shows a Western blot for NFATc3 protein from the various cotransfection conditions. (C) SDS-PAGE of an RNase protection assay from cultured cardiomyocytes infected with the indicated adenoviruses. The position of TNF-α, GAPDH, and L32 protected fragments is shown. (D) RT-PCR for TNF-α or GAPDH mRNA levels from cardiomyocytes infected with the indicated recombinant adenoviruses. (E) ChIP assay against the TNF-α and GAPDH promoters with antibodies against Mrj, HDAC4, IgG (nonspecific), or acetyl-histone 3 (molecular weight standard is shown in the far left lane). ChIP was performed from cardiomyocytes infected with AdΔNFATc3 or AdΔCnA, in conjunction with AdMrj or AdHDAC4-3SA. (F) ChIP for the TNF-α promoter after immunoprecipitation of NFATc3 from cardiomyocytes infected with the indicated adenoviruses with or without TSA treatment.
Mrj represses NFATc3-induced TNF-α expression through HDAC4.
Calcineurin-NFAT signaling functions as a prominent inducer of cytokine gene expression in lymphocytes, of which TNF-α is a well-characterized downstream effector that directly responds through NFAT DNA binding sites within its minimal promoter (42, 43), although this relationship has never been established in cardiomyocytes. Increased TNF-α expression is associated with a wide array of cardiac disease states, including hypertrophy, cardiomyopathy, and heart failure (25). Cotransfection of a TNF-α promoter-containing reporter plasmid demonstrated efficient upregulation in the presence of NFATc3 and activated-calcineurin in cardiomyocytes (Fig. 5B). The observed induction of TNF-α reporter activity was blocked by Mrj or HDAC4-3SA cotransfection, extending the identified repressor mechanism to a more relevant transcriptional response (Fig. 5B). Also of note, overexpression of HDAC4-S3A or Mrj did not alter expression of coinfected NFATc3, although overexpression of activated calcineurin (ΔCnA) did increase the mobility of NFATc3 protein as assessed by Western blotting (Fig. 5B, lower panel).
Adenoviral-mediated overexpression of activated calcineurin and NFATc3 in cultured primary cardiac myocytes potently induced endogenous TNF-α mRNA expression, as assessed by both RT-PCR and RNase protection assays (Fig. 5C and D). This calcineurin-NFAT-dependent induction of TNF-α expression was abrogated by coinfection with AdMrj or AdHDAC4-3SA, suggesting that the identified repressor mechanism functions on an endogenous gene (Fig. 5C and D).
Since recruitment of class II HDAC proteins can be associated with chromatin remodeling, we performed a chromatin immunoprecipitation assay (ChIP) to directly examine the configuration of the NFAT-inducible TNF-α promoter region in vivo. Cardiomyocytes were infected with recombinant adenoviruses encoding activated calcineurin (AdΔCnA), NFATc3, Mrj, or HDAC4-3SA. Soluble chromatin extracts were generated and immunoprecipitated with either NFATc3, Mrj, or HDAC4 antibody, followed by PCR amplification of the TNF-α promoter between nucleotides −202 and + 28 relative to the transcription start site. The data show occupancy of Mrj and HDAC4 on the TNF-α promoter but not on the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) promoter (Fig. 5E). Cardiomyocytes were also infected with the indicated adenoviruses and treated with trichostatin A (TSA) followed by ChIP to detect the presence of NFATc3. The data demonstrate that Mrj overexpression is associated with an 80% reduction in NFATc3 TNF-α promoter occupancy (lanes 2 and 3), while treatment with the HDAC inhibitor TSA reversed the inhibitory effect of Mrj on NFATc3 occupancy (Fig. 5F, lane 4).
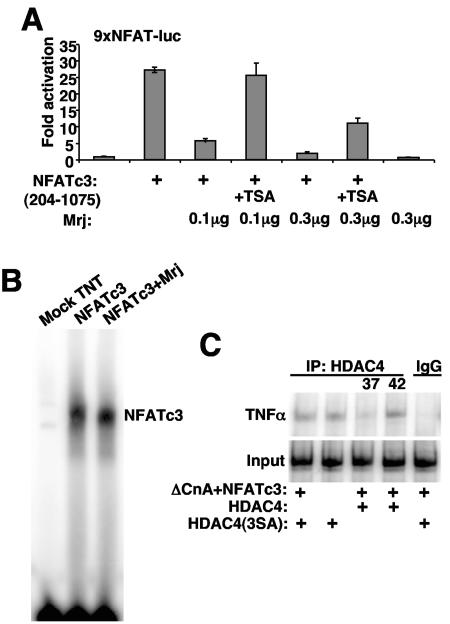
In support of these observations, TSA partially reversed Mrj-regulated repression of NFAT activation on the multimerized NFAT-dependent luciferase reporter in transient transfection assays (Fig. 6A). However, Mrj did not directly block the ability of NFAT to bind its DNA site, as assessed by electrophoretic mobility shift assay (Fig. 6B). Finally, heat shock increased the amount of HDAC4 that associated with the TNF-α promoter in cardiomyocytes, although a constitutively nuclear mutant of HDAC4 could still be recruited to the TNF-α promoter without NFATc3 overexpression (Fig. 6C). Thus, while Mrj does occupy the TNF-α promoter in coordination with NFATc3 and hence mediate repression through an HDAC-dependent mechanism, HDACs themselves likely have multiple interacting partners whereby they can alter TNF-α promoter activity.
