Abstract
Overexpression of mutant p53 is a common theme in tumors, suggesting a selective pressure for p53 mutation in cancer development and progression. To determine how mutant p53 expression may lead to survival advantage in human cancer cells, we generated stable cell lines expressing p53 mutants p53-R175H, -R273H, and -D281G by use of p53-null human H1299 (lung carcinoma) cells. Compared to vector-transfected cells, H1299 cells expressing mutant p53 showed a survival advantage when treated with etoposide, a common chemotherapeutic agent; however, cells expressing the transactivation-deficient triple mutant p53-D281G (L22Q/W23S) had significantly lower resistance to etoposide. Gene expression profiling of cells expressing transcriptionally active mutant p53 proteins revealed the striking pattern that all three p53 mutants induced expression of approximately 100 genes involved in cell growth, survival, and adhesion. The gene NF-κB2 is a prominent member of this group, whose overexpression in H1299 cells also leads to chemoresistance. Treatment of H1299 cells expressing p53-R175H with small interfering RNA specific for NF-κB2 made these cells more sensitive to etoposide. We have also observed activation of the NF-κB2 pathway in mutant p53-expressing cells. Thus, one possible pathway through which mutants of p53 may induce loss of drug sensitivity is via the NF-κB2 pathway.
Mutation in the p53 tumor suppressor gene is a common event in human cancer (6, 36, 40, 43, 44, 65, 71, 72). Unlike what is seen for other tumor suppressors, in the majority of human carcinomas with p53 mutations, a protein with one amino acid substitution is overexpressed, suggesting the existence of a selection pressure for maintaining expression of the mutant protein (6, 36, 40, 43, 44, 65, 71, 72). This also is perhaps indicative of an active role played by p53 mutants in oncogenesis and follows the gain-of-function hypothesis, which predicts not only that mutations in the p53 gene destroy the tumor suppressor function of the wild-type (WT) protein but that the mutant proteins may also gain oncogenic functions. The gain-of-function hypothesis also predicts that tumors with mutant p53 proteins may be more aggressive or that patients with tumors harboring mutant p53 have poorer prognoses than patients with tumors lacking the p53 protein. This has been found to be true for various types of cancers (10, 28, 29, 81, 82, 88).
WT p53 is a sequence-specific transactivator of promoters containing p53-binding sites. Elevated levels of WT p53 in response to cellular stress situations, such as DNA damage, can lead to apoptosis or induce cell cycle arrest (26, 53, 60, 68, 84, 89, 91) by inducing expression of genes involved in various aspects of cellular growth regulation (21, 26, 27, 49, 50, 53, 60, 63, 68, 84, 89, 91). A mutation in one allele of p53 generates a stable mutant protein with compromised tumor suppressor function. However, there is compelling evidence to suggest that apart from loss of growth suppressor function, p53 mutants can confer oncogenic properties even in the absence of WT p53 (reviewed in references 12 and 71).
Expression of mutant p53 in cells devoid of endogenous WT p53 has been shown to induce various growth-promoting functions that include tumorigenicity, metastasis, and colony-forming ability (1, 7, 13, 24, 37, 46, 51, 55, 56, 64, 67, 86). For example, p53-null 10(3) murine fibroblasts are normally nontumorigenic in nude mice; however, constitutive expression of mutant p53 in these cells makes them tumorigenic, showing a clear gain of function (24, 54).
We and others have shown that mutant p53 can transactivate cellular promoters of growth-related genes, such as human proliferating cell nuclear antigen (PCNA), multiple drug resistance gene 1 (MDR-1), and c-myc, among others, in vivo in p53-null cells, such as Saos-2 (derived from a human osteosarcoma) (13, 19, 24, 31, 54, 61; reviewed in references 12 and 71). Transactivation by tumor-derived p53 mutants does not require a WT p53 DNA-binding site on the promoter. Structure-function analysis indicates that domain requirements for mutant p53-mediated transactivation and WT p53-mediated transactivation are different, suggesting that the molecular mechanisms of transcriptional activation by WT and mutant p53 are not identical (18, 54, 58, 61).
In experiments with 10(3) fibroblasts, cells stably expressing a double mutant of the tumor-derived p53-D281G mutated at amino acids 22 and 23 (L22Q/W23S), eliminating the transactivation capability of the protein, were nontumorigenic when injected into nude mice, whereas cells expressing the transactivation-capable p53-D281G were tumorigenic (58). In similar experiments, we demonstrated that a deletion mutant of p53-D281G that eliminated the oligomerization domain (p53-D281G deletion 393 to 327) could not transactivate the MDR-1 promoter (transactivated by full-length p53-D281G) (54), and 10(3) cells expressing this mutant also failed to induce tumor formation when injected into nude mice. These experimental results, therefore, show that the transactivation function of p53 mutants is required for oncogenicity.
Tumor-derived p53 mutants have been shown to increase resistance of cultured cells to apoptosis induced by DNA-damaging agents, depending on the nature of the mutation and the drug used (7), suggesting that gain of function can influence the outcome of cancer therapy. Although the MDR-1 pathway cannot explain resistance against all the agents tested (14, 71), the transcriptional activation of genes involved in chemoresistance or antiapoptotic activity is likely to be a contributing factor (14). Using murine p53-null cells, it has also been shown that mutant p53 expression inhibits p53-independent apoptotic pathways; this activity is blocked, however, by the inclusion of actinomycin D, a potent inhibitor of transcription, suggesting further that mutant p53-mediated transcriptional activity may be involved in the antiapoptotic function (56). Thus, it would be very important to identify genes transcriptionally activated by tumor-derived p53 mutants to decipher whether antiapoptotic genes are targeted. Identifying mutant p53 target genes could also unearth genes that are involved in chemoresistance, eventually targeting those gene products for chemotherapy.
The transcription factor NF-κB has been implicated in cell survival and chemoresistance (3, 48, 77). NF-κB2 belongs to the NF-κB/Rel gene family. It is synthesized as an approximately 100-kDa protein that is processed before it can bind DNA either as a homodimer or as a heterodimer with other members of the NF-κB/Rel family. The unprocessed form behaves as an IκB-like protein, negatively regulating the activation of NF-κB2 (57). Earlier, we have demonstrated that in murine 10(3) cells, mutant p53 expression up-regulates NF-κB2 (18).
To test the hypothesis that mutations in p53 not only disable the growth suppressor function of p53 but also turn it into a transcriptional activator of genes that aid in aggressive growth of cancer cells under adverse conditions, we generated stable cell lines expressing p53 mutants p53-R175H, -R273H, and -D281G using human p53-null H1299 lung carcinoma and 21PT breast carcinoma cells. In contrast to what is seen for vector-transfected cells, the presence of mutant p53 induces chemoresistance. In order to define the molecular pathway by which mutant p53 achieves these effects, microarray analyses were conducted using Affymetrix U95Av2 arrays, which survey approximately 12,000 genes. When statistical significance analysis and clustering was used, a striking pattern emerged, revealing that all three p53 mutants induced expression of approximately 100 genes, including those involved in cell growth, survival, adhesion, and angiogenesis (e.g., E2F-5, NF-κB2, angiopoietin 1, integrin α6), which are inhibited or unaffected by WT p53. Quantitative real-time PCR (QPCR) analysis of representative genes verified the microarray data. Our evidence reveals that mutations in p53 not only disable the growth suppressor function of p53 but also evoke a pattern of transcriptional regulation that is the inverse of that for the WT protein, thus turning it into a transcriptional activator of potentially oncogenic genes. We demonstrate that NF-κB2 overexpression could substitute for mutant p53 overexpression in inducing a decreased sensitivity to etoposide in H1299 cells. Transfection of H1299 cells expressing p53-R175H with small interfering RNA (siRNA) specific for NF-κB2 made these cells more sensitive to etoposide. We also show that mutant p53-expressing cells have an activated NF-κB2 pathway. Thus, transactivation of NF-κB2 may be a step used by tumor-derived mutant p53 in inducing etoposide resistance in cancer cells.
MATERIALS AND METHODS
Generation of stable cell lines.
Stable cell lines were generated after transfection of p53-null H1299 and 21PT cells with mutant p53 expression plasmids (or expression vector alone) containing a neomycin resistance gene as described previously (54). The following p53 mutant cell lines were generated using G418 (400 μg/ml for H1299 cells and 500 μg/ml for 21PT cells) selection: p53-R175H, p53-R273H, and p53-D281G (clone numbers are indicated at the end of the cell line). The p53-D281G (L22Q/W23S) clones were generated only in H1299 cells. H1299 cells were also transfected with pIRES-puro3 vector alone or with pIRES-puro3 containing NF-κB2, p53-R175H, p53-R273H, p53-D281G, or p53-D281G (L22Q/W23S) cDNA. These are designated H-IRES. Cells were selected with 2.5 μg/ml of puromycin for 2 weeks. The resulting colonies were then pooled and used for further assays. 10(3) cell clones were generated earlier with 200 μg/ml G418 (54). Hip53, a wild-type p53-inducible cell line, was generated earlier (30). None of the cells lines used in our assays, i.e., Saos-2, H1299, 21PT, or 10(3), express detectable p53 (4, 7, 24, 58). 21PT expresses an N-terminally truncated and functionally inactive p53 (16).
Recombinant adenoviruses, adenoviral infection, and RNA preparation.
Recombinant adenoviruses expressing WT p53 and β-galactosidase were generated by the laboratory of Kristoffer Valerie, Massey Cancer Center (Richmond, Va.), and by the lab of Atsushi Miyanohara, UCSD Gene Therapy Program. In a 10-cm dish, 3 × 106 cells were infected with the recombinant adenoviruses expressing either WT p53 or β-galactosidase at a multiplicity of infection of 10 adenoviruses per cell. At 20 h, RNA was extracted for microarray analysis. Total RNA was isolated using Trizol (Invitrogen) reagent following the manufacturer's recommendations and checked by 1.2% agarose Tris-borate-EDTA gel electrophoresis.
Drug sensitivity assays.
