Abstract
The SOS response in bacteria includes a global transcriptional response to DNA damage. DNA damage is sensed by the highly conserved recombination protein RecA, which facilitates inactivation of the transcriptional repressor LexA. Inactivation of LexA causes induction (derepression) of genes of the LexA regulon, many of which are involved in DNA repair and survival after DNA damage. To identify potential RecA-LexA-regulated genes in Bacillus subtilis, we searched the genome for putative LexA binding sites within 300 bp upstream of the start codons of all annotated open reading frames. We found 62 genes that could be regulated by putative LexA binding sites. Using mobility shift assays, we found that LexA binds specifically to DNA in the regulatory regions of 54 of these genes, which are organized in 34 putative operons. Using DNA microarray analyses, we found that 33 of the genes with LexA binding sites exhibit RecA-dependent induction by both mitomycin C and UV radiation. Among these 33 SOS genes, there are 22 distinct LexA binding sites preceding 18 putative operons. Alignment of the distinct LexA binding sites reveals an expanded consensus sequence for the B. subtilis operator: 5′-CGAACATATGTTCG-3′. Although the number of genes controlled by RecA and LexA in B. subtilis is similar to that of Escherichia coli, only eight B. subtilis RecA-dependent SOS genes have homologous counterparts in E. coli.
Exposure of prokaryotes to DNA-damaging agents results in the induction of a diverse set of physiological responses collectively called the SOS response (8, 55). As first characterized in Escherichia coli, the SOS response includes an enhanced capacity for recombinational repair, enhanced capacity for excision repair, enhanced mutagenesis (due to error-prone repair), and inhibition of cell division (i.e., filamentation). Induction of the SOS response is due to the coordinate derepression of a number of SOS or din (for damage-inducible) genes. The SOS response to DNA damage in Bacillus subtilis is similar to that of E. coli (26, 56, 58), but unlike E. coli, the B. subtilis SOS system is also induced in competent cells in the absence of any DNA-damaging treatment (25, 57, 58). As in E. coli, SOS gene expression in B. subtilis is controlled by two proteins (which are themselves products of SOS genes): the LexA protein (also called DinR) (40, 54), which represses the transcription of din genes by binding to the SOS operator (31), and the RecA protein (30), which is activated by single-stranded DNA (29, 42) to stimulate the proteolytic autodigestion of LexA (24, 31). Thus, an SOS gene is defined by two criteria—RecA-dependent induction by DNA damage and a binding site for LexA overlapping its promoter.
By contrast with E. coli, where more than 30 SOS genes have been identified (7, 8), only 5 B. subtilis SOS genes have been shown to meet both SOS gene criteria thus far: recA, lexA, uvrB (formerly dinA), dinB, and dinC (also called tagC) (4, 9, 15, 25). The E. coli uvrB gene encodes part of the UvrABC endonuclease, which catalyzes nucleotide excision repair of a variety of DNA lesions (41). Both the E. coli uvrA and uvrB genes are damage-inducible SOS genes with LexA binding sites overlapping their promoters (8); the E. coli uvrC gene is not damage inducible (32), and LexA does not bind to its promoter region in vitro (12). Homologs of all three uvr genes are present in B. subtilis, and genetic and biochemical evidence indicate that the uvrA, uvrB, and uvrC genes are involved in excision repair (14, 23). The functions of the dinB and dinC genes are unknown. Because it is adjacent to the tag operon (which codes for enzymes involved in teichoic acid synthesis), the dinC gene has been named tagC; however, there is no evidence for its involvement in teichoic acid synthesis. We will refer to it as dinC in this report.
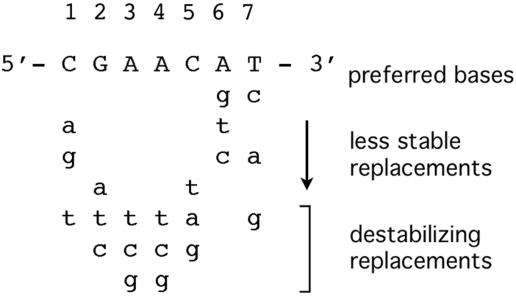
We report here the identification of 28 additional SOS genes in B. subtilis. They all have LexA binding sites, or SOS boxes, within their promoter regions, and they are induced by at least two distinct DNA-damaging treatments in RecA+, but not recA null, cells. The consensus operator sequence for the binding of a B. subtilis LexA dimer, 5′-CGAACN4GTTCG-3′, has been characterized by mutational analyses and DNA binding studies (4, 5, 27, 53). A study of LexA binding to recA operator mutants suggests the requirements for site-specific LexA binding summarized in Fig. 1 (E. S. Groban, N. Au, M. B. Johnson, P. Banky, P. G. Burnett, G. L. Calderon, E. C. Dwyer, S. N. Fuller, B. Gebre, L. M. King, I. N. Sheren, L. D. Von Mutius, T. M. O'Gara, and C. M. Lovett, submitted for publication). According to the study, the thermodynamically preferred half site sequence for LexA binding is 5′-CGAACAT-3′; certain substitutions do not reduce binding affinity significantly, while others (labeled destabilizing replacements) abolish binding altogether. Guided by these binding requirements, we searched the genome for sites within putative promoter regions that could potentially bind LexA. We assessed binding activity using mobility shift assays and we identified genes that show RecA-dependent induction by DNA damage using genomic microarrays.
FIG. 1.