FIG. 6.
Characterization of the NFATc3/Mrj/HDAC4 repressor mechanism. (A) Activation from a transient transfection assay in 10T1/2 cells with the NFAT-dependent luciferase reporter. TSA reversed Mrj-mediated repression of NFATc3 transactivation at 0.1 and 0.3 μg of constitutive active ΔNFATc3 (amino acids 204 to 1075) expression vector employed, although reversal was more effective at lower concentrations of Mrj. Cells were treated with or without 160 nM TSA for 16 h before harvest. (B) Electrophoretic mobility shift assay with a 32P-labeled NFAT binding site and in vitro-translated NFATc3 (amino acids 405 to 700) in the presence or absence of in vitro-translated Mrj. Mock-translated TNT lysates were used as a negative control. (C) ChIP assay to evaluate the amount of HDAC4 present at the TNF-α promoter. Cardiomyocytes were infected with AdHDAC4-3SA with or without AdNFATc3 and AdΔCnA, or infected with AdHDAC4, AdNFATc3, and AdΔCnA for 24 h and incubated continuously at 37°C or shifted to 42°C for 70 min. ChIP was performed with HDAC4 antibody.
Subcellular interactions between Mrj, HDAC4, and NFATc3 at rest or after heat shock.
Calcineurin-regulated translocation of NFAT transcription factors to the nucleus serves as the primary mechanism for controlling NFAT-dependent gene expression. Given this regulatory relationship and the ability of NFATs to shuttle through the nucleus, it was uncertain why NFATc3 would utilize a repressor mechanism involving Mrj and HDACs. However, we hypothesized that interaction with Mrj and HDAC might transform NFATc3 into a transcriptional repressor, thus adding another level of complexity underlying NFAT-dependent gene expression following a relevant stimulus, such as heat shock. Indeed, some of the chaperone proteins show intracellular translocation following heat shock stimulation, such as Hsp25 (3). Infection of primary neonatal rat cardiomyocytes with AdMrj resulted in efficient and widespread expression of Mrj that was within both the cytoplasm and nucleus, although most Mrj remained cytoplasmic in unstimulated cells (Fig. 7A). However, 70 min of heat shock induced a complete accumulation of Mrj in the nucleus (Fig. 7A).
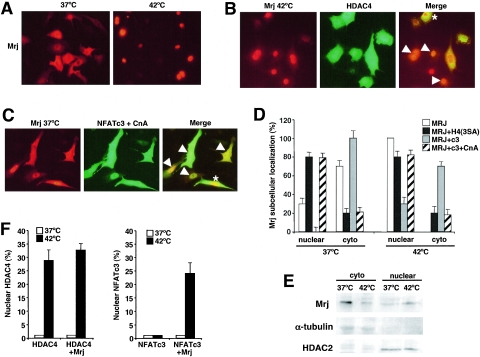
FIG. 7.
Mrj/HDAC4/NFATc3 subcellular localization. (A) Subcellular localization of Mrj in cultured cardiomyocytes at 37°C and 42°C. Myocytes were infected with an adenovirus encoding Mrj and subsequently immunostained for Mrj (red). (B) Subcellular localization of Mrj (red) and wild-type HDAC4 (green) in cardiomyocytes at 42°C (70 min of heat shock) after adenoviral overexpression of each protein together. The arrowheads show cells with colocalization, yet some cells fail to show colocalization as denoted by the asterisk. (C) Subcellular distribution of Mrj (red) and NFATc3 (green) in cardiomyocytes at 37°C previously infected with AdMrj, AdNFATc3, and AdΔCnA. (D) Graph showing quantitation of Mrj subcellular localization in either the nucleus or cytoplasm at 37°C or 42°C in cultured cardiomyocytes infected with the indicated combination of adenoviruses. (E) Western blot for Mrj, α-tubulin, and HDAC2 from cardiomyocytes infected with AdMrj. Nuclear and cytoplasmic extracts were generated from cells incubated at 37°C or 42°C (40 min of heat shock). (F) Graph showing percentages of cardiomyocytes with nuclear HDAC4 (left) or NFATc3 (right) at 37°C and 42°C after adenoviral infection to overexpress HDAC4 alone, HDAC4 with Mrj, or NFATc3 with or without Mrj. Subcellular localizations were quantified in 200 cells each in three independent experiments.