H1299 (or 21PT) cells stably expressing p53- R175H, -R175H (L22Q/W23S), -R273H, -D281G, and -D281G (L22Q/W23S) or transfected with vector alone (as indicated in the figures) were plated at equal densities and treated with final concentrations of 3 to 6 μM etoposide (Sigma) for 48 h as specified in the text. Chemosensitivities of the cells were then measured in two different ways: (i) by determining colony formation after 2 to 3 weeks and (ii) by determining the level of bromodeoxyuridine (BrdU) incorporation. For the colony formation assay, cells were plated at a density of 5 × 104 cells per 10-cm dish and exposed to a final concentration of 6 μM etoposide for 48 h. After treatment, plates were washed and the medium was replaced. The surviving cells were allowed to form colonies for 2 to 4 weeks with periodic changes of medium. Colonies were fixed with methanol, stained with methylene blue, and counted as described earlier (18). Control plates were plated at a density of 5 × 103 cells per 10-cm plate and treated with an equal amount of dimethyl sulfoxide (DMSO). Control plates were assessed for plating efficiency and DMSO effects on cell growth. For the BrdU incorporation assay, cells were plated at a density of 1 × 106 per 10-cm dish in the presence of etoposide (6 μM final concentration) or DMSO as the control for 48 to 72 h. Cells were incubated in the presence of 10 μM BrdU for 40 min. After washing with Dulbecco's phosphate buffer solution (DPBS), cells were trypsinized, counted, and fixed by vigorous vortexing, and dropwise addition of cold absolute ethanol was performed. Cells were then stored at 4°C for at least 18 h. Samples were washed with DPBS plus 0.5% bovine serum albumin to remove ethanol. Fixed cells were treated with 400 μl of 2N HCl for 20 min, which was followed by incubation in 0.1 M sodium borate, pH 8.5, for 2 min. Samples were washed and resuspended in DPBS plus 0.5% bovine serum albumin and 0.5% Tween 20, which was followed by incubation with anti-BrdU antibody coupled with fluorescein isothiocyanate for 1 h in the dark with tilting. After being washed to remove excess antibody, cells were resuspended in propidium iodide staining solution for at least 1 h prior to florescence-activated cell sorting analysis (11, 32). Samples were gated for sub-G1 DNA-containing cells.
siRNA transfection.
Mutant p53-R175H-expressing H1299 cells (or cells of the control cell line HC-5) were plated at equal densities in 12-well plates (3 × 105). Cells were transfected 0 and 24 h after plating using siRNA against NF-κB2 (or a nonspecific control). Sequences used were as follows: for the control, siControl, 5′-CAU GUC AUG UGU CAC AUC ACT T-3′ and 5′-GAG AUG UGA CAC AUG ACA UGT T-3′; for NF-κB2, siNFκB2 #1, 5′-GAC AAG GAA GAG GUG CAG CTT-3′ and 5′-GCU GCA CCU CUU CCU UGU CTT-3′; and siNFκB2 #2, 5′-GCC CUG AGU GCC UGA AUC U-3′ and 5′-AGA UCC AGG CAC UCA GGG CTT-3′. Twenty-four hours after the second transfection, cells were trypsinized, counted, and plated in 10-cm plates at a density of 5 × 104 cells per plate (control plates were plated at a density of 5 × 103 cells per plate). Cells were then treated with 6 μM etoposide (final concentration) or an equivalent vehicle control (DMSO). Cells were allowed to form colonies for 2 to 4 weeks. Colonies were then fixed with methanol, stained with methylene blue, and counted as described earlier (18).
DNA microarray hybridization, data management, and analysis.
Expression profiles of mutant p53-expressing cells were compared with H1299 (or 21PT) cells stably transfected with vector alone. Cells infected with recombinant adenovirus expressing WT p53 were compared to cells infected with adenovirus expressing β-galactosidase as a control. All microarray hybridization analyses were performed in duplicate with Affymetrix U95Av2 chips by either the GeneChip Core lab at UCSD or Virginia Commonwealth University's Institutional Nucleic Acid Research facility. The U95Av2 array represents ∼12,000 human gene sequences that have been previously characterized. The general procedures for microarray hybridization and analysis are described elsewhere (78). Data analysis was done using Affymetrix Microarray Analysis Suite 5. Changes in gene expression were detected in terms of the statistical significance (S-score) of the change in expression for a given gene between two compared microarrays. S-score analysis takes into account signals detected by 16 multiple probe pairs for individual genes, as well as intensity-dependent and -independent noise (52). Briefly, the files generated by Affymetrix software (Microarray Analysis Suite 5) were filtered to eliminate genes with average intensity values of less than 50 in at least one of the samples, resulting in 2,417 genes for further study. The filtered data were then analyzed with the S-score program. S-scores are derived to have a mean value of zero (representing no change) with a standard deviation of 1. The S-scores generated were then analyzed for significance across replicate experiments by using a permutation method performed with a significance analysis of microarray program from Stanford University (80). The settings for this analysis were as follows: unlogged data, 300 permutations, a k nearest neighbor imputer of 10, and random number seed 123,456,789. Once the program reported the list of ranked genes, the “delta” value was adjusted to a stringent false discovery rate of 0.3%, resulting in the identification of 149 genes upregulated by all three p53 mutants in both cell lines. Clustering analysis was done using the Cluster and TreeView programs (http://rana.lbl.gov/) to provide a graphical display of the expression patterns (34). Genes reported by significance analysis of microarray were analyzed by hierarchical clustering with average linkage grouping. For our analysis, the arrays were not clustered. Functional grouping of the identified genes was done by manual editing of gene ontology categories obtained through the DAVID annotation tool (http://david.niaid.nih.gov/david/ease.htm) (45).
QPCR.
QPCR was conducted using a LightCycler system (Roche) as described previously (73). cDNA was synthesized using a Thermoscript reverse transcription-PCR system (Invitrogen). Primers were designed using OLIGO 5 software (Molecular Biology Insights) and synthesized by Sigma Genosys. Reactions were performed in triplicate utilizing SYBR green dye, which exhibits a higher fluorescence upon binding of double-stranded DNA. The QPCR primers used were as follows: for NF-κB2, 5′-GGG GCA TCA AAC CTG AAG ATT TCT-3′ and 5′-TCC GGA ACA CAA TGG CAT ACT GT-3′; for NF-κB1, 5′-CAC TTA GCA ATC ATC CAC CTT-3′ and 5′-AGC CCT CAG CAA ATC CT-3′; for estrogen receptor-binding protein site-associated antigen 9 (EBAG9), 5′-TGC CTT TTA TTC ATC AGT CTT C-3′ and 5′-CGG CTG CTC TCT TTT CTC T-3′; for integrin α6 (ITGA6), 5′-GTC CAG AGC CAA GGT CCA G-3′ and 5′-CTC AAT CGC CCA TCA CAA AA-3′; for minichromosome maintenance protein 6 (MCM6), 5′-ATC CCT CTT GCC AAG GAT TT-3′ and 5′-GAA AAG TTC CGC TCA CAA GC-3′; for transcription factor E2F-5, 5′-CCC CCA CCT GAT GAC CTC AC-3′ and 5′-CTG CCG GGG TAG GAG AAA GC-3′; and for proto-oncogene c-Syn, 5′-TGA ACA GCT CGG AAG GA-3′ and 5′-CCC AAT CAC GGA TAG AAA GT-3′.
NF-κB2 promoter PCR.
NF-κB2 promoter PCR was performed using the conditions described by Lombardi et al. (59), with cDNA prepared from H1299 cells expressing mutant p53 (or with the control cell line HC-5). The promoter-specific primer pairs used were as follows: for presumptive promoter 1 (P1), 5′-AGA GCA GCA GCT GCA CAC AG-3′ (forward) and 5′-GCT CTG TCT AGT GGC TCC-3′ (reverse); and for P2, 5′-AAC TCC GGA TCT CGC TCT CC-3′ (forward) and 5′-GCT CTG TCT AGT GGC TCC-3′ (reverse). Note that the reverse primers for the P1 and P2 promoters are the same, while the forward primers are different, thus generating two distinct PCR products of 153 bp (P1) and 116 bp (P2).
Cloning of presumptive promoter.
The P2 sequence of NF-κB2 was cloned in the pGL3 basic vector upstream of the luciferase gene by use of the available genomic sequences in the NCBI database and genomic PCR using a commercial kit (Invitrogen). The primers used were as follows: 5′-CGC TAG CAA CTC GCG CCT GGT GTC CGT-3′ and 5′-CCA AGC TTG CGG CAT GAC TCA CTG GGT TGT AG-3′. These primers formed NheI and BglII sites, respectively, at the ends. These restriction enzymes indicate the locations of the inserts in the multiple cloning sites of the pGL3 reporter vector. The P2 promoter was cloned using Saos-2 cells and contains a single mismatch (A to G). We found the same mismatch in multiple analyzed clones and did not investigate any further. However, it should be noted that an analysis with the program TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) indicates that the sequence does not fall on any known transcription factor-binding site (data not shown).
Transient transcriptional assays.
Promoters were tested by transient transcriptional assays using p53-null Saos-2 (human osteosarcoma) cells. Transfections and luciferase assays were carried out as described previously (18). Saos-2 cells were plated at equal densities in 24-well plates and transfected with 200 ng of the promoter luciferase construct, 200 ng of Renilla luciferase control (TK.Renilla.luc, used for normalization of transfection), and 500 ng of the corresponding p53 expression plasmid 24 h after plating. H1299 cells stably expressing mutant p53 were plated at a density of 3 × 105 cells/well in 6-well plates and transfected with 500 ng of a reporter plasmid containing NF-κB sites upstream of the luciferase gene (kind gift from Valentine Andela of the University of Rochester) (2). Transfections were carried out using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's recommendations. Cell lysates were prepared 48 h after transfection using reporter lysis buffer (Promega). Luciferase activity was detected using a luminometer from Turner Designs. Transcriptional assays were repeated at least three times.
Electrophoretic mobility shift assay.
