Abstract
Little is known about cyclic AMP (cAMP) function in Mycobacterium tuberculosis, despite its ability to encode 15 adenylate cyclases and 10 cNMP-binding proteins. M. tuberculosis Rv3676, which we have designated CRPMt, is predicted to be a cAMP-dependent transcription factor. In this study, we characterized CRPMt's interactions with DNA and cAMP, using experimental and computational approaches. We used Gibbs sampling to define a CRPMt DNA motif that resembles the cAMP receptor protein (CRP) binding motif model for Escherichia coli. CRPMt binding sites were identified in a total of 73 promoter regions regulating 114 genes in the M. tuberculosis genome, which are being explored as a regulon. Specific CRPMt binding caused DNA bending, and the substitution of highly conserved nucleotides in the binding site resulted in a complete loss of binding to CRPMt. cAMP enhanced CRPMt's ability to bind DNA and caused allosteric alterations in CRPMt conformation. These results provide the first direct evidence for cAMP binding to a transcription factor in M. tuberculosis, suggesting a role for cAMP signal transduction in M. tuberculosis and implicating CRPMt as a cAMP-responsive global regulator.
Tuberculosis (TB) remains a serious global health problem that is growing at an estimated rate of 3% per year (49). This TB epidemic is exacerbated by an unexplained synergy with human immunodeficiency virus and steadily increasing rates of drug resistance that are a by-product of lengthy treatment regimens (15, 20). A better understanding of Mycobacterium tuberculosis biology is needed to improve treatment and develop a more effective vaccine. A key area of interest is how M. tuberculosis senses and responds to the environments it encounters during host infection.
Cyclic AMP (cAMP) is a critical signaling molecule in many bacterial and eukaryotic cells. The role of cAMP signal transduction in mediating catabolite repression has been well characterized in Escherichia coli, and this forms the paradigm for cAMP-mediated gene regulation in prokaryotes (7, 10, 11, 16, 33, 36). A class I adenylate cyclase (AC) in E. coli catalyzes the synthesis of cAMP, which then transduces the signal by binding cAMP receptor protein (CRP) and activating it as a transcription factor (18).
cAMP signaling is also critical for virulence in a diverse range of pathogens, including yeast, fungi, bacteria, and parasites (3, 12, 19, 25, 37, 38, 42, 50, 70). In some cases, cAMP regulates virulence genes within the pathogen (3, 38, 42). For example, CRP-cAMP signaling is essential for virulence in Salmonella enterica serovar Typhimurium (17) and has recently been shown to control virulence-associated type III secretion systems in Pseudomonas aeruginosa and Yersinia enterocolitica (50, 70).
The M. tuberculosis genome contains 15 putative class III adenylate cyclase genes (46). The activity of at least 10 of these cyclases has been confirmed with biochemical assays (13, 26, 40, 41, 61, 64), making it likely that cAMP contributes substantially to signal transduction in M. tuberculosis. We recently identified the first cAMP-regulated genes in M. tuberculosis by using an exogenous cAMP culture model (24). Some of these genes are upregulated during intracellular growth in macrophages (29), suggesting that cAMP signaling may be important to M. tuberculosis during its interaction with the host. This observation is intriguing in light of a previous study that reported elevated levels of cAMP in macrophages that showed an impairment of phagosome-lysosome fusion upon infection with Mycobacterium microti (44).
The mechanism of cAMP-mediated gene regulation in M. tuberculosis has not been explored. We previously reported that the M. tuberculosis Rv3676 protein belongs to a superfamily of proteins that contain both cAMP binding and helix-turn-helix (HTH) DNA binding motifs (46), and we hypothesized that the Rv3676 protein is a CRP-like transcriptional regulator in M. tuberculosis. For this study, we used experimental and computational approaches to define Rv3676's DNA binding sequence and characterize its interactions with DNA and cAMP. We designated the M. tuberculosis Rv3676 gene crp and the encoded protein CRPMt, based on the results. We also identified 114 members of a putative CRPMt regulon, implicating CRPMt as a biologically relevant global regulator of transcription in M. tuberculosis.
MATERIALS AND METHODS
Expression and purification of CRPMt.
crp was PCR amplified from M. tuberculosis H37Rv using primers KM977 (5′-GGAATTCGACGAGATCCTGGC CAGGGC) and KM963 (5′-AAGCTTGCGTGCGACTCTGTGTCTGC) with EcoRI and HindIII restriction sites (underlined). The amplified DNA fragment was cloned into pET28a+ (Novagen) using the EcoRI and HindIII sites to generate pMBC372, verified by sequencing, and maintained in E. coli BL21(DE3). CRPMt expression was induced from a bacterial culture (optical density at 600 nm, 0.6) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h and confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis using an anti-His6 monoclonal antibody (Clontech). For purification, a 200-ml culture was pelleted and resuspended in 1 ml lysis buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM dithiothreitol, and 1% protease inhibitor cocktail (Sigma). Bacteria were lysed by two freeze-thaw cycles (−70°C-37°C) and sonication for 5 min, followed by two additional freeze-thaw cycles, and the lysate was cleared by centrifugation at 13,000 × g for 10 min. CRPMt was purified using a Hitrap affinity column (Amersham Biosciences) per the manufacturer's instructions, and the eluted protein was dialyzed against phosphate-buffered saline with 10% glycerol. The protein concentration was measured with a NanoOrange protein quantitation kit (Molecular Probes) and diluted to 2 mg/ml before being stored in aliquots at −70°C.
Systematic evolution of ligands by exponential enrichment (SELEX)-based capture of CRPMt-binding ligand DNA.
Mycobacterium bovis BCG genomic DNA was digested to completion with Sau3AI and ligated to a linker sequence (5′CGAATTCAGGAAACAGCTATGTTAATTAA3′) prepared with a Sau3AI-compatible sticky end. A crude extract from 100 ml of His-CRPMt-expressing culture was applied to 100 μl His Mag agarose beads (Novagen), which were then washed and equilibrated with DNA binding buffer [10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM EDTA, 50 μg/ml bovine serum albumin, 1 mM dithiothreitol, 0.05% nonionic P-40 detergent, 100 μM cAMP, 20 μg/ml poly(dI-dC), and 10% glycerol] per the manufacturer's directions. Five micrograms of DNA fragments in 100 μl of DNA binding buffer was added to the CRPMt-bound beads for 15 min at room temperature. The beads were washed three times to remove nonspecific DNA before eluting CRPMt-DNA complexes in a volume of 100 μl. Ten microliters of this eluted protein was heated for 10 min at 95°C and used as a template for PCR with primer KM1040 (5′-CGAATTCAGGAAACAGCTATG). The resulting DNA product was cloned into the TA vector (Invitrogen). Individual E. coli DH5α transformants were picked into Luria broth containing 25 μg/ml kanamycin, grown overnight, and then used as templates for PCRs with primer KM1040 to amplify the captured insert fragments. PCR products of 200 to 300 bp were selected for further electrophoretic mobility shift assay (EMSA) analysis. The purity of the protein used for capturing DNA was also evaluated by SDS-PAGE.
EMSA.
A 33P-end-labeled DNA probe (0.05 pmol) was used in each 10-μl binding reaction mixture as described by others (60), with modifications. Briefly, purified His-CRPMt (at specified concentrations) and DNA probes were incubated for 30 min at room temperature in DNA binding buffer prior to electrophoresis on a nondenaturing 8% polyacrylamide gel for 2 to 3 h at 14 V/cm in 0.5× Tris-borate-EDTA. A 200- to 500-fold excess of unlabeled DNA fragments was used for competition experiments. Gels were vacuum dried, exposed on a phosphor screen, scanned with a Storm 860 PhosphorImager (Molecular Dynamics), and analyzed with ImageQuant software.