Coinfection of Mrj with wild-type HDAC4 in cardiomyocytes showed colocalization of both proteins in most cells examined at rest or following heat shock stimulation, suggesting that the observed interaction between these proteins might favor their comigration and association in vivo (Fig. 7B and D). However, some cells failed to show colocalization between Mrj and HDAC4, suggesting that each can be independently regulated (Fig. 7B and D). Similar profiles were observed for Mrj and NFATc3 colocalization. Adenovirus-mediated overexpression of Mrj with NFATc3 and activated calcineurin showed that the majority of cells had colocalization of NFATc3 with Mrj (in either the cytoplasm or nucleus) (Fig. 7C). Indeed, overexpression of Mrj with NFATc3 effectively diminished the population of cells that showed Mrj within the nucleus at rest (Fig. 7D), while coinfection with activated calcineurin, which sends most of the NFATc3 to the nucleus, dramatically increased the percentage of cells with nuclear Mrj (Fig. 7D). Since some cells showed partial localization of Mrj within the cytoplasm and nucleus before and after heat shock, the overall levels of Mrj in each compartment were quantified by Western blotting of nuclear and cytoplasmic protein extracts at 37°C and 42°C. The data show a significant redistribution of Mrj from the cytoplasm to the nucleus following heat shock stimulation (Fig. 7E). HDAC2 and α-tubulin were used as nuclear and cytoplasmic controls, respectively (Fig. 7E).
Overexpression of Mrj with NFATc3 after heat shock stimulation dramatically increased the amount of NFATc3 within the nucleus but not in the absence of Mrj (Fig. 7F). Thus, while NFATc3 and Mrj are independently regulated in their subcellular movements, overexpression of one can alter the localization of the other, further supporting the ability of these factors to interact in vivo. We also observed that heat shock stimulation induced HDAC4 nuclear translocation in approximately 30% of cardiomyocytes (Fig. 7F). However, overexpression of Mrj did not alter HDAC4 subcellular localization at baseline or following heat shock stimulation, suggesting that HDAC4 itself cannot be dominantly affected by Mrj (Fig. 7F), in contrast to the ability of the HDAC4 nuclear-localizing mutant to partially alter Mrj subcellular distribution when overexpressed (Fig. 7D). Thus, the ability of Mrj to interact with HDAC4 partially alters the subcellular distribution of Mrj to the location of HDAC4 but not vice versa. Collectively, these results define heat shock as at least one example of a physiologic stimulus whereby Mrj, HDAC4, and NFATc3 can affect one another in vivo at multiple levels.
Mrj regulates calcineurin-NFAT-induced cardiac hypertrophic growth.
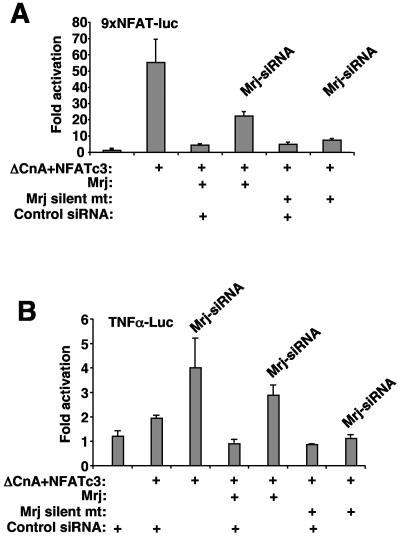
To further extend the potential biological relevance of the proposed NFAT-Mrj-HDAC regulatory complex, a cardiac growth assay was employed. Calcineurin-NFAT signaling has been previously shown to regulate the hypertrophic growth of both cultured cardiac myocytes and the intact heart (49). More importantly, class II HDACs have been shown to serve as global regulators of the cardiac hypertrophic response, although the downstream transcriptional targets are only partially defined (53). As previously observed by us, AdΔCnA infection promoted the hypertrophic growth of neonatal cardiac myocytes in culture characterized by enhanced sarcomeric organization and increased cell surface area (Fig. 8A and B). Coinfection with AdMrj blocked calcineurin-induced cardiac myocyte hypertrophy at the level of sarcomeric organization and the increase in cell size (Fig. 8A and B). Using an NFAT-dependent luciferase reporter in cultured cardiomyocytes, Mrj or HDAC4-3SA cotransfection was also shown to inhibit calcineurin-induced promoter upregulation (Fig. 8C). To analyze the antithetic regulatory relationship, a siRNA-based approach was utilized to reduce Mrj protein levels. Transfection of the Mrj siRNA-encoding expression vector enhanced NFATc3 driven expression of the NFAT-dependent luciferase reporter by 2.5-fold and reduced Mrj-mediated repression (Fig. 8D). To better control for the specificity of the Mrj siRNA, a control Mrj construct that contained silent mutations in the siRNA target sequence was generated (Fig. 9A). While Mrj siRNA partially reversed Mrj-mediated inhibition of the NFAT-dependent luciferase reporter, this same siRNA had no ability to reverse silent mutant Mrj-mediated repression (Fig. 9A).