Nuclear extracts were prepared in the following manner. Cells grown to confluence were washed twice in phosphate-buffered saline and lysed in 700 μl of Dignam buffer (23) (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 1% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride plus protease inhibitor [cocktail III from Calbiochem]) by adding the buffer directly onto the plate and incubating on ice for 20 min. Cells were then scraped and transferred into a prechilled Eppendorf tube and further incubated on ice for 20 min. Nuclei were then pelleted at 4,000 rpm at 4°C for 10 min, and the supernatant (cytoplasmic extract) was stored at −80°C. The nuclei were resuspended in 100 μl of nuclear extraction buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.1 mM EDTA, 1% Nonidet P-40 plus protease inhibitors) and incubated on ice for 1 h. The samples were pelleted at 10,000 rpm at 4°C for 10 min, and the supernatant (nuclear extract) was stored at −80°C. Protein concentrations were determined using Bio-Rad's protein assay reagent, and equal amounts of protein were used in each binding reaction. Electrophoretic mobility shift assays were performed as described previously (5) using 15 μg of nuclear extract protein per binding reaction. Reactions were preincubated at 25°C for 20 min prior to the addition of the oligonucleotide probe. After preincubation, approximately 20,000 counts of labeled probe were added to each reaction tube and incubated at 25°C for an additional 20 min. For the supershift experiments, antibodies (1 μg) were added after incubation with the probe and incubated for an additional 15 min. Samples were then loaded and complexes separated on a 6% 0.5× Tris-borate-EDTA gel run at 200 V for approximately 1.5 to 2 h. The gel was then fixed, dried, and exposed to film. Probes were annealed and [32P]dCTP labeled using Klenow fragment as described previously (69). The oligonucleotide sequences used were as follows: for the WT probe, 5′-GAT CCG AGG GCT GGG GAT TCC CAT CTC CCA CGT TTC ACT TCA-3′ and 5′-AGC TTG AAG TGA AAC GTG GGA GAT GGG AAT CCC CAG CCC TCG-3′; and for the mutant probe, 5′-GAT CCG AGG GCT TTT TAT GAA AAT CTC CCA CGT TTC ACT TCA-3′ and 5′-AGC TTG AAG TGA AAC GTG GGA GAT TTT CAT AAA AAG CCC TCG-3′. For competition studies, an unlabeled WT probe was used as the specific competitor and the mutant probe was used as the nonspecific competitor at both 20× and 40× molar excess. Hip53 cells were induced with 10 μM ponasterone A. NF-κB1 (p50)-, NF-κB2 (p52)-, and p53-specific antibodies were used for the supershift experiment (sc-114X from Santa Cruz, product number 4185 from the Rockland catalog, and Ab-6 from Oncogene, respectively).
Western blotting.
NF-κB2 and Sp1 levels were detected using antibodies from Santa Cruz Biotechnologies (sc-7386 and sc-59, respectively). Actin levels were detected using an antibody from Sigma (AC-15). NF-κB1 (p105/p50), RelA, and c-Rel were detected using an antibody kit from Calbiochem (catalog number ASK20). p53 was detected using the p53 antibody PAb 1801. Western blots were developed by the ECL method (Amersham). In Western blotting for the siRNA and in the colony-forming assay with the H-IRES NF-κB2 (p100/p52) cell line, NF-κB2 protein was detected using an antibody from Upstate Biosciences (catalog number 05-361).
RESULTS
H1299 cells expressing p53-R175H, p53-R273H, and p53-D281G withstand etoposide better than those stably transfected with vector alone.
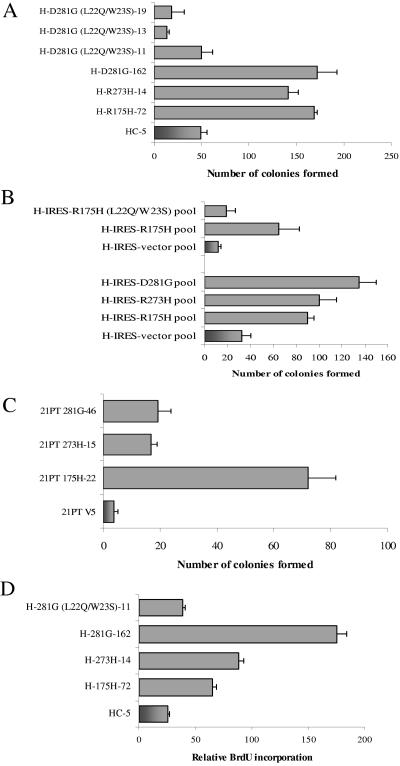
Earlier work has shown that expression of mutant p53 in cells results in a lessened sensitivity to chemotherapeutic drugs (7). To determine the mechanism by which mutant p53 induces this decreased chemosensitivity and to test whether this is dependent on mutant p53-mediated transactivation, H1299 lung cancer and 21PT breast cancer cells stably expressing p53-R175H, -R273H, or -D281G (or transfected with vector alone) were generated. The mutant p53-expressing H1299 cells were then tested for sensitivity to etoposide by use of a colony formation assay as described in Materials and Methods. Data shown in Fig. 1A indicate that cells expressing p53 mutants p53-R175H, -R273H, and -D281G are less sensitive to etoposide than the cells stably transfected with vector alone. This is in accord with results published by others using H1299 and other cell lines (7). Expression of a transactivation-deficient mutant p53 (L22Q/W23S) in H1299 cells did not decrease the sensitivity of the cells to etoposide. These results are recapitulated with H1299 pooled cell lines expressing the same p53 mutants (Fig. 1B) and with 21PT (p53−/−) breast cancer cells (Fig. 1C). Furthermore, a BrdU incorporation assay using the H1299 stable transfectants shows increased DNA replication in cells expressing mutant p53 (but not the L22Q/W23S transactivation-deficient protein) after drug exposure (Fig. 1D). As shown, cell lines expressing the transactivation-deficient mutant have a disadvantage in etoposide-induced cell death, suggesting a correlation between mutant p53-mediated transactivation and the loss of chemosensitivity observed in cancer cells expressing p53 mutants.
FIG. 1.
Expression of mutant p53 in H1299 and 21PT cells imparts decreased sensitivity to etoposide. (A to C) Cells were plated at 50,000 cells/plate and treated with 6 μM etoposide (final concentration) for 48 h. After treatment, cells were washed with Hanks balanced salt solution, and the surviving cells were allowed to recuperate for 3 weeks with periodic changes of medium. Colonies formed were fixed with methanol, stained with methylene blue, and counted. The data shown are representative of three independent experiments run simultaneously; colony numbers were adjusted to account for plating differences based on control plates. Control plates were plated at 1/10 the density and treated with DMSO for 48 h. Clones of H1299 cells expressing mutant p53-R175H, -R273H, -D281G, p53-D281G (L22Q/W23S), or vector alone (panel A), pools of H1299 cells expressing p53-R175H, -R175H (L22Q/W23S), -R273H, -D281G, or vector alone (panel B), and clones of 21PT cells expressing mutant p53-R175H, -R273H, -D281G, or vector alone (panel C) were used. (D) H1299 cells expressing mutant p53-R175H, -R273H, -D281G, the transactivation-deficient p53-D281G (L22Q/W23S), or vector alone were exposed to 6 μM etoposide (final concentration) for 48 h. After treatment, cells were allowed to incorporate BrdU for 40 min, washed, and fixed. Prior to florescence-activated cell sorting analysis, cells were stained with a fluorescein isothiocyanate-coupled anti-BrdU antibody and propidium iodide as described previously (32).
H1299 and 21PT cells expressing p53-R175H, -R273H, and -D281G overexpress a common set of about 150 genes, including NF-κB2.
Since data described earlier suggest that mutant p53-mediated transactivation is necessary for its growth-promoting functions, we planned to identify genes that are transactivated by common p53 mutants in an effort to decipher the pathway used by the mutants to perturb drug sensitivity, which may impact chemotherapy. Microarray hybridization analysis of three H1299-derived and three 21PT-derived cell clones expressing p53 mutants, i.e., p53-R175H, -R273H, and -D281G, was performed. The gene expression profiles of vector-transfected H1299 and 21PT cells were used as controls. Affymetrix U95Av2 arrays (HG-U95Av2) representing ∼12,000 sequences were used. Changes in gene expression were detected in terms of the statistical significance (S-score) of the change in expression for a given gene between two compared microarrays. S-score analysis takes into account signals detected by 16 multiple probe pairs for individual genes, as well as intensity-dependent and -independent noise (52). A common set of approximately 150 genes was found to be significantly upregulated by all three p53 mutants in both the cell lines.
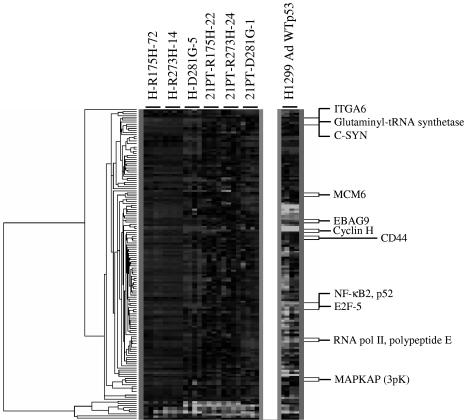
We have also performed microarray analysis after infection of H1299 cells with recombinant adenoviruses expressing WT p53 or β-galactosidase as described in Materials and Methods with the RNA harvested 20 h postinfection. In order to further focus on the genes implicated in oncogenesis, the common set of 150 genes identified above was compared to the transcriptional profile of WT p53 (Fig. 2). Approximately 100 of those genes were found to be inhibited or unaffected by WT p53 but up-regulated in the presence of mutant p53 (Table 1 shows a partial list). This common set of genes includes genes involved in cell growth, survival, and adhesion (e.g., E2F-5, NF-κB2, and integrin α6, etc.), which are often implicated in cancer. The up-regulation of gene expression observed in microarray analysis was verified by QPCR for a representative group of gene targets (Fig. 3A). As expected from earlier transient transcriptional analysis, expression of the triple mutant p53-D281G (L22Q/W23S) in H1299 cells did not result in up-regulation of the same genes (58). Combined data presented in Fig. 1 and 3 indicate that transactivation by tumor-derived p53 mutants is related to the mutant protein's ability to decrease sensitivity toward chemotherapeutic drugs.
FIG. 2.
Microarray clustering of H1299 and 21PT cells expressing mutant p53. Stable cell lines were generated from H1299 and 21PT to express mutant p53-R175H, -R273H, -D281G, or vector alone. RNA from selected stable clones was extracted and analyzed for gene expression using an Affymetrix U95Av2 array chip. Expression data were analyzed as described in Materials and Methods. The false discovery rate was 0.3%. Clustering of the identified genes reveals a common set of genes up-regulated by all three p53 mutants in both cell lines, suggesting the existence of a common pathway through which mutant p53 aids in oncogenesis. Genes and gene products listed in the figure are also listed in Table 1.