Additional assays.
The effects of cAMP on CRPMt conformation were examined by treating 5 μg of CRPMt with 0.2 or 1 μg trypsin (Sigma) for 10 min at 37°C, as described by others (35). Half of the digested protein was assayed in a 15% SDS-PAGE gel, and a portion of the remainder was diluted for use in EMSA.
For DNA-bending experiments, five 156-bp DNA fragments were PCR amplified from different locations within the Rv0884c-Rv0885 intergenic region. Fragment end points were as follows: F1, positions 982623 to 982778; F2, positions 982590 to 982745; F3, positions 982554 to 982709; F4, positions 982528 to 982683; and F5, positions 982495 to 982650. The forward primer of each fragment was phosphorylated with [γ-33P]ATP to generate end-labeled probes using PCR. CRPMt (35 nM) was used for EMSA to compare the mobilities of the binding complex for each DNA fragment.
Sequence analysis.
We identified the set of promoter-containing regions in the M. tuberculosis genome as those sequences that were upstream of a gene and contained at least 20 bp of intergenic sequence, as defined by M. tuberculosis H37Rv annotation (GenBank accession no. NC_000962.1) (14). This set of M. tuberculosis intergenic promoter regions consists of 2,066 sequences, totaling 346,025 bp of searchable sequence after masking a 43-bp repeat sequence.
Regulatory motifs were predicted using the Gibbs sampler (67). For applications to subsets of intergenic sequences or DNA fragments from CRPMt trap experiments, where most sequences were believed to contain a common pattern, the Gibbs recursive sampler was used with the following parameters: either one or two motif models were specified, where each model was 16 bp allowed to fragment to 24 bp and allowed to choose an even- or odd-width palindromic model, based on the sequence evidence; a position-specific background model (43) was incorporated; uninformed priors (i.e., no prior information on the motif models) were used; a maximum of two sites per sequence was allowed; and run parameters consisted of 5,000 iterations with a plateau period of 500 iterations and reinitializing 50 times using a random seed. For applications to the genome-scale data set, the Gibbs motif sampler was used with the following parameters: the motif model was 16 bp allowed to fragment to 24 bp and allowed to choose an even- or odd-width palindromic model, a position-specific background model (43) was incorporated, prior information was specified by providing a motif model and was weighted to provide three to five pseudocounts per motif position, the estimated number of sites was 50, and run parameters were as described above. The two motif models used as prior information were (i) an alignment of 87 experimentally verified (DNase I footprinted) E. coli CRP binding sites, weighted to provide five pseudocounts per motif position; and (ii) an alignment of putative CRPMt binding sites from the M. tuberculosis genome, weighted to provide either three, four, or five pseudocounts per motif position.
RESULTS
Identification of CRPMt binding sites.
Experimental and computational approaches were combined to identify CRPMt binding sites. One hundred sixty-six M. bovis BCG genomic DNA fragments were captured using a SELEX-based technique and then screened by EMSA with purified CRPMt, as described in Materials and Methods. Five of these DNA fragments showed obvious mobility retardation in the presence of CRPMt (data not shown). These DNA sequences were analyzed using the Gibbs sampler (see Materials and Methods), and a common sequence pattern consistent with the E. coli CRP motif model was identified in all five DNA fragments (Table 1; Fig. 1A).
TABLE 1.
Sequence alignment of DNA ligands of CRPMt captured by SELEX and EMSA
| Clone | Aligned sequencea | Genome coordinatesb |
|---|---|---|
| 1D1 | ccgtc TGTGAGCAAGATCACA tagct | 4110823-4110835 (cr) |
| 1B5 | gtatc TGTGACTAAGGTCACA gacgc | 13463-13478 (cr) |
| 2D3 | ctcta TGTGACGAAGCCCACA tcgac | 4329936-4329951 (nc) |
| S16 | tgcaa TGTGATATTAGCCTCA ttttg | 1178962-1178977 (nc) |
| 3G1 | gttgt GGTGACCGCCGTCACA gcgac | 2518157-2518172 (cr) |
Sites detected by Gibbs sampling in each clone are shown in capital letters; the underlined sequence for clone 1D1 is from the DNA linker.
M. tuberculosis (entry NC_000962.1) genome coordinates of the aligned sites (capital letters). The locations of these regions with respect to annotated protein coding regions or noncoding regions are indicated by “(cr)”and “(nc),”respectively.
FIG. 1.
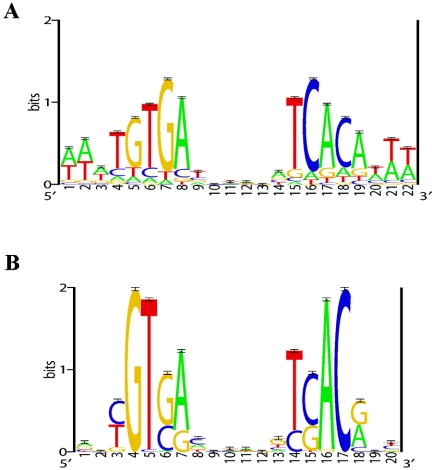
Sequence logos of the E. coli CRP motif model (A) and the putative CRPMt motif model (B). The E. coli CRP motif model represents 87 experimentally identified (DNase I footprinted) binding sites; this motif model was used as prior information in the prediction of CRPMt binding sites (see Materials and Methods). The putative CRPMt motif model represents 58 predicted sites (see Table 2) from the set of intergenic regions from the M. tuberculosis genome. Sequence logos depict the relative frequency of each base at each position of the motif. The y axis indicates the information content measured in bits, and error bars represent standard deviations at each position due to the limited sample size (58).
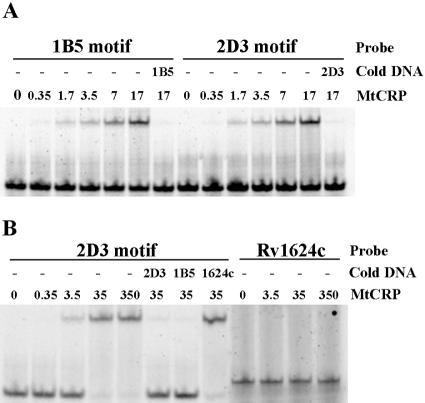
Binding of CRPMt to the predicted sites within the recovered 1B5 and 2D3 DNA fragments (Table 1) was confirmed by EMSA. A 40-bp intergenic DNA fragment upstream of Rv1624c was used as a nonspecific control. The presence of CRPMt retarded, in a dose-dependent manner, the migration of 28-bp synthetic oligonucleotides containing the predicted DNA binding site of either 1B5 or 2D3 (Fig. 2A). The reappearance of the free probes in the presence of a 500-fold excess of unlabeled probe DNA confirmed the specificity of these CRPMt-DNA interactions (Fig. 2A). In addition, binding of the 2D3 probe could be competed with an unlabeled 2D3 or 1B5 oligonucleotide, but not by the same concentration of the nonspecific DNA control (Fig. 2B).
FIG. 2.
EMSA experiment showing binding of CRPMt with motifs identified in captured DNA sequences. (A) The mobility of 28-bp synthesized probes containing motifs of 1B5 and 2D3 fragments was retarded by CRPMt, and the free probes reappeared in the presence of a 500-fold excess of unlabeled probe DNA, as specified. (B) The shift of the 2D3 28-bp motif probe by CRPMt could be competed by either 2D3 or 1B5 unlabeled motif DNA, but not by the 40-bp intergenic DNA upstream of Rv1624c (1624c) that was used as a negative control. This control Rv1624c DNA probe also failed to bind to CRPMt. The labeled DNA probe, unlabeled competitor DNA (cold DNA), and amount of CRPMt (nM) that was used are specified for each lane.