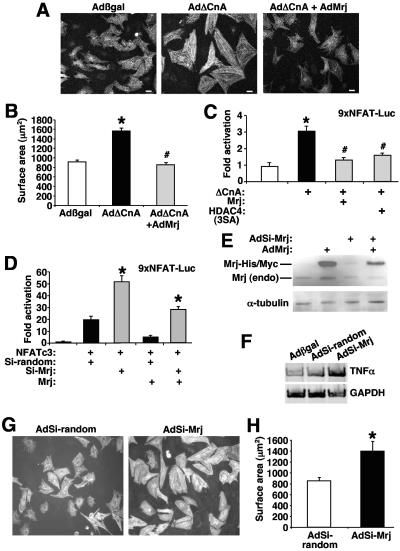
FIG. 8.
Mrj represses calcineurin-induced cardiomyocyte hypertrophy. (A) Pictures of α-actinin-stained neonatal cardiomyocytes infected with the indicated recombinant adenoviruses. (B) Quantification of cardiomyocyte size. *, P < 0.05 versus Adβgal; #, P < 0.05 versus AdΔCnA. (C) Relative luciferase activity from cardiomyocytes transfected with the NFAT-dependent luciferase reporter plasmid along with expression plasmids encoding activated calcineurin, Mrj, and HDAC4-3SA. *, P < 0.05 versus reporter only; #, P < 0.05 versus ΔCnA. (D) Relative luciferase activity from 10T1/2 cells transfected with the NFAT-dependent luciferase reporter plasmid and expression plasmids encoding NFATc3, Mrj, siMrj (SiMrJ), or a random siRNA (Si-random) sequence. *, P < 0.05 versus NFATc3 with random siRNA sequence. (E) Western blot analysis of Mrj protein from adenoviral infected neonatal cardiomyocytes expressing siRNA-Mrj or Mrj. Endo, endogenous Mrj protein. The adenoviral-encoded Mrj is larger due to epitope tags. α-Tubulin, as a loading control, is shown. (F) RT-PCR for TNF-α or GAPDH mRNA levels from cardiomyocytes infected with Adβgal (lane 1), AdΔCnA, AdNFATc3, and AdSi-random (lane 2), and AdΔCnA, AdNFATc3, and AdSi-Mrj (lane 3). (G) Pictures of α-actinin-stained neonatal cardiomyocytes infected with the indicated recombinant adenoviruses. (H) Quantification of cardiomyocyte size. *, P < 0.05 versus AdSi-random.
FIG. 9.
(A) Activation (n-fold) from a transient transfection assay in 10T1/2 cells with the NFAT-dependent luciferase reporter. The experiment shows the specificity of Mrj-siRNA by cotransfecting a vector that encodes wild-type Mrj with silent mutations in the area recognized by the Mrj-siRNA. Mrj-siRNA partially reverses Mrj-dependent inhibition of NFAT activity, but this partial reversal was not observed with cotransfected silent mutant Mrj. Luciferase activity was determined 60 h after transfection. (B) Relative luciferase activity in 10T1/2 cells transfected with a TNF-α-luciferase reporter in conjunction with NFATc3 and ΔCnA in the presence of a negative control siRNA, Mrj-siRNA, or the silent mutant Mrj as in panel A. Mrj-siRNA up-regulated TNF-α reporter activity.
To assess the biologic effects associated with reduced Mrj activity in myocytes we generated an adenovirus expressing the siRNA-Mrj cassette or a random siRNA as a control. Infection of neonatal cardiomyocytes with AdMrj-siRNA produced a 74% reduction of endogenous Mrj protein and a 65% reduction of coexpressed Mrj protein by adenoviral coinfection (Fig. 8E). Remarkably, AdSi-Mrj infection induced cardiomyocyte growth in culture in the absence of any other stimulation and produced a twofold increase in endogenous TNF-α expression, while a nonspecific control adenovirus expressing a random siRNA had no effect (Fig. 8F through H). Mrj-siRNA also increased expression of the TNF-α-luciferase reporter activity in 10T1/2 cells (Fig. 9B). These results show that inhibition of Mrj can induce expression of an endogenous NFAT target gene, the TNF-α gene and that the Mrj-HDAC inhibitory mechanism can function as a regulator of cellular hypertrophy, consistent with observations made with HDAC9−/− and HDAC4−/− mice (46, 53).