TABLE 1.
Microarray hybridization analysis of RNA from H1299 and 21PT cells expressing p53-R175H, -R273H and, -D281G in comparison to RNA from vector-transfected cells
| Function(s) | Gene or gene producta | Average S-score | GenBank no. | Effect of WT p53 |
|---|---|---|---|---|
| Amino acid and protein synthesis | Glutaminyl-tRNA synthetase | 1.677 | X54326 | Not affected |
| Spermidine synthase | 1.766 | M64231 | Repressed | |
| General metabolism | Human poly(ADP-ribose) synthetase | 1.277 | J03473 | Repressed |
| Ku (p70/p80) subunit | 1.081 | M30938 | Repressed | |
| Cell cycle | HsMcm6 | 1.960 | D84557 | Repressed |
| Cyclin B2 | 2.790 | AL080146 | Repressed | |
| Oncogenesis, transformation, invasion, metastasis | Cell adhesion molecule (CD44) | 1.841 | M59040 | Repressed |
| Developmentally regulated GTP-binding protein 1 | 1.877 | AJ005940 | Repressed | |
| Human cdc25A | 1.838 | M81933 | Repressed | |
| c-Syn proto-oncogene | 1.240 | M14333 | Repressed | |
| EBAG9 | 1.435 | AB007619 | Repressed | |
| B-myb | 1.304 | X13293 | Repressed | |
| Integrin alpha 6 | 2.747 | X53586 | Repressed | |
| DNA replication | DNA polymerase alpha | 1.971 | L24559 | Repressed |
| HsMcm6 | 1.960 | D84557 | Repressed | |
| HsMcm3 | 1.315 | D38073 | Repressed | |
| NAP (nucleosome assembly protein) | 1.205 | M86667 | Repressed | |
| Signal transduction | MAP kinase-activated protein (3pK) | 1.692 | U09578 | Not affected |
| MAP kinase-activated protein kinase 2 | 1.312 | U12779 | Not affected | |
| Survival, apoptosis | NF-κB, p52 | 1.964 | S76638 | Repressed |
| EBAG9 | 1.435 | AB007619 | Repressed | |
| Transcription | NF-κB, p52 | 1.964 | S76638 | Repressed |
| Transcription factor E2F-5 | 1.262 | D82348 | Repressed | |
| RNA polymerase II, polypeptide E | 2.012 | D38251 | Repressed |
Only up-regulated genes and gene products are listed. Functional classification of genes was performed by manual editing of the output file obtained using the DAVID (http://david.niaid.nih.gov/david/ease.htm) annotation tool (45). Bold lettering indicates up-regulation has been confirmed by QPCR (in this publication or previously [70]). A complete list of identified genes can be found at http://www.people.vcu.edu/∼sdeb. MAP, mitogen-activated protein.
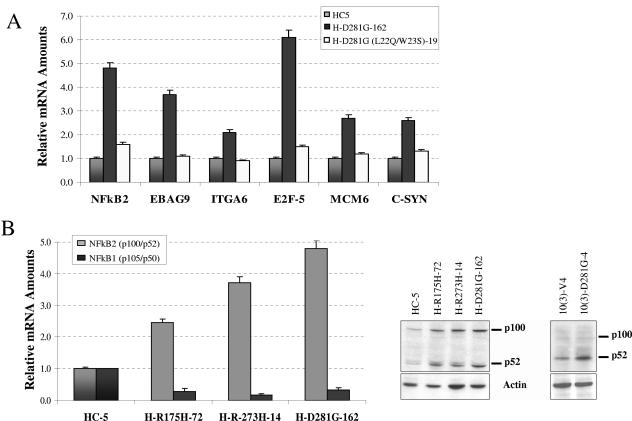
FIG. 3.
Expression of mutant p53 up-regulates genes in H1299 cells. (A) mRNA was extracted from exponentially growing plates of the indicated cell lines, and cDNA was prepared. The cDNA was analyzed by QPCR using gene-specific primers for various genes identified by microarray analysis. The degree of expression was quantitated using a relative standard curve and normalized to an internal control (brome mosaic virus) corresponding to the cDNA batch as described previously (69, 73). The normalized mRNA levels in the HC-5 control cell line were arbitrarily set to 1, and the relative differences (n-fold) were calculated. Additional mutations at amino acids 22 and 23 decrease up-regulation by the mutant p53 protein. Expression levels of NF-κB2, estrogen receptor binding site-associated antigen 9 (EBAG9), integrin α6 (ITGA6), the transcription factor E2F-5, minichromosome maintenance protein 6 (MCM6), and the proto-oncogene c-Syn have been tested. (B) On the left, results from an analysis of cDNA from mutant p53-expressing H1299 cells by QPCR using gene-specific primers for NF-κB1 and NF-κB2 expression are shown. NF-κB2 but not NF-κB1 is up-regulated by mutant p53. Shown at the right is a Western blot demonstrating up-regulation of NF-κB2 in mutant p53-expressing H1299 cell lines and in a murine cell line [10(3)] stably transfected to express vector alone (V4) or mutant p53-D281G. Cell extracts were prepared using reporter lysis buffer (Promega). NF-κB2 was detected using a specific antibody as specified in Materials and Methods.
Interestingly, the group of up-regulated genes includes a member of the NF-κB family, NF-κB2 (p100/p52), which we have shown earlier to be up-regulated in murine 10(3) cells constitutively expressing tumor-derived p53-D281G (18). Figure 3B (left panel) shows QPCR analysis demonstrating up-regulation of expression of NF-κB2 (p100/p52) in different mutant p53-expressing cell clones. This up-regulation was also confirmed by Western blot analysis using a specific antibody against NF-κB2 (p52) in the H1299 mutant p53-expressing cell lines (Fig. 3B, right panel) (18). Expression levels of NF-κB1 (p105/p50) were also determined by QPCR and found to be unaffected (Fig. 3B). NF-κB2 has been implicated in survival and antiapoptosis functions of cells (3, 48, 77). It is therefore possible that a pathway used by mutant p53 to reduce drug sensitivity utilizes the NF-κB2 route.
Specific NF-κB2 promoters are upregulated by mutant p53 in H1299.
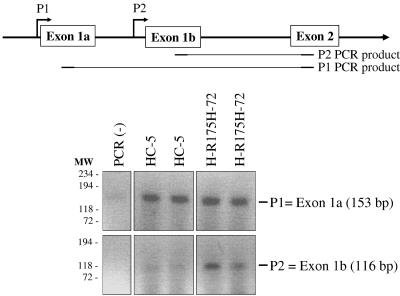
NF-κB2 has been shown to have two different promoters (P1 and P2), both of which can be active in a cell (59). Using promoter-specific primers, we have performed PCR using the conditions described by Lombardi et al. (59) with cDNA prepared from H1299 cells expressing p53-R175H or vector control (HC-5). The data shown in Fig. 4 demonstrate that in H1299 cells, mutant p53 up-regulates NF-κB2 (p100/p52) expression using the P2 promoter, whereas the P1 promoter remains unaffected.
FIG. 4.
Mutant p53 up-regulates NF-κB2 using the P2 promoter in H1299 cells. Using promoter-specific primers, PCR was conducted using the conditions described by Lombardi et al. (59) with cDNA prepared from clones of H1299 cells expressing mutant p53-R175H (or the vector control cell line HC-5). PCR primers are designed to detect mRNA products resulting from the two NF-κB2 promoters. PCRs were performed in duplicate. The data shown demonstrate that in H1299 cells, mutant p53 up-regulates NF-κB2 using the P2 promoter, whereas the P1 promoter remains unaffected.
Mutants of p53 transactivate the NF-κB2 P2 promoter in vivo in transient transcriptional analysis.
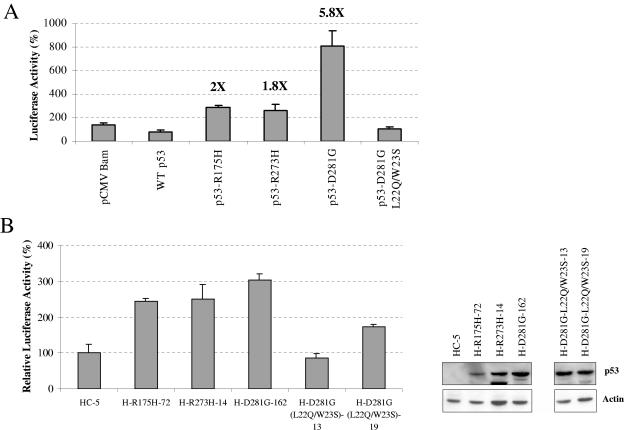
After ascertaining that mutant p53 up-regulates the P2 promoter of NF-κB2 (p100/p52), we tested whether the promoter is transactivated by p53 mutants in transient transcriptional analysis. We cloned the promoter sequences upstream of the luciferase gene in pGL3 vector as described in Materials and Methods using published sequence information (59). Transient transcriptional assays were carried out in p53-null Saos-2 cells as described in Materials and Methods. Transcriptional results shown in Fig. 5A demonstrate that tumor-derived p53 mutants p53-R175H, -R273H, and -D281G transactivate the NF-κB2 P2 promoter, while WT p53 represses, in agreement with our in vivo expression data. Furthermore, transfection of cells with mutant p53-D281G (L22Q/W23S) did not show up-regulation of the NF-κB2 P2 promoter (Fig. 5A).
FIG. 5.
Mutant p53 up-regulates NF-κB2 P2 promoter. (A) Saos-2 cells were transfected with 200 ng of pGL3 vector containing the NF-κB2 P2 promoter upstream of the luciferase reporter gene, 200 ng of TK.Renilla.luc control plasmid, and 600 ng of the indicated p53 expression plasmid. Cells lysates were prepared 48 h after transfection, and luciferase activity was detected using a dual luciferase reporter assay system (Promega). Reporter luciferase readings were normalized based on TK.Renilla.luc control plasmid readings. Inductions (n-fold) for p53-R175H, -273H, and-D281G are shown in bold. (B) H1299 cells expressing mutant p53-R175H, -R273H, -D281G, -D281G (L22Q/W23S), or vector alone were transfected with 500 ng of a reporter plasmid containing five NF-κB sites. Cells were harvested 48 h after transfection, lysates were prepared, and luciferase activity was detected. Cell extracts were normalized based on total protein concentration; the vector reading was arbitrarily set to 100. At the right, an inset shows Western blot analysis demonstrating similar levels of p53 expression in the cell lines. Protein amounts were normalized based on total protein concentration.