Together, these results indicate that specific CRPMt binding sites are contained within the 28-bp oligonucleotides and that the same protein domain was responsible for CRPMt binding to both 1B5 and 2D3 DNA. However, only two of the predicted binding sites in the SELEX-trapped fragments are within annotated intergenic regions, so we pursued additional computational analyses to refine the CRPMt binding motif and identify potential regulon members.
An alignment of the helix-turn-helix regions of CRPMt and E. coli CRP and fumarate nitrate reductase regulator (FNR) (Pfam 17.0, PF00325) (4) showed several conserved amino acids in the DNA recognition helix (Fig. 3). We also found using EMSA that CRPMt could bind the CRP binding site in the E. coli lac promoter (5′-TAATGTGAGTTAGCTCACTCAT-3′), but not the FNR binding site in the E. coli ndh1 promoter (5′-AAACTTGATTAACATCAATTTT-3′) (data not shown). These observations were consistent with our identification of a CRP-like pattern for the CRPMt SELEX DNA fragments and suggested that information on the E. coli CRP binding motif could be used to predict the CRPMt binding motif.
FIG. 3.
Sequence alignment of helix-turn-helix DNA recognition domains of CRPMt and E. coli CRP (Ec CRP) and FNR (Ec FNR) (Pfam 17.0, PF00325) (4). CRP and FNR amino acid residues that form hydrogen bonds with DNA bases are underlined.
We used the Gibbs motif sampler (see Materials and Methods) with prior information on the E. coli CRP motif model (Fig. 1A) to detect a putative CRPMt binding motif in the intergenic regions of the M. tuberculosis genome. We initially identified a subset of 29 M. tuberculosis intergenic regions that contained CRP-like sites in at least 3 of 20 independent Gibbs sampling runs. The Gibbs recursive sampler was then used to detect a motif model de novo (i.e., without prior information) for this set of 29 intergenic regions. This preliminary CRPMt model was then used as prior information to analyze the intergenic regions of the M. tuberculosis genome again (see Materials and Methods). These analyses provided a set of 55 M. tuberculosis intergenic regions that contained a site in at least 1 of 30 independent Gibbs sampling runs, and the Gibbs recursive sampler was used to detect a motif model de novo for these 55 sequences (Fig. 1B).
The resulting motif model consists of 58 sites from the 55 input sequences and resembles the central core of the E. coli CRP binding motif, with a palindrome of two 5-bp half-sites separated by a 6-bp spacer (Fig. 2B; Table 2). Several of these 55 intergenic regions are positioned between divergently transcribed genes, such that a total of 73 genes are immediately flanking and downstream of these intergenic regions. Furthermore, several of these genes are likely to be the first gene of a polycistronic operon, suggesting that the 55 intergenic regions contain promoters that regulate the expression of ∼114 genes. These regulon candidates include genes reported to be starvation (34 genes) or hypoxia (16 genes) regulated, members of the PE and PPE families (10 genes), or essential for M. tuberculosis growth in culture medium (9 genes) (5, 8, 57, 62). The remaining 55 genes are mostly uncharacterized.
TABLE 2.
CRPMt binding sites at the intergenic region of the M. tuberculosis H37Rv genome
| Probabilitya | Sampling frequencyb | Site sequence | Genomic coordinates | Flanking gene(s)c | Distance(s) upstreamd |
|---|---|---|---|---|---|
| 0.92 | 1.00 | CAGGTCACACCAGTCACAGA | 1571-1590 | Rv0002 (dnaN) | −462 |
| 0.98 | 1.00 | CACGTCAGCAAAGTCACGAT | 23788-23807 | Rv0019c | −51 |
| 0.97 | 0.69 | ACCGTGAATCTGGCGACGCC | 161521-161540 | Rv0134 (ephF) | −229 |
| 0.59 | 1.00 | AATGTGATCCTCTTCGCGTG | 301656-301675 | Rv0249c | −3 |
| 0.92 | 1.00 | ACGGTCGCAACAGTCACATG | 309913-309932 | Rv0256c (PPE2) | −368 |
| 0.99 | 0.91 | CGCGTCGCAGGATTCACACT | 518531-518550 | Rv0429c (def) / Rv0430 | −137/−181 |
| 0.92 | 1.00 | AGTGTAACGCATATCACGTG | 542072-542091 | Rv0451c (mmpS4) /Rv0452 | −164/−49 |
| 0.97 | 1.00 | TAGGTGACCAAACTCACGCT | 542901-542920 | Rv0453 (PPE11) | −252 |
| 0.46 | 1.00 | AGTGTGAGCTGTATTACATG | 549630-549649 | Rv0457c/Rv0458 | −25/−24 |
| 0.97 | 0.53 | ATTGTGAATCTGGCGACGCG | 747949-747968 | Rv0651 (rplJ) | −306 |
| 0.98 | 0.60 | GGCGTCGCGGGATTCACACT | 747981-748000 | Rv0651 (rplJ) | −274 |
| 0.78 | 1.00 | ATCGTGGGCTTGCTGACGCG | 754621-754640 | Rv0658c | −214 |
| 0.96 | 1.00 | GATGTCAGCTACTCCACAAC | 780543-780562 | Rv0680c/Rv0681 | −129/−157 |
| 0.97 | 1.00 | ATCGTGGAACGGCTGACAAC | 898731-898750 | Rv0805 | −79 |
| 0.95 | 1.00 | AGCGTGATTCTGGCGACGCC | 948516-948535 | Rv0851c/Rv0852 (fadD16) | −50/−22 |
| 0.93 | 0.86 | TATGTGGGTAATGTCACATG | 965958-965977 | Rv0867c (rpfA) | −425 |
| 1.00 | 1.00 | AGTGTGAACAAGCTCACATG | 982625-982644 | Rv0884c (serC)/Rv0885 | −73/−116 |
| 0.99 | 1.00 | ATTGTGAGTTGGATCACGTT | 1061787-1061806 | Rv0950c/Rv0951 (sucC) | −135/−156 |
| 0.97 | 1.00 | ACTGTGACTGGCGCGACACG | 1112239-1112258 | Rv0996 | −124 |
| 0.70 | 1.00 | CTCGTGACCTACGTGGCAGC | 1220408-1220427 | Rv1092c (coaA)/Rv1093 (glyA1) | −224/−145 |