DISCUSSION
NFAT factors are regulated at multiple levels.
While calcium is certainly a pivotal regulator of NFAT nuclear occupancy through calcineurin, several protein kinases are known to counterregulate NFAT nuclear occupancy by rephosphorylation of sites within the N-terminal regulatory domain (6, 16). NFAT factors are also subjected to regulation within the nucleus through their ability to interact with other transcriptional regulatory factors, as well as more global-acting transcriptional cofactors. Indeed, the ability of NFAT factors to regulate the differentiation of T lymphocytes, as well as other diverse tissues, has been hypothesized to require chromatin remodeling (16). For example, NFATc2 associates with the histone acetyltransferases p300 and CBP (12). NFATc2 was also shown to interact with MEF-2, which together recruit p300 to synergistically activate Nur77 transcription (52). Moreover, CBP/p300 were shown to directly bind to the N-terminal transcriptional activation domain of NFATc1, resulting in augmented gene expression and T-cell activation (1).
Here we showed that NFATc3 is subjected to yet another level of regulation through Mrj in association with class II HDACs, an event that essentially transforms it into a transcriptional repressor. While the full biologic context underlying NFAT transcriptional repression through Mrj and HDAC is uncertain, heat shock was identified as one potential stimulus whereby such an interaction may be of physiologic relevance. Other types of stresses might also mobilize Mrj and HDAC to mediate NFATc3 inhibition, which might underlie the ability of Mrj and HDAC to inhibit the cardiac hypertrophic response in neonatal cardiac myocytes. The proposed repressor function underlying this NFATc3-Mrj/HDAC4 interaction is consistent with a previous report showing that calcineurin-NFAT signaling negatively regulates cyclin-dependent kinase 4 (CDK4) expression through a histone deacetylase-sensitive mechanism mediated by an NFAT binding site in the CDK4 promoter (2). Indeed, calcineurin Aα or NFATc2 gene-targeted mice were shown to have elevated CDK4 expression (2). Taken together, these results suggest a novel paradigm whereby NFAT transcriptional responses are also regulated at the level of chromatin remodeling through class II HDACs. Our results further suggest that the regulated interaction between Mrj and NFATc3 defines a novel mechanism for determining the transcriptional regulatory properties of NFAT factors, especially given the ability of Mrj and HDACs to translocate to the nucleus following heat shock and possibly other stresses.
Alternate effectors whereby HDACs regulate transcription.
While class I HDACs are largely constitutively nuclear, class II HDACs are able to translocate between the cytoplasm and nucleus in a regulated manner (reviewed in references 8 and 51). In general, both classes of HDACs function as repressors of gene expression by directly deacetylating histones and promoting greater DNA condensation into higher order chromatin structure. HDAC4, HDAC5, and HDAC7 are capable of associating with two other transcriptional repressor-like factors, nuclear receptor corepressor, and silencing mediator for retinoid and thyroid receptors (8, 27). The interacting complex between HDAC4, HDAC5, HDAC7, and nuclear receptor corepressor/silencing mediator for retinoid and thyroid receptors was also shown to contain HDAC3, suggesting that class I and class II HDACs are part of a larger repressor complex (10, 50). More significantly, class II HDACs can associate with transcription factors such as MEF-2 and GATA2, suggesting a mechanism whereby expressed genes could be targeted for inactivation through specific transcription factor interactions (24, 29, 35, 39). However, it is uncertain how interaction with only the two characterized transcription factors can mediate such diverse biological functions attributed to HDACs, suggesting that additional transcription factors are also likely subjected to HDAC regulation through direct or indirect interactions.
Functional analysis of class II HDACs has demonstrated a prominent role in regulating skeletal muscle myogenesis and cardiac hypertrophy (24, 53). NFAT transcription factors have been implicated in directly mediating the cardiac hypertrophic program, consistent with the observation that HDAC9 gene-targeted mice show significantly greater cardiac growth in the presence of the activated calcineurin transgene (53). This result might also suggest that nuclear NFAT factors are normally restrained at some level by HDACs when translocated into the nucleus following calcineurin activation. Loss of HDAC9 eliminates part of this basal repressor function potentially permitting greater hypertrophic gene activation by NFATs, as well as MEF-2 factors. NFAT transcription factors have also been implicated in regulating skeletal muscle development (20), consistent with the observation that class II HDACs must be exported/inactivated by calmodulin-dependent kinase to permit efficient myogenic differentiation (24). Taken together, these previous results discussed here, in conjunction with our observations, suggest that class II HDACs can also regulate gene expression through an NFAT-dependent mechanism.