NF-κB pathway is activated in cells expressing mutant p53.
To determine whether expression of mutant p53 results in activation of the NF-κB pathway, we assayed NF-κB-specific transcription with a synthetic construct containing five NF-κB binding sites upstream of a TATA box cloned upstream of a luciferase reporter gene (2). If the NF-κB pathway is active, more NF-κB2 should result in a higher level of promoter activity in cells expressing the p53 mutants. As expected, cells expressing mutant p53-R175H, -R273H, and -D281G had levels of promoter activity higher than that of the HC-5 cell line (stably transfected with vector alone) (Fig. 5B). This demonstrates that the NF-κB pathway is more active in cells expressing mutant p53 and that this activation is dependent on transactivation by tumor-derived p53 mutants. Western blot analysis showed similar levels of expression of p53 in these cells (Fig. 5B, right inset). Interestingly, at least one of the mutant p53 target genes, c-myc, has also been found to be a target of NF-κB, suggesting the possibility that mutant p53 may be transactivating c-myc through NF-κB (25). However, it is also possible that both mutant p53 and NF-κB independently up-regulate c-myc expression.
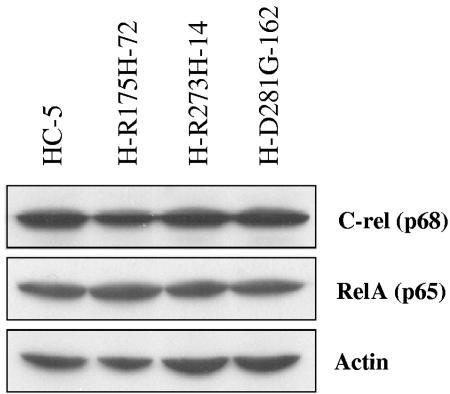
Since QPCR data showed that mutant p53 up-regulates NF-κB2 but not NF-κB1, we have also looked at protein expression levels of RelA and c-Rel, two other members of the NF-κB protein family. Western blot analyses using extracts from vector control H1299 cells (HC-5) and H1299 cells expressing mutant p53 show no appreciable difference at the protein level for either of these proteins (Fig. 6). The experimental results so far show that p53 mutants specifically up-regulate NF-κB2 gene expression.
FIG. 6.
Mutant p53 does not up-regulate other members of the NF-κB family. H1299 cells expressing the various p53 mutants were grown to confluence and harvested using reporter lysis buffer (Promega) following the manufacturer's recommendations. RelA and c-Rel were detected in these cell lines to determine expression levels and found to remain unchanged. Actin was used as a loading control.
H1299 cells expressing mutant p53 show increased NF-κB DNA binding.
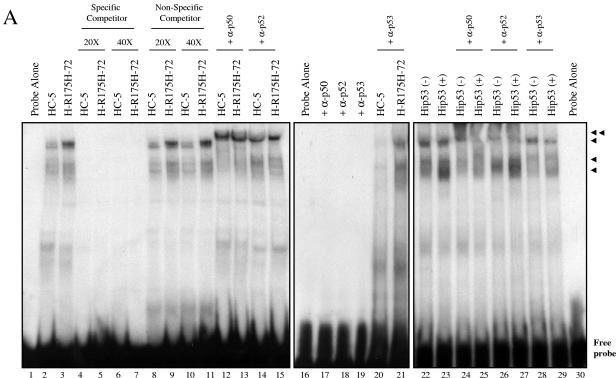
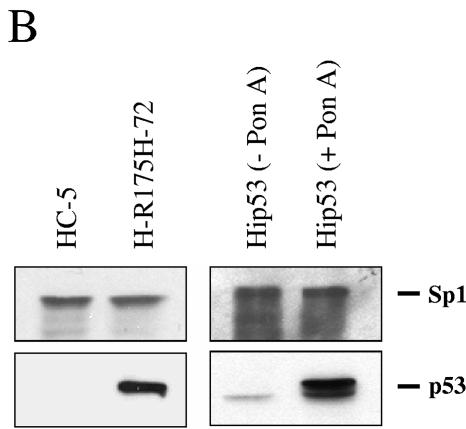
We performed a DNA-binding assay to determine whether NF-κB binding activity in cells expressing mutant p53 was different from that in the control cells. Nuclear extracts from HC-5 control and mutant p53-R175H-expressing H1299 cells were prepared, and binding reactions were performed as described previously (5, 23, 69, 83) using double-stranded DNA oligonucleotide probe containing the NF-κB DNA-binding site. The experimental data suggest that there is an increase in DNA-binding activity in cells expressing the mutant p53-R175H (Fig. 7). Antibody supershift using specific antibodies against p50 or p52 suggests that the complexes formed contain p50 or p52 (Fig. 7). This supershift is not observed in the presence of antibody alone (lanes 17 to 19). Thus, data shown so far indicate that mutant p53 expression leads to an activation of the NF-κB pathway. In parallel, using a WT p53 ecdysone (ponasterone A)-inducible cell line (30), similar DNA-binding reactions were carried out. Induction of WT p53 resulted in increased DNA binding. Unlike in the case of mutant p53-expressing cells, these complexes contained p50, in agreement with the results reported by Bohuslav et al. (8), but did not contain p52 (compare lanes 12 to 15 with lanes 24 to 27). Sp1 and p53 were detected to determine equal loading of protein and proper WT p53 induction using specific antibodies as indicated (Fig. 7B).
FIG. 7.
H1299 cells expressing mutant p53-R175H show increased binding to the NF-κB site. (A) Nuclear extracts of HC-5 and H-R175H were incubated as described in Materials and Methods with a 32P-labeled probe containing the sequence of the NF-κB DNA-binding site. Competition studies were done using a specific competitor (lanes 4 to 7) and a nonspecific competitor (lanes 8 to 11) at both 20× (lanes 4, 5, 8, and 9) and 40× (lanes 6, 7, 10, and 11) molar excess. The single arrows indicate the DNA complexes containing NF-κB complexes. Increased NF-κB activity is observed in the presence of mutant p53 (lanes 2, 3, and 8 to 11). The double arrow indicates the supershifted complex in the presence of antibodies specific for NF-κB1 (p50), NF-κB2 (p52), and p53 (lanes 12 to 15, 20 to 21, and 24 to 29, respectively). Equal amounts of protein were added to each lane. (B) In a parallel experiment, nuclear fractions prepared for the DNA-binding experiment were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Sp1 and p53 were detected by Western blotting to evaluate the equal loading of protein and the presence of p53 during the shift assay experiment.
Overexpression of NF-κB2 in H1299 cells leads to loss of sensitivity to etoposide.
Since the transcription factor NF-κB has been implicated in cell survival and antiapoptotic activity induced by chemotherapeutic drugs (3, 48, 77), we tested whether NF-κB2 overexpression in H1299 cells can substitute for mutant p53 and lead to a relative loss of sensitivity to chemotherapeutic drugs.
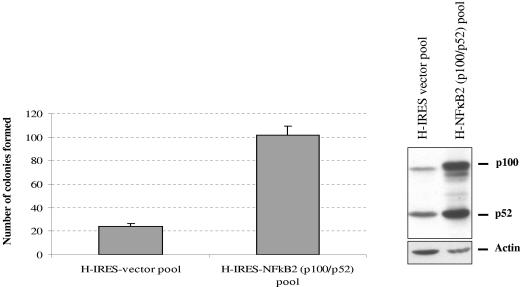
We cloned the human NF-κB2 (p100/p52) gene (42) in pIRESpuro3 (Clontech), a bicistronic expression vector under the cytomegalovirus immediate-early promoter, in which the NF-κB2 gene shares the same promoter with the puromycin resistance gene. Because of the bicistronic nature of this cloning vector, most of the puromycin-resistant colonies express the gene of interest. Using this NF-κB2 expression plasmid and the empty expression vector, we generated pools of H1299 cells expressing NF-κB2 (or stably transfected with vector alone) (Fig. 8, right panel). Cells were exposed to 6 μM etoposide (final concentration) for 48 h. Overexpression of NF-κB2 in these cells led to an increase in the number of surviving cells over that for the vector control cell line (Fig. 8, left panel). The data presented demonstrate that NF-κB2 overexpression in H1299 cells imparts a lack of sensitivity to etoposide. That mutant p53 expression in H1299 cells leads to induction of NF-κB2 expression as well as a lack of sensitivity to etoposide suggests that mutant p53 desensitizes the cells to etoposide via the NF-κB2 signal transduction pathway.
FIG. 8.
Expression of NF-κB2 confers H1299 cells with a decreased sensitivity to etoposide. H1299 cells stably expressing NF-κB2 were treated with 6 μM etoposide (final concentration) for 48 h. After treatment, the surviving cells were washed and allowed to form colonies for 3 weeks with periodic changes of medium. Colonies were then fixed in methanol, stained, and counted. The data shown are representative of three independent experiments; colony numbers were adjusted to account for plating differences based on control plates. Control plates were plated at 1/10 the density and treated with DMSO. Expression of NF-κB2 protein was checked by Western blotting (right). Actin was used as a loading control.
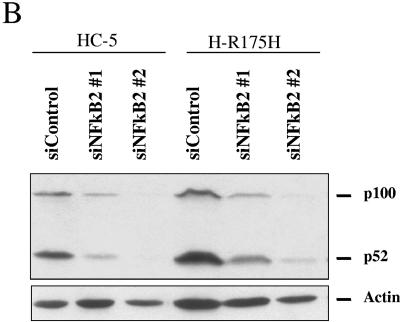
NF-κB2-specific siRNA reduces the level of NF-κB2 in H1299 cells expressing p53 and inhibits mutant p53-induced lack of sensitivity towards etoposide.