| 1.00 | 1.00 | GATGTGACTCAAGTGACACG | 1284911-1284930 | Rv1158c/Rv1159 | −51/−60 |
| 0.98 | 1.00 | GCCGTGAGGGGCGTCACGGT | 1327660-1327679 | Rv1185c (fadD21) | −150 |
| 0.47 | 0.66 | TTCGTGAACTAGATCACCAT | 1374256-1374275 | Rv1230c | −61 |
| 0.97 | 0.85 | ATTGTGAATCTGGTGACGCG | 1404643-1404662 | Rv1256c (cyp-130) | −42 |
| 0.97 | 0.55 | CGCGTCGTGCGGTTCACACT | 1404675-1404694 | Rv1256c (cyp-130) | −74 |
| 0.90 | 1.00 | CGTGTGACCAAACTCACCGC | 1561327-1561346 | Rv1386 (PE15) | −116 |
| 0.96 | 1.00 | ATTGTGGCCCACGCCACGTC | 1582120-1582139 | Rv1405c/Rv1406 (fmt) | −153/−25 |
| 1.00 | 1.00 | AATGTGATCTAGGTCACGTG | 1757393-1757412 | Rv1552 (frdA) | −267 |
| 0.99 | 0.98 | GTTGTGATTTCAGTCACGCG | 1774835-1774854 | Rv1566c | −217 |
| 0.99 | 0.83 | GCCGTGACGCATGTGACGTT | 1784400-1784419 | Rv1581c | −101 |
| 0.99 | 0.68 | TGCGTCACATGCGTCACGTT | 1784468-1784487 | Rv1581c | −169 |
| 0.99 | 1.00 | ACCGTCACAGCTGTCACACG | 1992785-1992804 | Rv1759c (wag-22)/Rv1760 | −210/−347 |
| 0.91 | 1.00 | CTTGTGGTGTTGTTCACAAC | 2014548-2014567 | Rv1779c/Rv1780 | −71/−130 |
| 0.99 | 1.00 | GCCGTGAGATTCGTCACGTC | 2052776-2052795 | Rv1810 | −136 |
| 0.96 | 1.00 | AATGTCAGTGATCTGACGGC | 2219565-2219584 | Rv1976c/Rv1977 | −316/−168 |
| 0.99 | 1.00 | ATCGTCACGCACGTCACGGC | 2234038-2234057 | Rv1990c | −403 |
| 0.95 | 0.63 | ACCGTCACCTGCGTCACCGC | 2334300-2334319 | Rv2077c/Rv2078 | −8/−738 |
| 0.97 | 1.00 | GACGTCGCATCGGTCACAGC | 2386129-2386148 | Rv2124c (metH)/Rv2125 | −64/−143 |
| 0.97 | 1.00 | GCTGTCAAATCCGTCACGAA | 2610746-2610765 | Rv2336 | −70 |
| 0.85 | 1.00 | GTCGTGAATTGGGTGACCAA | 2620124-2620143 | Rv2342 | −127 |
| 0.91 | 1.00 | CGGGTCACTTCGATCACGCG | 2677687-2677706 | Rv2385 (mbtJ) | −21 |
| 0.98 | 1.00 | AGTGTGGCCAAGGTGACAGC | 2692237-2692256 | Rv2396 (PE_PGRS41) | −541 |
| 0.97 | 1.00 | GTGGTGAGCTGGTTCACACC | 2704451-2704470 | Rv2406c/Rv2407 | −16/−225 |
| 0.96 | 1.00 | GCGGTGATCGGCGTCACGCC | 2705800-2705819 | Rv2408 (PE24) | −196 |
| 1.00 | 1.00 | CGCGTGACATGTGTCACATG | 2921509-2921528 | Rv2591 (PE_PGRS44) | −21 |
| 0.97 | 1.00 | AGTGTGATTTACATCACATA | 3015060-3015079 | Rv2699c/Rv2700 | −98/−121 |
| 0.91 | 0.95 | AATGTGAGGTTCACGACGCG | 3412018-3412037 | Rv3050c | −64 |
| 0.99 | 1.00 | GGCGTCGCCAGATTCACGCT | 3427147-3427166 | Rv3063 (cstA) | −74 |
| 0.96 | 1.00 | GACGTCGGCCGCGTCACGTT | 3675815-3675834 | Rv3295 | −228 |
| 0.99 | 1.00 | AACGTCATCTTTGCCACGTT | 3874764-3874783 | Rv3454 | −35 |
| 0.99 | 1.00 | GATGTGATGCACTTGACATC | 4091145-4091164 | Rv3650 (PE33) | −66 |
| 0.99 | 1.00 | GTCGTGAACGGTTTGACGGT | 4092932-4092951 | Rv3652 (PE_PGRS60) | −678 |
| 0.92 | 1.00 | GCTGTGAACCCTGCGACGCA | 4169322-4169341 | Rv3724A (cut5a) | −262 |
| 0.99 | 1.00 | ACTGTGACCACGGCCACGCT | 4178090-4178109 | Rv3729 | −173 |
| 0.97 | 1.00 | GTTGTGACTTACCTCACCGT | 4317080-4317099 | Rv3843c/Rv3844 | −487/−1673 |
| 0.96 | 1.00 | TATGTGACGAAGCCCACATC | 4329934-4329953 | Rv3857c | −323 |
| 0.99 | 1.00 | CATGTCAAAACATTGACAAT | 4360091-4360110 | Rv3879c | −312 |
| 0.98 | 1.00 | GATGTGAAATCTGCGACGCC | 4376153-4376172 | Rv3893c (PE36) | −161 |
Probability that the site belongs in the alignment.
Frequency with which the site was sampled.
Gene(s) encoded downstream of the site; for sites found between divergently transcribed genes, the two genes are listed separated by a slash.
Distance upstream of the start codon of the gene listed in the previous column.
CRPMt binding to computationally predicted sites within the M. tuberculosis genome.
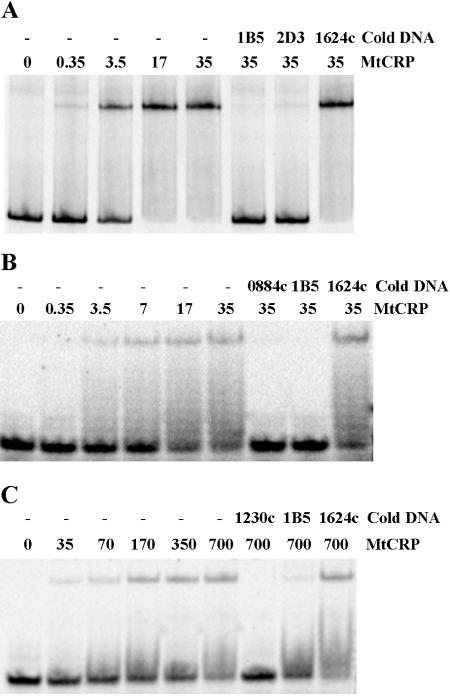
Two of the 58 sites that were computationally predicted in the M. tuberculosis genome (Table 2), including a “good” site and a “poor” site, were examined for CRPMt binding. The good site in the Rv0884c-Rv0885 intergenic region was selected because of its reproducibility. This site was consistently sampled (frequency, 1.00) during 5,000 Gibbs sampling iterations and had a high probability (1.00) of belonging to the CRPMt motif model (Fig. 1B). In contrast, the poor site upstream of Rv1230c was sampled much less frequently (0.66) and had a relatively low probability (0.47) of belonging to the model, making it the weakest of the putative regulon members. Both sites were capable of binding to CRPMt when tested by EMSA as 20-bp DNA probes, although the Rv0884c-Rv0885 site bound CRPMt more efficiently (Fig. 4).
FIG. 4.
EMSA showing CRPMt interactions with representative DNA binding sites identified by Gibbs sampling. DNA probes are as follows: A, the full-length Rv0884c-Rv0885 intergenic region, which contains a motif with a high probability of belonging to the CRPMt motif model; B, the 20-bp predicted binding site in the Rv0884c-Rv0885 intergenic region; and C, the 20-bp predicted binding site upstream of the Rv1230c open reading frame, which has a low probability of belonging to the motif model. For each lane, the CRPMt concentration (nM) is noted, and “cold DNA” specifies unlabeled competitor DNA fragments used at a 500-fold excess relative to the labeled probe DNA. The 40-bp Rv1624c nonspecific DNA was used as a negative control throughout.
Substitution of highly conserved nucleotides in CRPMt-binding DNA motif.