Another issue that requires discussion is the potential interplay between calcium signals and heat shock stimulation with the coordination of HDAC and NFAT nuclear shuttling. Stimuli that increase intracellular calcium are known to activate calcineurin and calcium-calmodulin-dependent protein kinases. Such stimuli would cause nuclear import of NFAT through calcineurin activation yet nuclear export of HDAC4 and HDAC5 through the action of calcium-calmodulin-dependent protein kinases (26). However, more recent investigation suggests that calcium-independent signaling pathways are potentially more important physiologic regulators of class II HDAC nuclear shuttling (45). Thus, the interplay between calcium signals and heat shock stimulation could still potentially coordinate NFAT repression through Mrj and class II HDACs. Indeed, careful examination of nuclear and cytoplasmic shuttling of NFATc3 suggested that it could be partially influenced by Mrj overexpression. For example, heat shock stimulation had no effect on NFATc3 nuclear translocation by itself, yet overexpression of Mrj promoted significant NFATc3 nuclear occupancy after heat shock. Moreover, overexpression of NFATc3 within the cytoplasm (in the absence of activated calcineurin) maintained more Mrj within the cytoplasm of cardiomyocytes, while induction of NFATc3 nuclear translocation with activated calcineurin similarly increased Mrj nuclear occupancy (Fig. 7D). By comparison, overexpression of the nuclear HDAC4 mutant (3SA) also increased the percentage of cells with Mrj within the nucleus at rest, although Mrj had no effect on HDAC4 subcellular distribution. The observation that HDAC4 and NFATc3 could alter Mrj subcellular localization, in part, most likely reflects the ability of these factors to interact when overexpressed, rather than demonstrating a mechanism of coordinate regulation. Indeed, Mrj, HDAC4, and NFATc3 subcellular distribution appear to be dominantly regulated by independent effector mechanisms, although heat shock can send both Mrj and HDAC4 to the nucleus, possibly through a similar mechanism involving larger complexes of heat shock proteins (30).
Chaperones as novel regulators of gene expression.
Recently, chaperone proteins have been shown to serve as important regulators of gene expression through direct disassembly of transcriptional complexes (11). For example, the molecular chaperone factor p23 was shown to localize to genomic response elements in a hormone-sensitive manner, thereby disrupting nuclear receptor-mediated transcriptional activation. Hsp90 was also shown to possess a similar ability to disrupt transcription, albeit with less strength (11). Hsp90 also links to epigenetic regulation by interacting with key components of chromatin remodeling factors (54). In Drosophila melanogaster, the repressor function underlying the Polycomb group of genes is regulated by HSC4, an HSP70 homologue (32). This is intriguing, given that the most important products of the Polycomb group of genes serve as regulators of gene expression through modulation of chromatin structure. More remarkably, the polyhomeotic proximal (PHP) protein, which is the major isoform of the Polycomb locus, directly interacts with the Hsp70 homologue HSC4, as well as a novel DnaJ domain-containing chaperone, Droj2 (47). Mutation in either HSC4 or Droj2 enhanced the homeotic transformations in Polycomb genes, indicating a role in silencing. Interestingly, characterization of the human Polycomb repressive complex from HeLa cells, which efficiently blocked remodeling of nucleosomal arrays, was also associated with Hsp70 (22). Moreover, transcriptional repression by human Polycomb also involves histone deacetylation (44). A more recent study showed that loss of a single DnaJ member (DjA1) in mice lead to severe defects in spermatogenesis that involved enhanced androgen receptor transcriptional activity, although the precise mechanism was not determined (41). Collectively, these results suggest a novel function for molecular chaperones as repressors of gene expression by either disassembly of active transcriptional complexes, or by facilitating active repression in association with chromatin remodeling factors.
The identification of Mrj as an NFATc3 and HDAC interacting factor suggests a unique mechanism for controlling gene expression in response to environmental stress. Heat shock and/or elevation of Hsp70 protein levels both have growth inhibitory functions, presumably as a means of conserving cellular resources following stress stimulation (38). Heat shock induction of Mrj nuclear translocation would serve a repressor function by more efficiently recruiting HDAC proteins to NFAT transcriptional complexes, thus diminishing calcium-dependent and differentiated gene expression. Consistent with this interpretation, overexpression of Mrj repressed calcineurin-NFAT-induced gene expression and the biologic process of hypertrophic growth through a mechanism involving the inhibition of NFATc3 recruitment to target gene promoters. That some Mrj is present within the nucleus of unstimulated cells suggests that HDAC recruitment might also function to potentially “fine-tune” NFAT transcriptional responses so that only a large degree of calcineurin activation would be likely to elicit a dramatic change in gene expression. However, our data do not exclude the possibility that class II HDACs might regulate NFAT transcription factors independent of Mrj, such as through recruitment of MEF2, which strongly interacts with class II HDACs. Alternatively, class II HDACs might directly deacetylate NFAT transcription factors or HDACs might alter NFAT's ability to interact with activator protein-1 or GATA transcription factors.