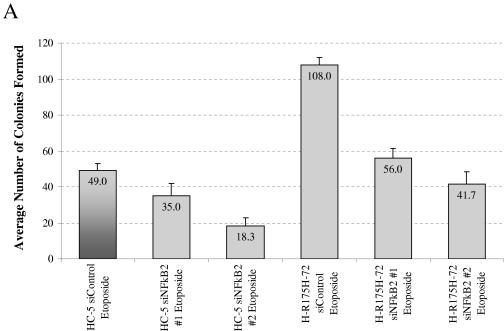
Observations reported in Fig. 1, 3, 5, and 7 suggest that mutant p53 may increase chemoresistance of cells by inducing overexpression of NF-κB2. We tested this notion by using siRNA capable of reducing expression of NF-κB2 specifically and determined whether this would lead to increased etoposide sensitivity in H1299 cells expressing mutant p53 (-R175H). We reasoned that a decrease in NF-κB2 protein levels during the incubation period with etoposide would increase the sensitivity of the cells to the drug. Two different siRNAs against NF-κB2 were used. After transfection of siRNA specific for NF-κB2, the level of NF-κB2 protein is significantly reduced (Fig. 9B). The parallel nonspecific siRNA had little or no influence (data not shown). A reduction in the NF-κB2 level led to a reduction in the number of surviving cells (colonies formed) after etoposide treatment, regardless of the p53 status in the cells. The average number of colonies formed is depicted in the figure. In parallel, cells transfected with the same siRNAs were plated and treated with vehicle (DMSO) as indicated in Materials and Methods. Plating efficiencies differed by less than 10% (data not shown). The siRNA experimental data corroborate our idea that at least one pathway through which mutant p53 induces chemoresistance in cancer cells is by the up-regulation of the expression of genes such as NF-κB2.
FIG. 9.
siRNA against NF-κB2 increases the sensitivity of cells to etoposide. H1299 cells expressing mutant p53-R175H (or the control cell line HC5) were plated at equal densities in 12-well plates. Cells were transfected 24 and 48 h after plating by use of siRNAs against NF-κB2 (or a nonspecific control) and plated, as described in Materials and Methods, 24 h after the second transfection. Cells were then exposed to 6 μM etoposide (final concentration) or vehicle control (DMSO) for 48 h. Cells were allowed to grow and form colonies. Colonies were then fixed with methanol, stained with methylene blue, and counted. (A) Graphical representation of the results. The average numbers of colonies formed are depicted in the figure. Plating efficiencies differed by less than 10% (data not shown). (B) Western blot analysis of cells from a parallel experiment transfected with siRNAs to evaluate the effectiveness of the RNA interference constructs. Actin was used as a loading control.
DISCUSSION
The microarray analysis (Table 1) revealed a common set of genes modulated by p53 mutants p53-R175H, -R273H (two of the most common p53 mutants found in human cancer), and -D281G in H1299 and 21PT cells (6, 36, 40, 43, 44, 65, 71, 72). Our finding that all three p53 mutants transactivate a common set of genes in the absence of WT p53 provides highly significant information toward understanding the contribution of mutant p53 in the progression of cancer and suggests the existence of a common pathway utilized by p53 mutants.
A look at the sample of common genes up-regulated by the three mutants shown in Table 1 reveals that different groups of genes are activated; perhaps the most important and interesting ones, from the point of view of cancer research, are those involved in cell cycle control, oncogenesis, invasion, metastasis, DNA replication, cell survival, and transcription. The gene expression profile hints at which genes may be responsible for the aggressive phenotypes observed for human cancers exhibiting p53 mutations.
We confirm and extend the finding that the expression of mutant p53 in lung (Fig. 1A and B) and breast (Fig. 1C) cancer cells makes them less sensitive to the chemotherapeutic drug etoposide (7) and that this property of mutant p53 depends on its transactivation function (Fig. 1 to 3). We have identified NF-κB2 as a target gene up-regulated in mutant p53-expressing cells (Table 1). H1299 cells overexpressing NF-κB2 are less sensitive to etoposide (Fig. 8), indicating that NF-κB2 is involved in etoposide sensitivity. This is further corroborated by the observation that siRNAs directed against NF-κB2 increased etoposide sensitivity (Fig. 9A).
NF-κB2 is induced by p53-R175H, -R273H, and -D281G and repressed by WT p53 (Table 1). As mentioned earlier, NF-κB2 is also up-regulated by p53-D281G in murine 10(3) cells (18). 10(3) cells do not express p53 and are not tumorigenic when injected subcutaneously in nude mice; however, on constitutive expression of tumor-derived p53 mutants, these cells become tumorigenic (24). In general, NF-κB is a family of sequence-specific DNA-binding transcription factors (22). Although initially discovered to control gene expression of the immune system, its importance in regulating cell growth, development, apoptosis, and oncogenesis has become apparent. NF-κB acts as an antiapoptotic factor (39, 66). In agreement with this, it has been observed that inhibition of NF-κB leads to an increase of chemotherapeutic drug sensitivity. NF-κB2 belongs to this group of transcription factors, has been implicated in malignancies, and is involved in cell survival in the context of chemotherapeutic drugs (3, 17, 48, 57, 77, 85).
Dejardin et al. (20) studied the expression of NF-κB2 (p100/p52) in human breast cancer cell lines as well as in primary breast tumors and reported that p100 was overexpressed in tumor cells relative to its expression in human mammary epithelial cells. By use of coimmunoprecipitation, they showed that p100 interacts with p50 (NF-κB1)/p65 (RelA) in the cytoplasm and suggested that NF-κB2 could be involved in carcinogenesis by sequestering other NF-κB family member proteins. Thus, transactivation of NF-κB2 gene expression could be one crucial step by which mutant p53 induces oncogenic progression along with the observed chemoresistance reported here.
Although the exact molecular mechanism behind mutant p53 gain of function is yet to be completely clarified, transactivation of antiapoptotic genes remains a strong possibility. Tumor-derived p53 mutants p53-R175H, -R273H, and -D281G confer chemoresistance in the absence of WT p53. Since the mutant p53-D281G (L22Q/W23S) is defective in up-regulation of endogenous genes (Fig. 3A) and fails to decrease sensitivity to the chemotherapeutic drug etoposide (Fig. 1), the overall conclusion from our work is a direct relationship between chemoresistance induced by mutant p53 and its transactivation ability. This implies that p53 mutants decrease sensitivity towards drugs by transactivating expression of genes such as NF-κB2. Our work also demonstrates that expression of mutant p53 activates the NF-κB pathway (Fig. 5B and 7). The excess NF-κB2 produced may enter the nucleus and act on its target genes. This transcription factor has been shown to protect against a variety of cellular stress conditions (reviewed in reference 41). The exact mechanism by which NF-κB may help in generating resistance against chemotherapeutic drugs is not known. Furthermore, the mechanism by which NF-κB2 is activated in cells overexpressing mutant p53 is not clear at the moment. However, our work suggests that significant amounts of p100 become processed into p52 that may result in up-regulation of NF-κB-responsive genes. One implication of our work is that NF-κB2 may play an important role in the mutant p53-mediated oncogenic pathway in general.
The lack of sensitivity of cells expressing mutant p53 towards chemotherapeutic drugs has been explained by the neutralization of p73 and p63 (which are induced upon drug treatment) by tumor-derived p53 mutants (33, 47, 62, 74-76). Strong supporting evidence has been presented in this regard. Our data do not contradict these results but suggest an additional pathway. It is also possible that mutant p53, through its ability to hetero-oligomerize with p73 and p63, may somehow use them to transactivate genes such as NF-κB2. It is interesting to note that p53-R273H also induces chemoresistance in our hands, although this mutant apparently does not have an altered conformation of its DNA-binding domain (15) and therefore, according to some reports, does not bind to p73 or p63 efficiently (33, 47, 74, 75). The fact that the NF-κB2 pathway is activated in mutant p53-expressing cells (Fig. 5B) suggests that mutant p53 may be using this pathway also to decrease sensitivity of the cells to etoposide.
The mechanism of transactivation by p53 mutants is not yet clear. Studies done by others and us show that mutant p53-mediated transactivation does not require WT p53 DNA-binding sites, demonstrating that the mechanism of mutant p53 transactivation is distinct from that of the WT protein (18, 31, 54, 58, 61, 90). We envision at least three molecular explanations for the up-regulation of genes in the presence of mutant p53. (i) It is possible that mutant p53-mediated induction of mRNA, as judged by microarray or QPCR analysis, is the result of stabilization of RNA by a posttranscriptional mechanism. (ii) It may be that p53 mutants differ in their abilities to recognize and bind DNA-responsive elements on promoters, although there is no strong evidence so far for any mutant p53 response elements in the literature (71). In this case, mutant p53 proteins may possess a DNA-binding ability altogether distinct from that of WT. (iii) Alternatively, mutant p53 may become associated with other transcription factors, such as Sp1 or ATF/CREB, to activate promoters. Interestingly, WT p53 already has been shown to interact with these factors (9, 35, 38, 79). As such, differences in levels of transcription can be accounted for by the identity of the transcription factors involved and/or by the relative affinity of the mutant for the transcription factor. Recently, at least two groups have demonstrated localization of mutant p53 on the upstream sequences of genes modulated by mutant p53, suggesting the possibility that mutant p53 modulates gene transcription directly by being nucleated on the upstream regulatory sequences (69, 87). However, it is still not clear whether it binds DNA directly or not.
The identification of targets of transcriptional activation common to multiple p53 mutants is indicative of the existence of common or perhaps overlapping pathways to mutant p53-mediated tumorigenesis. Definition of this pathway and the molecular mechanisms dictating it will provide strong candidates for therapy development.
Acknowledgments
This work was supported by grants from NIH to Sumitra Deb (CA70712), Swati Palit Deb (CA74172), and Mike F. Miles (AA13678). Research described in this article was also supported by Philip Morris USA Inc. and by Philip Morris International (04-I176-01). Katherine E. R. Stagliano is supported by a dissertation award from the Susan G. Komen Breast Cancer Foundation (DISS0201749). Mariano J. Scian is supported by a predoctoral fellowship from NCI (F31 CA97520).
We thank Arnold Levine, Bert Vogelstein, Vimla Band, Satoshi Inoue, Valentine Andela, and Warner C. Greene for providing us with cells, plasmids, and technical advice.