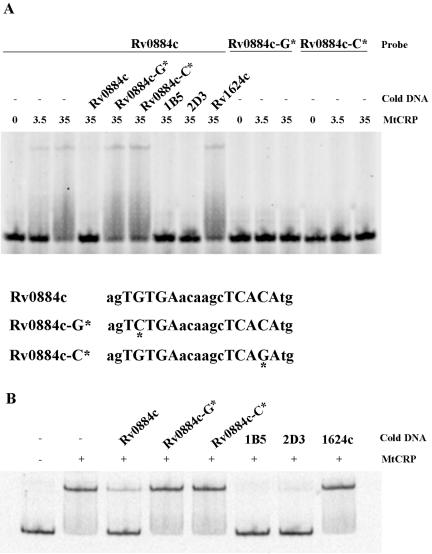
Contact between CRP and specific nucleotides within the DNA binding site is crucial for CRP-DNA interactions (30), and we reasoned that this might also be true for CRPMt. We used the sequence of the Rv0884c-Rv0885 site to examine the specificity of CRPMt binding when the strongly conserved nucleotides at positions 4 and 17 (Fig. 1B) were altered. Native and modified sites within 20-bp oligonucleotides and a 250-bp DNA fragment encompassing the entire Rv0884c-Rv0885 intergenic region were labeled and subjected to EMSA with CRPMt.
A complete loss of binding to CRPMt occurred with modified probes that contained either a G-to-C change at position 4 or a C-to-G substitution at position 17 (Fig. 5A). These modified DNA probes also failed to compete CRPMt binding to either the 20-bp native site probe (Fig. 5A) or the 250-bp intergenic DNA probe (Fig. 5B). These results indicate that in the context of the Rv0884c-Rv0885 site, these bases are essential for specific binding by CRPMt, and suggest that the highly conserved G4 and C17 positions in the motif model reflect the importance of these positions for CRPMt-DNA interactions.
FIG. 5.
Binding of CRPMt to native and modified sequences in the Rv0884c-Rv0885 intergenic sequence. (A) Sequences of the 20-bp native and modified sites are shown below the figure. Modified sites are marked with asterisks. The mobility of the native probe was retarded by CRPMt, and the reappearance of free probe was observed when a 500-fold excess of unlabeled native DNA or 1B5 or 2D3 DNA was present. The same concentration of unlabeled modified DNA and the Rv1624c 40-bp nonspecific DNA failed to compete the binding of the probe. Modified probes failed to bind CRPMt. (B) Binding of CRPMt to the Rv0884c-Rv0885 full-length intergenic DNA could be competed by a 500-fold excess of unlabeled 1B5 or 2D3 DNA, as well as by the native Rv0884c probe, but not the modified probes, as shown in panel A. “Cold DNA” refers to the competitor. The CRPMt concentration (nM) is noted for each lane in panel A and was 20 nM for panel B.
Evidence for allosteric alteration of CRPMt upon cAMP binding.
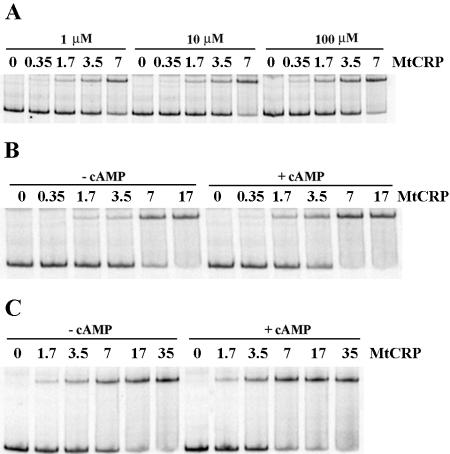
cAMP binding to CRP induces conformational changes that affect CRP's ability to interact specifically with DNA, resulting in cAMP-CRP-mediated gene regulation in E. coli (33). In this study, we used several approaches to test the ability of CRPMt to bind cAMP and to examine the effect of cAMP on CRPMt's DNA binding ability. The effect of the cAMP concentration on the CRPMt-DNA interaction was first measured by EMSA, using cAMP levels from 1 to 100 μM in the binding reaction buffer. The affinity of the protein-DNA complex was highest with 100 μM of cAMP (Fig. 6A).
FIG. 6.
Effect of cAMP on affinity of CRPMt-DNA interaction, as measured by EMSA. (A) Binding of CRPMt to the 2D3 intergenic DNA probe in the presence of different amounts of cAMP in the binding buffer, as specified. (B and C) Comparison of CRPMt binding affinities in the presence and absence of 100 μM cAMP. (B) 2D3 probe; (C) Rv0884c-Rv0885 intergenic region probe. CRPMt concentrations are shown in nM.
Complex formation was further examined with several additional probes in the presence or absence of 100 μM cAMP. CRPMt bound approximately twofold more efficiently to each of the DNA probes in the presence of 100 μM cAMP than in its absence (Fig. 6B and C). Similar results were obtained when 100 μM cAMP was also added to the gel and running buffer (data not shown). These results indicate that cAMP improves the binding of CRPMt to specific DNA sequences but is not absolutely required with any of the probes we tested.
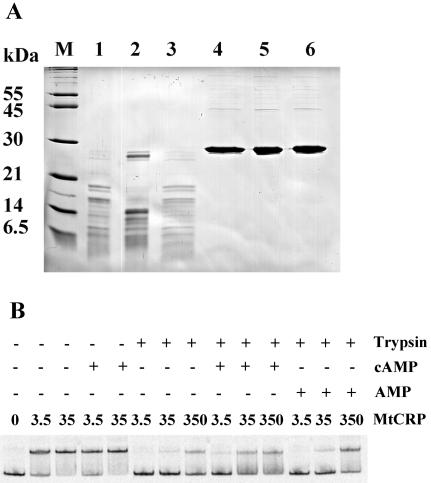
CRP loses DNA binding activity after limited proteolysis with trypsin in the presence, but not the absence, of cAMP (35). This is due to the conformational change induced in CRP upon cAMP binding. We examined the effect of cAMP on CRPMt's conformation by using limited proteolysis of CRPMt with trypsin. Surprisingly, SDS-PAGE analysis of trypsin-digested samples showed less proteolysis in the presence of 100 μM cAMP than in the absence of cAMP (Fig. 7A). In particular, a large protein fragment was retained in the presence of cAMP that was missing when digestion was performed without cAMP. The addition of AMP had no protective effect, confirming the specificity of this effect for cAMP.
FIG. 7.
Evidence of allosteric alteration of CRPMt by cAMP. (A) SDS-PAGE of CRPMt after limited proteolysis with trypsin in the presence or absence of cAMP or AMP. M, molecular weight marker (Molecular Probes). CRPMt treatment was as follows: lanes 1 to 3, trypsin digestion; lanes 4 to 6, undigested controls; lanes 2 and 5, addition of 100 μM cAMP; lanes 3 and 6, supplementation with 100 μM AMP. (B) EMSA of CRPMt using intergenic Rv0884c-Rv0885 DNA probe after limited proteolysis with trypsin in the presence or absence of 100 μM cAMP or 100 μM AMP. The CRPMt treatment is shown at the top, with the digested CRPMt concentration indicated in mM. An EMSA with all samples was performed with 100 μM cAMP in the binding reaction buffer. The figure is representative of three experimental repeats.
On a functional level, trypsin digestion of CRPMt in the absence of cAMP nearly abolished its ability to bind the Rv0884c probes (Fig. 7B). However, CRPMt's DNA binding ability was partially protected when 100 μM cAMP was present during the digestion procedure. Together, these results indicated that the conformation of CRPMt was altered in the presence of cAMP, resulting in an increased resistance of CRPMt to trypsin cleavage and partial protection of its DNA binding domain.