While Mrj is a member of a large family of DnaJ domain-containing proteins, not all are likely to interact with NFAT transcription factors or HDACs. Indeed, a more distant relative, Hsp40, was incapable of mediating these interactions. The C-terminal HDAC and NFATc3 interaction domains of Mrj are relatively conserved among its four closest relatives, suggesting the potential for functional redundancy. Despite this conservation, Mrj gene-targeted mice are not viable and show embryonic lethality due to placental abnormalities (17). However, it is likely that other members of the Mrj subfamily also serve a similar repressor function. The 70-amino-acid DnaJ domain is conserved from Escherichia coli through Saccharomyces cerevisiae and mammals, whereas in higher organisms it appears to be specialized for mediating Hsp70 interactions (21). Thus, proteins containing the DnaJ domain have been classified as cochaperones, although their individual functions are likely very diverse and largely unknown. Indeed, the function of the five “Mrj-like” subfamily members is essentially unknown, other than the observation that Mrj can also associate with keratin 8/18 in epithelial cells and regulate intracellular filament organization (19). Here we identified a novel function for Mrj as an NFATc3 corepressor and HDAC interacting factor. This function for Mrj extends the overall paradigm discussed above, whereby chaperones serve as important regulators of gene expression through transcriptional modulatory functions.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health and by an American Heart Association Established Investigator grant (J.D.M.)
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. [DOI] [PubMed] [Google Scholar]
- 2.Baksh, S., H. R. Widlund, A. A. Frazer-Abel, J. Du, S. Fosmire, D. E. Fisher, J. A. DeCaprio, J. F. Modiano, and S. J. Burakoff. 2002. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10:1071-1081. [DOI] [PubMed] [Google Scholar]
- 3.Bryantsev, A. L., S. A. Loktionova, O. P. Ilyinskaya, E. M. Tararak, H. H. Kampinga, and A. E. Kabakov. 2002. Distribution, phosphorylation, and activities of Hsp25 in heat-stressed H9c2 myoblasts: a functional link to cytoprotection. Cell Stress Chaperones 7:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang, J. Z., H. Zhou, M. Zhu, S. H. Li, X. J. Li, and C. H. Sung. 2002. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J. Biol. Chem. 277:19831-19838. [DOI] [PubMed] [Google Scholar]
- 5.Crabtree, G. R. 1999. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96:611-614. [DOI] [PubMed] [Google Scholar]
- 6.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 7.de la Pompa, J. L., L. A. Timmerman, H. Takimoto, H. Yoshida, A. J. Elia, E. Samper, J. Potter, A. Wakeham, L. Marengere, B. L. Langille, G. R. Crabtree, and T. W. Mak. 1998. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392:182-186. [DOI] [PubMed] [Google Scholar]
- 8.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Windt, L. J., H. W. Lim, S. Haq, T. Force, and J. D. Molkentin. 2000. Calcineurin promotes protein kinase C and c-Jun NH2-terminal kinase activation in the heart. Cross-talk between cardiac hypertrophic signaling pathways. J. Biol. Chem. 275:13571-13579. [DOI] [PubMed] [Google Scholar]
- 10.Fischle, W., F. Dequiedt, M. Fillion, M. J. Hendzel, W. Voelter, and E. Verdin. 2001. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 276:35826-35835. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, B. C., and K. R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232-2235. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graef, I. A., F. Chen, L. Chen, A. Kuo, and G. R. Crabtree. 2001. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105:863-875. [DOI] [PubMed] [Google Scholar]
- 14.Graef, I. A., F. Wang, F. Charron, L. Chen, J. Neilson, M. Tessier-Lavigne, and G. R. Crabtree. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113:657-670. [DOI] [PubMed] [Google Scholar]
- 15.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, P. J., B. J. Swanson, M. A. Haendel, G. E. Lyons, and J. C. Cross. 1999. Mrj encodes a DnaJ-related co-chaperone that is essential for murine placental development. Development 126:1247-1258. [DOI] [PubMed] [Google Scholar]
- 18.Hunton, D. L., P. A. Lucchesi, Y. Pang, X. Cheng, L. J. Dell'Italia, and R. B. Marchase. 2002. Capacitative calcium entry contributes to nuclear factor of activated T-cells nuclear translocation and hypertrophy in cardiomyocytes. J. Biol. Chem. 277:14266-14273. [DOI] [PubMed] [Google Scholar]
- 19.Izawa, I., M. Nishizawa, K. Ohtakara, K. Ohtsuka, H. Inada, and M. Inagaki. 2000. Identification of Mrj, a DnaJ/Hsp40 family protein, as a keratin 8/18 filament regulatory protein. J. Biol. Chem. 275:34521-34527. [DOI] [PubMed] [Google Scholar]