REFERENCES
- 1.Agapova, L. S., G. V. Ilyinskaya, N. A. Turovets, A. V. Ivanov, P. M. Chumakov, and B. P. Kopnin. 1996. Chromosome changes caused by alterations of p53 expression. Mutat. Res. 354:129-138. [DOI] [PubMed] [Google Scholar]
- 2.Andela, V. B., T. J. Sheu, E. J. Puzas, E. M. Schwarz, R. J. O'Keefe, and R. N. Rosier. 2002. Malignant reversion of a human osteosarcoma cell line, Saos-2, by inhibition of NFkappaB. Biochem. Biophys. Res. Commun. 297:237-241. [DOI] [PubMed] [Google Scholar]
- 3.Arlt, A., and H. Schafer. 2002. NFkappaB-dependent chemoresistance in solid tumors. Int. J. Clin. Pharmacol. Ther. 40:336-347. [DOI] [PubMed] [Google Scholar]
- 4.Band, V., D. Zajchowski, K. Swisshelm, D. Trask, V. Kulesa, C. Cohen, J. Connolly, and R. Sager. 1990. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 50:7351-7357. [PubMed] [Google Scholar]
- 5.Bentires-Alj, M., V. Barbu, M. Fillet, A. Chariot, B. Relic, N. Jacobs, J. Gielen, M. P. Merville, and V. Bours. 2003. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 22:90-97. [DOI] [PubMed] [Google Scholar]
- 6.Beroud, C., and T. Soussi. 1998. p53 gene mutation: software and database. Nucleic Acids Res. 26:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blandino, G., A. J. Levine, and M. Oren. 1999. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18:477-485. [DOI] [PubMed] [Google Scholar]
- 8.Bohuslav, J., L. F. Chen, H. Kwon, Y. Mu, and W. C. Greene. 2004. p53 induces NF-kappaB activation by an IkappaB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J. Biol. Chem. 279:26115-26125. [DOI] [PubMed] [Google Scholar]
- 9.Borellini, F., and R. I. Glazer. 1993. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J. Biol. Chem. 268:7923-7928. [PubMed] [Google Scholar]
- 10.Borresen-Dale, A. L., R. A. Lothe, G. I. Meling, P. Hainaut, T. O. Rognum, and E. Skovlund. 1998. TP53 and long-term prognosis in colorectal cancer: mutations in the L3 zinc-binding domain predict poor survival. Clin. Cancer Res. 4:203-210. [PubMed] [Google Scholar]
- 11.Brown, D. R., C. A. Thomas, and S. P. Deb. 1998. The human oncoprotein MDM2 arrests the cell cycle: elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 17:2513-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadwell, C., and G. P. Zambetti. 2001. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene 277:15-30. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Y., P. L. Chen, and W. H. Lee. 1994. Hot-spot p53 mutants interact specifically with two cellular proteins during progression of the cell cycle. Mol. Cell. Biol. 14:6764-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin, K. V., K. Ueda, I. Pastan, and M. M. Gottesman. 1992. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science 255:459-462. [DOI] [PubMed] [Google Scholar]
- 15.Cho, Y., S. Gorina, P. D. Jeffrey, and N. P. Pavletich. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265:346-355. [DOI] [PubMed] [Google Scholar]
- 16.Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. [DOI] [PubMed] [Google Scholar]
- 17.Cusack, J. C., Jr., R. Liu, M. Houston, K. Abendroth, P. J. Elliott, J. Adams, and A. S. Baldwin, Jr. 2001. Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor- kappaB inhibition. Cancer Res. 61:3535-3540. [PubMed] [Google Scholar]
- 18.Deb, D., M. Scian, K. E. Roth, W. Li, J. Keiger, A. S. Chakraborti, S. P. Deb, and S. Deb. 2002. Hetero-oligomerization does not compromise “gain of function” of tumor-derived p53 mutants. Oncogene 21:176-189. [DOI] [PubMed] [Google Scholar]
- 19.Deb, S., C. T. Jackson, M. A. Subler, and D. W. Martin. 1992. Modulation of cellular and viral promoters by mutant human p53 proteins found in tumor cells. J. Virol. 66:6164-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dejardin, E., G. Bonizzi, A. Bellahcene, V. Castronovo, M. P. Merville, and V. Bours. 1995. Highly-expressed p100/p52 (NFKB2) sequesters other NF-kappa B-related proteins in the cytoplasm of human breast cancer cells. Oncogene 11:1835-1841. [PubMed] [Google Scholar]
- 21.Del Sal, G., M. Murphy, E. Ruaro, D. Lazarevic, A. J. Levine, and C. Schneider. 1996. Cyclin D1 and p21/waf1 are both involved in p53 growth suppression. Oncogene 12:177-185. [PubMed] [Google Scholar]
- 22.de Martin, R., J. A. Schmid, and R. Hofer-Warbinek. 1999. The NF-kappaB/Rel family of transcription factors in oncogenic transformation and apoptosis. Mutat. Res. 437:231-243. [DOI] [PubMed] [Google Scholar]
- 23.Dignam, J. D. 1990. Preparation of extracts from higher eukaryotes. Methods Enzymol. 182:194-203. [DOI] [PubMed] [Google Scholar]
- 24.Dittmer, D., S. Pati, G. Zambetti, S. Chu, A. K. Teresky, M. Moore, C. Finlay, and A. J. Levine. 1993. Gain of function mutations in p53. Nat. Genet. 4:42-46. [DOI] [PubMed] [Google Scholar]
- 25.Duyao, M. P., D. J. Kessler, D. B. Spicer, C. Bartholomew, J. L. Cleveland, M. Siekevitz, and G. E. Sonenshein. 1992. Transactivation of the c-myc promoter by human T cell leukemia virus type 1 tax is mediated by NF kappa B. J. Biol. Chem. 267:16288-16291. [PubMed] [Google Scholar]
- 26.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 27.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 28.Erber, R., C. Conradt, N. Homann, C. Enders, M. Finckh, A. Dietz, H. Weidauer, and F. X. Bosch. 1998. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene 16:1671-1679. [DOI] [PubMed] [Google Scholar]
- 29.Falette, N., M. P. Paperin, I. Treilleux, A. C. Gratadour, N. Peloux, H. Mignotte, N. Tooke, E. Lofman, M. Inganas, A. Bremond, M. Ozturk, and A. Puisieux. 1998. Prognostic value of P53 gene mutations in a large series of node-negative breast cancer patients. Cancer Res. 58:1451-1455. [PubMed] [Google Scholar]
- 30.Flatt, P. M., L. J. Tang, C. D. Scatena, S. T. Szak, and J. A. Pietenpol. 2000. p53 regulation of G2 checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 20:4210-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frazier, M. W., X. He, J. Wang, Z. Gu, J. L. Cleveland, and G. P. Zambetti. 1998. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol. Cell. Biol. 18:3735-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frum, R., and S. P. Deb. 2003. Flow cytometric analysis of MDM2-mediated growth arrest, vol. 234. Methods in molecular biology. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 33.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasch, A. P., and M. B. Eisen. 2002. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome. Biol. 3:research0059. [Online.] [DOI] [PMC free article] [PubMed]
- 35.Giebler, H. A., I. Lemasson, and J. K. Nyborg. 2000. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol. Cell. Biol. 20:4849-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenblatt, M. S., W. P. Bennett, M. Hollstein, and C. C. Harris. 1994. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 54:4855-4878. [PubMed] [Google Scholar]
- 37.Gualberto, A., K. Aldape, K. Kozakiewicz, and T. D. Tlsty. 1998. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc. Natl. Acad. Sci. USA 95:5166-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gualberto, A., and A. S. Baldwin, Jr. 1995. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J. Biol. Chem. 270:19680-19683. [DOI] [PubMed] [Google Scholar]
- 39.Hacker, H., and M. Karin. 2002. Is NF-kappaB2/p100 a direct activator of programmed cell death? Cancer Cell 2:431-433. [DOI] [PubMed] [Google Scholar]
- 40.Hainaut, P., T. Hernandez, A. Robinson, P. Rodriguez-Tome, T. Flores, M. Hollstein, C. C. Harris, and R. Montesano. 1998. IARC database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 26:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 42.Heusch, M., L. Lin, R. Geleziunas, and W. C. Greene. 1999. The generation of nfkb2 p52: mechanism and efficiency. Oncogene 18:6201-6208. [DOI] [PubMed] [Google Scholar]
- 43.Hollstein, M., M. J. Marion, T. Lehman, J. Welsh, C. C. Harris, G. Martel-Planche, I. Kusters, and R. Montesano. 1994. p53 mutations at A:T base pairs in angiosarcomas of vinyl chloride-exposed factory workers. Carcinogenesis 15:1-3. [DOI] [PubMed] [Google Scholar]
- 44.Hollstein, M., K. Rice, M. S. Greenblatt, T. Soussi, R. Fuchs, T. Sorlie, E. Hovig, B. Smith-Sorensen, R. Montesano, and C. C. Harris. 1994. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22:3551-3555. [PMC free article] [PubMed] [Google Scholar]
- 45.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsiao, M., J. Low, E. Dorn, D. Ku, P. Pattengale, J. Yeargin, and M. Haas. 1994. Gain-of-function mutations of the p53 gene induce lymphohematopoietic metastatic potential and tissue invasiveness. Am. J. Pathol. 145:702-714. [PMC free article] [PubMed] [Google Scholar]
- 47.Irwin, M. S., K. Kondo, M. C. Marin, L. S. Cheng, W. C. Hahn, and W. G. Kaelin, Jr. 2003. Chemosensitivity linked to p73 function. Cancer Cell 3:403-410. [DOI] [PubMed] [Google Scholar]