DNA bending by CRPMt.
The binding of CRP to specific DNA sequences induces a sharp bend in the DNA (59, 71). This induced DNA bending is an integral part of the mechanism by which CRP activates gene transcription (37, 39). Bent DNA fragments, including those bound by CRP, typically display lower electrophoretic mobilities when the bend occurs near the center of the DNA molecule (34).
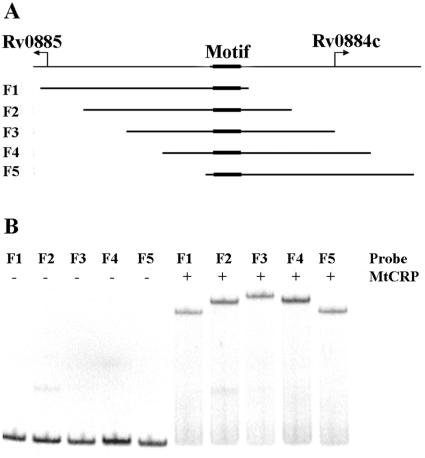
We used Rv0884c-Rv0885 intergenic DNA sequences containing the CRPMt binding site to test the possibility that CRPMt binding causes DNA bending. Five identically sized (156-bp) DNA fragments were PCR amplified from overlapping regions of the Rv0884c-Rv0885 intergenic sequence, placing the motif at different positions relative to the center of each DNA molecule (Fig. 8A). The mobilities of the DNA fragments were compared by EMSA with CRPMt.
FIG. 8.
DNA bending by CRPMt. (A) Graphic showing the genetic structure of the Rv0884c-Rv0885 intergenic region. Five 156-bp subregions, designated F1 to F5, were amplified by PCR, with the binding site at a different location within each fragment, as shown. (B) Fragments F1 through F5 were labeled and used for EMSA with 35 nM of CRPMt. Unbound probes showed similar mobilities (left half of gel), while the mobility of each protein-DNA complex varied depending on the position of the CRPMt binding site within the fragment (right side of gel).
The mobilities of all five labeled DNA fragments were quite similar in the absence of CRPMt (Fig. 8B). In contrast, the mobility of the DNA-protein complexes was affected by the position of the motif within the fragment. Complexes formed with F3, in which the motif is centrally located, had the slowest mobility, while F1 and F5 complexes, which contain a motif within 6 bp of an end of the fragment, migrated the fastest. The mobility retardation of the complexes formed with F2 or F4 was intermediate, consistent with the location of their binding sites midway between the middle and end of each fragment (Fig. 8). These results indicated that CRPMt binding caused DNA bending and are consistent with CRPMt being a CRP-like gene regulatory factor.
DISCUSSION
Combination of genomic SELEX and Gibbs sampler for identifying DNA binding sites.
The identification of regulons and their cognate regulatory factors is one of the major challenges of the current genomic era. This study shows the power of combining experimental and computational approaches. SELEX has been successfully used on a genome-wide scale to identify transcription factors and their cognate binding sites in a variety of organisms (22, 32, 51, 55, 63, 68). However, a limitation of SELEX is that its tendency to bias towards the strongest binding sequences can reduce the diversity of the DNA sequences that are recovered. Although our SELEX studies provided a critical experimental foundation early on in this investigation, these results alone were not sufficient for making genome-scale predictions about CRPMt regulon membership. The computational approach was essential for extending the DNA binding data to make regulon predictions.
Computational methods designed to detect matches to a consensus sequence or motif model suffer from poor specificity or poor sensitivity (or both) and thus are of limited use when searching genome-scale sequence data. Therefore, we employed a sequence alignment (i.e., motif discovery) algorithm, Gibbs sampling, to predict CRPMt binding sites in the M. tuberculosis genome. Gibbs sampling has been widely used to detect transcription factor binding sites and their motif models when the model is not known beforehand, where the sequence data analyzed are typically from coregulated or coexpressed genes from a single species (21) or from the promoter regions of a single gene from several closely related species (i.e., phylogenetic footprinting) (45, 47). We took a novel approach in the advanced application of Gibbs sampling described here, using this motif prediction algorithm to detect a single regulon in genome-scale data in the absence of gene expression information, i.e., to detect a single, relatively rare, sequence pattern in nearly 350,000 bp of intergenic sequence data, using Bayesian prior information on an E. coli motif.
Given the size of the sequence search space and the stochastic nature of Gibbs sampling, we do not expect to have completely delineated the CRPMt regulon. For example, the electrophoretic mobility of the relatively low-probability Rv1230c binding-site probe was obviously retarded in the presence of CRPMt (Fig. 4C), suggesting that additional binding sites remain to be discovered. It is also important that the EMSA experiments were performed in vitro with purified protein and DNA and that additional cofactors and/or competing paralogs could alter CRPMt's behavior in vivo. Future in vivo expression-based studies are expected to refine and extend these predictions in a biological context. The identification of several additional CRPMt regulon candidates from a recent expression-based study is consistent with this prediction (54).
It remains possible that the predicted regulon represents a mixture of binding sites for paralogous transcription factors, but we think this is unlikely based on our experimental results. For example, such a Gibbs sampling approach with the E. coli genome might detect a mixed model containing CRP and FNR sites. However, all of the predicted DNA sites that we tested bound specifically to CRPMt, and CRPMt was able to readily discriminate between various closely related sequences (e.g., CRP and FNR or mutant CRPMt sites). In addition, the genome sequence of M. tuberculosis H37Rv encodes relatively few transcription factors that are likely to cause the type of mixed-model regulon predictions described above. Specifically, there are only three predicted transcription factors with putative CRP/FNR-family HTH regions (Rv3676, Rv1675c, and Rv1719). Two of these proteins (Rv3676 and Rv1675c) are likely paralogs with similar domain structures (cAMP-binding domain followed by an HTH domain), whereas the third (Rv1719) contains an N-terminal HTH domain followed by an IclR-type effector binding domain (Pfam PF01614) (4). This information and our unique Gibbs sampling approach provide a foundation for generating testable hypotheses to address such issues in future studies.
Putative CRPMt regulon membership in M. tuberculosis.
The putative CRPMt regulon is consistent with the size of the CRP regulon, which contains >100 experimentally verified and newly predicted genes (9, 33, 66, 72). Despite the resemblance of CRPMt and CRP binding motif models (Fig. 2), there does not appear to be a correspondence in regulon membership. The two largest groups of genes in the predicted CRPMt regulon are both associated with in vitro dormancy models, including 14% which are linked to hypoxia (62) and 30% from a nutrient starvation model (5). crp is upregulated in this starvation model (5), suggesting that CRPMt may be important for regulating this starvation response. However, it is difficult to predict a specific role for CRPMt in M. tuberculosis latency from these data. Both previous studies reported large numbers of genes, and the proportional representation of each group in our study is similar to the proportions of these genes in the overall genome. Nonetheless, future studies on the possible role of CRPMt in latency are clearly warranted.
Another gene of interest in the predicted regulon is Rv0805, which is annotated as a homolog of E. coli cpdA, encoding a 3′,5′-cyclic-nucleotide phosphodiesterase (http://genolist.pasteur.fr/TubercuList/). Rv0805 contains a strong CRPMt motif in its upstream intergenic region and is a putative member of our CRPMt regulon. This suggests a role for CRPMt in the regulation of cAMP levels within the cell, and we are currently exploring this possibility.