- 20.Kegley, K. M., J. Gephart, G. L. Warren, and G. K. Pavlath. 2001. Altered primary myogenesis in NFATC3(-/-) mice leads to decreased muscle size in the adult. Dev. Biol. 232:115-126. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, W. L. 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23:222-227. [DOI] [PubMed] [Google Scholar]
- 22.Levine, S. S., A. Weiss, H. Erdjument-Bromage, Z. Shao, P. Tempst, and R. E. Kingston. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 25.Mann, D. L. 2002. Tumor necrosis factor-induced signal transduction and left ventricular remodeling. J. Card. Fail. 8:S379-S386. [DOI] [PubMed] [Google Scholar]
- 26.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 28.Michels, A. A., B. Kanon, O. Bensaude, and H. H. Kampinga. 1999. Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J. Biol. Chem. 274:36757-36763. [DOI] [PubMed] [Google Scholar]
- 29.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto, Y., T. Saiwaki, J. Yamashita, Y. Yasuda, I. Kotera, S. Shibata, M. Shigeta, Y. Hiraoka, T. Haraguchi, and Y. Yoneda. 2004. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J. Cell Biol. 165:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollaaghababa, R., L. Sipos, S. Y. Tiong, O. Papoulas, J. A. Armstrong, J. W. Tamkun, and W. Bender. 2001. Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc. Natl. Acad. Sci. USA 98:3958-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 34.Ohtsuka, K., and M. Hata. 2000. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5:98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozawa, Y., M. Towatari, S. Tsuzuki, F. Hayakawa, T. Maeda, Y. Miyata, M. Tanimoto, and H. Saito. 2001. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 98:2116-2123. [DOI] [PubMed] [Google Scholar]
- 36.Ranger, A. M., L. C. Gerstenfeld, J. Wang, T. Kon, H. Bae, E. M. Gravallese, M. J. Glimcher, and L. H. Glimcher. 2000. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp. Med. 191:9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranger, A. M., M. J. Grusby, M. R. Hodge, E. M. Gravallese, F. C. de la Brousse, T. Hoey, C. Mickanin, H. S. Baldwin, and L. H. Glimcher. 1998. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392:186-190. [DOI] [PubMed] [Google Scholar]
- 38.Song, J., M. Takeda, and R. I. Morimoto. 2001. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 39.Sparrow, D. B., E. A. Miska, E. Langley, S. Reynaud-Deonauth, S. Kotecha, N. Towers, G. Spohr, T. Kouzarides, and T. J. Mohun. 1999. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 18:5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayanagi, H., S. Kim, T. Koga, H. Nishina, M. Isshiki, H. Yoshida, A. Saiura, M. Isobe, T. Yokochi, J. Inoue, E. F. Wagner, T. W. Mak, T. Kodama, and T. Taniguchi. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3:889-901. [DOI] [PubMed] [Google Scholar]
- 41.Terada, K., K. Yomogida, T. Imai, H. Kiyonari, N. Takeda, T. Kadomatsu, M. Yano, S. Aizawa, and M. Mori. 2005. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 24:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfeld. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsytsykova, A. V., and A. E. Goldfeld. 2002. Inducer-specific enhanceosome formation controls tumor necrosis factor alpha gene expression in T lymphocytes. Mol. Cell. Biol. 22:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 45.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y. J., and H. W. Brock. 2003. Polyhomeotic stably associates with molecular chaperones Hsc4 and Droj2 in Drosophila Kc1 cells. Dev. Biol. 262:350-360. [DOI] [PubMed] [Google Scholar]
- 48.Wilkins, B. J., L. J. De Windt, O. F. Bueno, J. C. Braz, B. J. Glascock, T. F. Kimball, and J. D. Molkentin. 2002. Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol. Cell. Biol. 22:7603-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkins, B. J., and J. D. Molkentin. 2002. Calcineurin and cardiac hypertrophy: where have we been? Where are we going? J. Physiol. 541:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, W. M., S. C. Tsai, Y. D. Wen, G. Fejer, and E. Seto. 2002. Functional domains of histone deacetylase-3. J. Biol. Chem. 277:9447-9454. [DOI] [PubMed] [Google Scholar]
- 51.Yang, X. J., and S. Gregoire. 2005. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25:2873-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Youn, H. D., T. A. Chatila, and J. O. Liu. 2000. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 19:4323-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, R., M. Davey, Y. C. Hsu, P. Kaplanek, A. Tong, A. B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, C. Boone, A. Emili, and W. A. Houry. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120:715-727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.