- 48.Jones, D. R., R. M. Broad, L. D. Comeau, S. J. Parsons, and M. W. Mayo. 2002. Inhibition of nuclear factor kappaB chemosensitizes non-small cell lung cancer through cytochrome c release and caspase activation. J. Thorac. Cardiovasc. Surg. 123:310-317. [DOI] [PubMed] [Google Scholar]
- 49.Kannan, K., N. Amariglio, G. Rechavi, J. Jakob-Hirsch, I. Kela, N. Kaminski, G. Getz, E. Domany, and D. Givol. 2001. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225-2234. [DOI] [PubMed] [Google Scholar]
- 50.Kannan, K., N. Kaminski, G. Rechavi, J. Jakob-Hirsch, N. Amariglio, and D. Givol. 2001. DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene 20:3449-3455. [DOI] [PubMed] [Google Scholar]
- 51.Kawamura, M., T. Yamashita, K. Segawa, M. Kaneuchi, M. Shindoh, and K. Fujinaga. 1996. The 273rd codon mutants of p53 show growth modulation activities not correlated with p53-specific transactivation activity. Oncogene 12:2361-2367. [PubMed] [Google Scholar]
- 52.Kerns, R., L. Zhang, and M. Miles. 2003. Application of the S-score algorithm for analysis of oligonucleotide microarrays. Methods 4:274-281. [DOI] [PubMed] [Google Scholar]
- 53.Lane, D. P. 1994. p53 and human cancers. Br. Med. Bull. 50:582-599. [DOI] [PubMed] [Google Scholar]
- 54.Lanyi, A., D. Deb, R. C. Seymour, J. H. Ludes-Meyers, M. A. Subler, and S. Deb. 1998. “Gain of function” phenotype of tumor-derived mutant p53 requires the oligomerization/nonsequence-specific nucleic acid-binding domain. Oncogene 16:3169-3176. [DOI] [PubMed] [Google Scholar]
- 55.Lassus, P., C. Bertrand, O. Zugasti, J. P. Chambon, T. Soussi, D. Mathieu-Mahul, and U. Hibner. 1999. Anti-apoptotic activity of p53 maps to the COOH-terminal domain and is retained in a highly oncogenic natural mutant. Oncogene 18:4699-4709. [DOI] [PubMed] [Google Scholar]
- 56.Li, R., P. D. Sutphin, D. Schwartz, D. Matas, N. Almog, R. Wolkowicz, N. Goldfinger, H. Pei, M. Prokocimer, and V. Rotter. 1998. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene 16:3269-3277. [DOI] [PubMed] [Google Scholar]
- 57.Liao, G., and S. C. Sun. 2003. Regulation of NF-kappaB2/p100 processing by its nuclear shuttling. Oncogene 22:4868-4874. [DOI] [PubMed] [Google Scholar]
- 58.Lin, J., A. K. Teresky, and A. J. Levine. 1995. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene 10:2387-2390. [PubMed] [Google Scholar]
- 59.Lombardi, L., P. Ciana, C. Cappellini, D. Trecca, L. Guerrini, A. Migliazza, A. T. Maiolo, and A. Neri. 1995. Structural and functional characterization of the promoter regions of the NFKB2 gene. Nucleic Acids Res. 23:2328-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe, S. W., S. Bodis, N. Bardeesy, A. McClatchey, L. Remington, H. E. Ruley, D. E. Fisher, T. Jacks, J. Pelletier, and D. E. Housman. 1994. Apoptosis and the prognostic significance of p53 mutation. Cold Spring Harbor Symp. Quant. Biol. 59:419-426. [DOI] [PubMed] [Google Scholar]
- 61.Ludes-Meyers, J. H., M. A. Subler, C. V. Shivakumar, R. M. Munoz, P. Jiang, J. E. Bigger, D. R. Brown, S. P. Deb, and S. Deb. 1996. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol. Cell. Biol. 16:6009-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marin, M. C., and W. G. Kaelin, Jr. 2000. p63 and p73: old members of a new family. Biochim. Biophys. Acta 1470:M93-M100. [DOI] [PubMed] [Google Scholar]
- 63.Maxwell, S. A., and G. E. Davis. 2000. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc. Natl. Acad. Sci. USA 97:13009-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muller, B. F., D. Paulsen, and W. Deppert. 1996. Specific binding of MAR/SAR DNA-elements by mutant p53. Oncogene 12:1941-1952. [PubMed] [Google Scholar]
- 65.Ozbun, M. A., and J. S. Butel. 1995. Tumor suppressor p53 mutations and breast cancer: a critical analysis. Adv. Cancer Res. 66:71-141. [DOI] [PubMed] [Google Scholar]
- 66.Pahl, H. L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 67.Pohl, J., N. Goldfinger, A. Radler-Pohl, V. Rotter, and V. Schirrmacher. 1988. p53 increases experimental metastatic capacity of murine carcinoma cells. Mol. Cell. Biol. 8:2078-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 69.Scian, M. J., K. E. Stagliano, D. Deb, M. A. Ellis, E. H. Carchman, A. Das, K. Valerie, S. P. Deb, and S. Deb. 2004. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene 23:4430-4443. [DOI] [PubMed] [Google Scholar]
- 70.Scian, M. J., K. E. Stagliano, M. A. Ellis, S. Hassan, M. Bowman, M. F. Miles, S. P. Deb, and S. Deb. 2004. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 64:7447-7454. [DOI] [PubMed] [Google Scholar]
- 71.Sigal, A., and V. Rotter. 2000. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 60:6788-6793. [PubMed] [Google Scholar]
- 72.Soussi, T., K. Dehouche, and C. Beroud. 2000. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum. Mutat. 15:105-113. [DOI] [PubMed] [Google Scholar]
- 73.Stagliano, K. E., E. Carchman, and S. Deb. 2003. Real-time polymerase chain reaction quantitation of relative expression of genes modulated by p53 using SYBR Green I. Methods Mol. Biol. 234:73-91. [DOI] [PubMed] [Google Scholar]
- 74.Strano, S., and G. Blandino. 2003. p73-mediated chemosensitivity: a preferential target of oncogenic mutant p53. Cell Cycle 2:348-349. [PubMed] [Google Scholar]
- 75.Strano, S., G. Fontemaggi, A. Costanzo, M. G. Rizzo, O. Monti, A. Baccarini, G. Del Sal, M. Levrero, A. Sacchi, M. Oren, and G. Blandino. 2002. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 277:18817-18826. [DOI] [PubMed] [Google Scholar]
- 76.Strano, S., M. Rossi, G. Fontemaggi, E. Munarriz, S. Soddu, A. Sacchi, and G. Blandino. 2001. From p63 to p53 across p73. FEBS Lett. 490:163-170. [DOI] [PubMed] [Google Scholar]
- 77.Tamatani, T., M. Azuma, Y. Ashida, K. Motegi, R. Takashima, K. Harada, S. Kawaguchi, and M. Sato. 2004. Enhanced radiosensitization and chemosensitization in NF-kappaB-suppressed human oral cancer cells via the inhibition of gamma-irradiation- and 5-FU-induced production of IL-6 and IL-8. Int. J. Cancer 108:912-921. [DOI] [PubMed] [Google Scholar]
- 78.Thibault, C., L. Wang, L. Zhang, and M. F. Miles. 2001. DNA arrays and functional genomics in neurobiology. Int. Rev. Neurobiol. 48:219-253. [DOI] [PubMed] [Google Scholar]
- 79.Torgeman, A., N. Mor-Vaknin, E. Zelin, Z. Ben-Aroya, M. Lochelt, R. M. Flugel, and M. Aboud. 2001. Sp1-p53 heterocomplex mediates activation of HTLV-I long terminal repeat by 12-O-tetradecanoylphorbol-13-acetate that is antagonized by protein kinase C. Virology 281:10-20. [DOI] [PubMed] [Google Scholar]
- 80.Tusher, V., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ushio, Y., K. Tada, S. Shiraishi, T. Kamiryo, N. Shinojima, M. Kochi, and H. Saya. 2003. Correlation of molecular genetic analysis of p53, mdm2, p16, pten, and egfr and survival of patients with anaplastic astrocytoma and glioblastoma. Front. Biosci. 8:E281-E288. [DOI] [PubMed] [Google Scholar]
- 82.van Slooten, H. J., M. J. van De Vijver, A. L. Borresen, J. E. Eyfjord, R. Valgardsdottir, S. Scherneck, J. M. Nesland, P. Devilee, C. J. Cornelisse, and J. H. van Dierendonck. 1999. Mutations in exons 5-8 of the p53 gene, independent of their type and location, are associated with increased apoptosis and mitosis in invasive breast carcinoma. J. Pathol. 189:504-513. [DOI] [PubMed] [Google Scholar]
- 83.Viatour, P., M. Bentires-Alj, A. Chariot, V. Deregowski, L. de Leval, M. P. Merville, and V. Bours. 2003. NF- kappa B2/p100 induces Bcl-2 expression. Leukemia 17:1349-1356. [DOI] [PubMed] [Google Scholar]
- 84.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 85.Wang, C. Y., J. C. Cusack, Jr., R. Liu, and A. S. Baldwin, Jr. 1999. Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat. Med. 5:412-417. [DOI] [PubMed] [Google Scholar]
- 86.Wang, X. J., D. A. Greenhalgh, A. Jiang, D. He, L. Zhong, B. R. Brinkley, and D. R. Roop. 1998. Analysis of centrosome abnormalities and angiogenesis in epidermal-targeted p53172H mutant and p53-knockout mice after chemical carcinogenesis: evidence for a gain of function. Mol. Carcinog. 23:185-192. [DOI] [PubMed] [Google Scholar]
- 87.Weisz, L., A. Zalcenstein, P. Stambolsky, Y. Cohen, N. Goldfinger, M. Oren, and V. Rotter. 2004. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 64:8318-8327. [DOI] [PubMed] [Google Scholar]
- 88.Wen, W. H., A. Reles, I. B. Runnebaum, J. Sullivan-Halley, L. Bernstein, L. A. Jones, J. C. Felix, R. Kreienberg, A. el-Naggar, and M. F. Press. 1999. p53 mutations and expression in ovarian cancers: correlation with overall survival. Int. J. Gynecol. Pathol. 18:29-41. [DOI] [PubMed] [Google Scholar]
- 89.Yu, J., L. Zhang, P. M. Hwang, C. Rago, K. W. Kinzler, and B. Vogelstein. 1999. Identification and classification of p53-regulated genes. Proc. Natl. Acad. Sci. USA 96:14517-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zambetti, G. P., and A. J. Levine. 1993. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 7:855-865. [DOI] [PubMed] [Google Scholar]
- 91.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]