Recently, Spreadbury et al. proposed 15 genes as potential Rv3676 regulon candidates, although Rv3676 binding sites were not specified. Only Rv0867c (rpfA) and Rv1552 (frdA) are predicted by both their study and the present study to be members of the putative CRPMt regulon (65). The regulation of rpfA by CRPMt has also recently been experimentally confirmed (54). While both studies made use of information from the E. coli CRP regulon, differences between studies in the approaches used for regulon prediction could account for this limited overlap. In particular, we restricted our regulon predictions by searching for putative binding sites only within intergenic regions upstream of annotated open reading frames because we expect that these sequences are most likely to be involved in transcriptional regulation. In addition, our search was genome-wide, that is, not restricted by putative gene function, because we expect that regulon membership has likely diverged between these two very distantly related bacterial species.
Functional comparison of CRPMt with CRP.
In E. coli, activation of the lac promoter involves the binding of cAMP-CRP to its cognate site, which induces DNA bending that facilitates direct contact between CRP and RNA polymerase (23, 31, 53). An amino acid sequence alignment has shown that the cAMP binding (46) and DNA binding (Fig. 1) contact residues found in CRP are conserved in CRPMt. Our results experimentally confirm CRPMt's interactions with both cAMP and DNA, including a cAMP-induced conformational change and an ability to bend DNA upon binding. Although CRPMt undergoes a conformational change in the presence of cAMP, structural differences between CRPMt and CRP were also indicated by trypsin digestion experiments. Whereas cAMP binding opens up the CRP molecule, making it more vulnerable to proteolytic cleavage (35), cAMP binding decreased CRPMt's sensitivity to trypsin digestion. This suggests that the overall topologies of these two proteins differ, at least when bound to cAMP, and this may have functional implications.
CRPMt bears some similarity to CRP*, in that CRPMt does not require cAMP for specific DNA binding in vitro, although the presence of cAMP did enhance CRPMt's DNA binding ability. Native E. coli CRP requires a cAMP-dependent conformational change to bind DNA specifically and function as a transcriptional regulator. However, mutant crp* alleles encode cAMP-independent proteins that are functionally active in vitro and in vivo in the absence of cAMP (2, 6). In addition, both CRP* and CRPMt are highly susceptible to proteolysis in the absence of cAMP but are protected from cleavage when bound to cAMP (28, 52). In contrast, unliganded CRP is resistant to digestion but is easily degraded when in complex with cAMP. CRPMt's interaction with cAMP during gene regulation remains to be explored in vivo.
It was recently reported that CRPMt was not functional in E. coli (65), and we have made similar observations (G. Bai and K. A. McDonough, unpublished). Such interspecies functional studies can be difficult to interpret. For example, the P. aeruginosa vfr gene product, a homolog of E. coli CRP, complements the β-galactosidase- and tryptophanase-deficient phenotypes of an E. coli crp mutant (69), but the E. coli crp gene does not complement the vfr mutation. These results have been interpreted as a failure of CRP to interact properly with P. aeruginosa RNA polymerase. We hypothesize that functional interactions of CRPMt may be restricted to mycobacterial RNA polymerase, and this warrants further investigation.
Despite its apparent functional similarity with CRP, the regulation of CRPMt expression differs from that of CRP and does not appear to be autoregulatory. M. tuberculosis crp does not contain a CRPMt motif in its upstream intergenic region, and its intergenic sequences failed to bind CRPMt in EMSA (data not shown). In addition, the mechanism of crp (1, 27, 48) and vfr (56) autoregulation involves competitive expression of a divergently transcribed gene, and there is no divergent open reading frame annotated upstream of M. tuberculosis crp (http://genolist.pasteur.fr). We expect that the regulation of M. tuberculosis crp will be an interesting area of future investigation.
In summary, DNA binding sites of CRPMt were identified by multiple approaches, including genome-scale experimental and computational approaches. We identified a palindromic DNA motif with similarity to the E. coli CRP binding motif and predicted a CRPMt regulon containing ∼114 genes. The interaction of CRPMt with cAMP and specific DNA binding sites was experimentally confirmed, providing the first direct evidence for cAMP interaction with a transcription factor in M. tuberculosis. These results indicate an important role for cAMP signal transduction during global gene regulation in M. tuberculosis. Future studies of cAMP-mediated gene regulation are likely to contribute to an understanding of M. tuberculosis's response to the host environment during infection.
Acknowledgments
We gratefully acknowledge Victoria Derbyshire and Qingqing Liu for many helpful discussions and Eric Smith for excellent technical assistance. We thank the Wadsworth Center's Molecular Genetics Core for DNA sequencing and Computational Molecular Biology and Statistics Core for assistance with data analysis.
This work was supported in part by National Institute of Health grants AI4565801 and AI063499.
REFERENCES
- 1.Aiba, H. 1983. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell 32:141-149. [DOI] [PubMed] [Google Scholar]
- 2.Aiba, H., T. Nakamura, H. Mitani, and H. Mori. 1985. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 4:3329-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1: 75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 6.Blazy, B., and A. Ullmann. 1986. Properties of cyclic AMP-independent catabolite gene activator proteins of Escherichia coli. J. Biol. Chem. 261:11645-11649. [PubMed] [Google Scholar]
- 7.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan, M. J., and G. Delogu. 2002. The PE multigene family: a “molecular mantra” for mycobacteria. Trends Microbiol. 10:246-249. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. T., and C. G. Callan, Jr. 2004. Evolutionary comparisons suggest many novel cAMP response protein binding sites in Escherichia coli. Proc. Natl. Acad. Sci. USA 101:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busby, S., and H. Buc. 1987. Positive regulation of gene expression by cyclic AMP and its receptor protein in Escherichia coli. Microbiol. Sci. 4:371-375. [PubMed] [Google Scholar]
- 11.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 12.Caler, E. V., R. E. Morty, B. A. Burleigh, and N. W. Andrews. 2000. Dual role of signaling pathways leading to Ca2+ and cyclic AMP elevation in host cell invasion by Trypanosoma cruzi. Infect. Immun. 68:6602-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cann, M. J., A. Hammer, J. Zhou, and T. Kanacher. 2003. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J. Biol. Chem. 278:35033-35038. [DOI] [PubMed] [Google Scholar]
- 14.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 15.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 16.Crasnier, M. 1996. Cyclic AMP and catabolite repression. Res. Microbiol. 147:479-482. [DOI] [PubMed] [Google Scholar]
- 17.Curtiss, R., III, R. M. Goldschmidt, N. B. Fletchall, and S. M. Kelly. 1988. Avirulent Salmonella typhimurium delta cya delta crp oral vaccine strains expressing a streptococcal colonization and virulence antigen. Vaccine 6:155-160. [DOI] [PubMed] [Google Scholar]
- 18.de Crombrugghe, B., S. Busby, and H. Buc. 1984. Cyclic AMP receptor protein: role in transcription activation. Science 224:831-838. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25: 349-364. [DOI] [PubMed] [Google Scholar]
- 20.Espinal, M. A. 2003. The global situation of MDR-TB. Tuberculosis (Edinburgh) 83:44-51. [DOI] [PubMed] [Google Scholar]
- 21.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freede, P., and S. Brantl. 2004. Transcriptional repressor CopR: use of SELEX to study the copR operator indicates that evolution was directed at maximal binding affinity. J. Bacteriol. 186:6254-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaston, K., A. Bell, A. Kolb, H. Buc, and S. Busby. 1990. Stringent spacing requirements for transcription activation by CRP. Cell 62:733-743. [DOI] [PubMed] [Google Scholar]
- 24.Gazdik, M. A., and K. A. McDonough. 2005. Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J. Bacteriol. 187:2681-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross, A., M. Bouaboula, P. Casellas, J. P. Liautard, and J. Dornand. 2003. Subversion and utilization of the host cell cyclic adenosine 5′-monophosphate/protein kinase A pathway by Brucella during macrophage infection. J. Immunol. 170:5607-5614. [DOI] [PubMed] [Google Scholar]
- 26.Guo, Y. L., T. Seebacher, U. Kurz, J. U. Linder, and J. E. Schultz. 2001. Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J. 20:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanamura, A., and H. Aiba. 1991. Molecular mechanism of negative autoregulation of Escherichia coli crp gene. Nucleic Acids Res. 19:4413-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harman, J. G., and W. J. Dobrogosz. 1983. Mechanism of CRP-mediated cya suppression in Escherichia coli. J. Bacteriol. 153:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobson, R. J., A. J. McBride, K. E. Kempsell, and J. W. Dale. 2002. Use of an arrayed promoter-probe library for the identification of macrophage-regulated genes in Mycobacterium tuberculosis. Microbiology 148:1571-1579. [DOI] [PubMed] [Google Scholar]
- 30.Jansen, C., A. M. Gronenborn, and G. M. Clore. 1987. The binding of the cyclic AMP receptor protein to synthetic DNA sites containing permutations in the consensus sequence TGTGA. Biochem. J. 246:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joung, J. K., L. U. Le, and A. Hochschild. 1993. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc. Natl. Acad. Sci. USA 90:3083-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, S., H. Shi, D. K. Lee, and J. T. Lis. 2003. Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 31:1955-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62: 749-795. [DOI] [PubMed] [Google Scholar]
- 34.Kolb, A., A. Spassky, C. Chapon, B. Blazy, and H. Buc. 1983. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 11:7833-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krakow, J. S., and I. Pastan. 1973. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc. Natl. Acad. Sci. USA 70:2529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson, C. L., D. Swigon, K. S. Murakami, S. A. Darst, H. M. Berman, and R. H. Ebright. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14:10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, N., C. A. D'Souza, and J. W. Kronstad. 2003. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 41:399-427. [DOI] [PubMed] [Google Scholar]
- 38.Li, C. C., D. S. Merrell, A. Camilli, and J. B. Kaper. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43:1577-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, S. H., and J. C. Lee. 2003. Determinants of DNA bending in the DNA-cyclic AMP receptor protein complexes in Escherichia coli. Biochemistry 42:4809-4818. [DOI] [PubMed] [Google Scholar]
- 40.Linder, J. U., A. Hammer, and J. E. Schultz. 2004. The effect of HAMP domains on class IIIb adenylyl cyclases from Mycobacterium tuberculosis. Eur. J. Biochem. 271:2446-2451. [DOI] [PubMed] [Google Scholar]
- 41.Linder, J. U., A. Schultz, and J. E. Schultz. 2002. Adenylyl cyclase Rv1264 from Mycobacterium tuberculosis has an autoinhibitory N-terminal domain. J. Biol. Chem. 277:15271-15276. [DOI] [PubMed] [Google Scholar]
- 42.Liu, H. 2002. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292:299-311. [DOI] [PubMed] [Google Scholar]
- 43.Liu, J. S., and C. E. Lawrence. 1999. Bayesian inference on biopolymer models. Bioinformatics 15:38-52. [DOI] [PubMed] [Google Scholar]
- 44.Lowrie, D. B., P. S. Jackett, and N. A. Ratcliffe. 1975. Mycobacterium microti may protect itself from intracellular destruction by releasing cyclic AMP into phagosomes. Nature 254:600-602. [DOI] [PubMed] [Google Scholar]
- 45.McCue, L., W. Thompson, C. Carmack, M. P. Ryan, J. S. Liu, V. Derbyshire, and C. E. Lawrence. 2001. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 29:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10:204-219. [DOI] [PubMed] [Google Scholar]
- 47.McCue, L. A., W. Thompson, C. S. Carmack, and C. E. Lawrence. 2002. Factors influencing the identification of transcription factor binding sites by cross-species comparison. Genome Res. 12:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto, K., and M. Freundlich. 1986. Mechanism for the autogenous control of the crp operon: transcriptional inhibition by a divergent RNA transcript. Proc. Natl. Acad. Sci. USA 83:5000-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen, A. W., and P. Andersen. 2003. A novel TB vaccine; strategies to combat a complex pathogen. Immunol. Lett. 85:207-211. [DOI] [PubMed] [Google Scholar]
- 50.Petersen, S., and G. M. Young. 2002. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70: 3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren, Y. L., S. Garges, S. Adhya, and J. S. Krakow. 1988. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc. Natl. Acad. Sci. USA 85:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reznikoff, W. S. 1992. Catabolite gene activator protein activation of lac transcription. J. Bacteriol. 174:655-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menendez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roulet, E., S. Busso, A. A. Camargo, A. J. Simpson, N. Mermod, and P. Bucher. 2002. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 20:831-835. [DOI] [PubMed] [Google Scholar]
- 56.Runyen-Janecky, L. J., A. K. Sample, T. C. Maleniak, and S. E. West. 1997. A divergently transcribed open reading frame is located upstream of the Pseudomonas aeruginosa vfr gene, a homolog of Escherichia coli crp. J. Bacteriol. 179:2802-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 58.Schneider, T. D., G. D. Stormo, L. Gold, and A. Ehrenfeucht. 1986. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188:415-431. [DOI] [PubMed] [Google Scholar]
- 59.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 60.Seoh, H. K., and P. C. Tai. 1999. Catabolic repression of secB expression is positively controlled by cyclic AMP (cAMP) receptor protein-cAMP complexes at the transcriptional level. J. Bacteriol. 181:1892-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenoy, A. R., N. P. Sreenath, M. Mahalingam, and S. S. Visweswariah. 2005. Characterization of phylogenetically distant members of the adenylate cyclase family from mycobacteria: Rv1647 from Mycobacterium tuberculosis and its orthologue ML1399 from M. leprae. Biochem. J. 387:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer, B. S., T. Shtatland, D. Brown, and L. Gold. 1997. Libraries for genomic SELEX. Nucleic Acids Res. 25:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha, S. C., M. Wetterer, S. R. Sprang, J. E. Schultz, and J. U. Linder. 2005. Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c. EMBO J. 24:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spreadbury, C. L., M. J. Pallen, T. Overton, M. A. Behr, S. Mostowy, S. Spiro, S. J. Busby, and J. A. Cole. 2005. Point mutations in the DNA- and cNMP-binding domains of the homologue of the cAMP receptor protein (CRP) in Mycobacterium bovis BCG: implications for the inactivation of a global regulator and strain attenuation. Microbiology 151:547-556. [DOI] [PubMed] [Google Scholar]
- 66.Tan, K., G. Moreno-Hagelsieb, J. Collado-Vides, and G. D. Stormo. 2001. A comparative genomics approach to prediction of new members of regulons. Genome Res. 11:566-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, W., L. A. McCue, and C. E. Lawrence. 2005. Using the Gibbs motif sampler to find conserved domains in DNA and protein sequences, p. 2.8.1-2.8.42. In A. D. Baxevanis and D. B. Davison (ed.), Current protocols in bioinformatics. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 68.Wen, J. D., and D. M. Gray. 2004. Selection of genomic sequences that bind tightly to Ff gene 5 protein: primer-free genomic SELEX. Nucleic Acids Res. 32:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.West, S. E., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 71.Wu, H. M., and D. M. Crothers. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature 308:509-513. [DOI] [PubMed] [Google Scholar]
- 72.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]