Sequence requirements for LexA binding. The preferred half site sequence based on a thermodynamic analysis of LexA binding to recA operator mutants. Base substitutions labeled as destabilizing abolish LexA binding to the recA operator (Groban et al., submitted).
MATERIALS AND METHODS
Materials.
The B. subtilis LexA protein was purified as described previously (31). Oligonucleotide primers were purchased from Sigma Genosys. Pfu polymerase (Stratagene), T4 kinase (Promega Corp.), and SuperScript II RNase H- reverse transcriptase (Invitrogen) were used as recommended by the manufacturers. Microarrays covering >99% of the B. subtilis open reading frames were prepared as previously described and spotted onto GAPS II slides from Corning (16).
Preparation of promoter regions for mobility shift assays.
DNA containing putative SOS operators was prepared by PCR amplification of B. subtilis YB886 (59) DNA (10 ng/ml) using synthetic oligonucleotide primers (2 μM) with a Peltier PTC-200 thermal cycler (MJ Research). Samples of amplified DNA were electrophoresed alongside DNA samples of known concentration; gels were analyzed by densitometry with an Alpha Innotech imaging system, and the concentration of amplified DNA was interpolated from DNA standard curves. The promoter regions prepared by PCR amplification were radiolabeled with [γ-32P]ATP using T4 kinase. Radiolabeled DNA was purified by electrophoresis on an 8% nondenaturing polyacrylamide gel.
Mobility shift assays.
For competition experiments, purified LexA was incubated with radiolabeled recA promoter DNA (5 to 10 nM) and a 5- to 50-fold molar excess of competitor DNA for 30 min at 25°C in mobility shift buffer, which consisted of 12 mM HEPES-NaOH (pH 7.9), 4 mM Tris-Cl (pH 7.9), 12% glycerol, 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 2 μg poly(dI-dC) · poly(dI-dC), and 0.3 mg/ml bovine serum albumin. This incubation mixture (10 μl) was loaded on a 4% (acrylamide:bisacrylamide ratio of 80:1) nondenaturing polyacrylamide gel, and electrophoresis was begun immediately. The buffer within the gel and the running buffer were both 25 mM Tris-Cl (pH 8.5), 250 mM glycine, and 1 mM EDTA. Samples were electrophoresed, and the dried gel was subjected to densitometric analysis using a Bio-Rad Molecular Imager FX phosphorimaging system.
For determination of the apparent binding constant, Kd, purified LexA was incubated with radiolabeled promoter DNA (5 to 10 nM) for 30 min at 25°C in mobility shift buffer, and 10 μl of this incubation mixture was subjected to electrophoresis and phosphorimaging analysis as described above. For Kd determinations using promoter fragments as competitive inhibitors of LexA binding to the recA operator, purified LexA was incubated with radiolabeled recA promoter DNA (5 to 10 nM) and competitor DNA (50 nM) for 30 min at 25°C in mobility shift buffer and analyzed as described above.
Microarray analyses.
DNA microarrays were prepared using PCR products from >99% of the annotated B. subtilis open reading frames spotted onto Corning GAPS II slides, essentially as described previously (16). Prior to hybridization with biological samples, arrays were prehybridized for at least 45 min at 42°C in 1% bovine serum albumin, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), washed in water, and dried (16).
Cultures (25 ml) of strain YB886 (metB5 trpC2 xin-1 SPβ− amyE sigB) or YB3000 (YB886 recA260) were grown in defined minimal medium to an optical density at 600 nm of 0.3, treated with 1 μg/ml mitomycin C (MC) or 25 mJ/m2 UV radiation, incubated for 1 h at 37°C, and harvested by centrifugation. For UV treatment, cells were pelleted, resuspended in SMS minimal medium (16a), transferred to sterile petri plates, and exposed to a 254-nm germicidal lamp to a dose of 25 mJ/m2. Survival at this dose was between 20% and 50%. Untreated cells were handled similarly except they were not UV irradiated. Culture samples were immediately mixed with an equal volume of methanol (prechilled to −20°C). Samples were then spun to pellet the cells, the supernatant was discarded, and cell pellets were frozen at −80°C until further use. RNA was extracted using QIAGEN RNeasy kits combined with on-column DNase treatment according to the manufacturer's recommendations. RNA was then precipitated with ethanol and LiCl and resuspended in RNase-free water to a desired concentration, usually ≥1.0 mg/ml. The quality of RNA was checked on agarose gels by visualizing the integrity of the 23S and 16S rRNA.
To generate cDNA, RNA from the different experimental conditions was reverse transcribed in the presence of amino-allyl-dUTP, followed by coupling to Cy5 for all experimental samples or Cy3 for all reference RNAs. Reference RNA was made by pooling RNA samples from various strains grown under conditions similar to the experimental conditions. For reverse transcriptase reactions, 10 μg RNA template was mixed with 2.5 μg random hexamers (in 18 μl) and incubated at 70°C for 10 min and on ice for 5 min. Reverse transcription reactions were then started by the addition of a cocktail resulting in a final mix of RNA template, random hexamer primers, and 300 U SuperScript II reverse transcriptase, 1× reverse transcription buffer, 10 mM DTT, and deoxyribonucleoside triphosphates (0.5 mM each dATP, dCTP, and dGTP; 0.1 mM dTTP; 0.4 mM aminoallyl-dUTP) in a final volume of 30 μl. The labeling reaction mixtures were incubated at 25°C for 10 min and at 42°C for 70 min and then shifted to 70°C for 15 min to stop the reactions. RNA in the reaction mixtures was degraded by adding NaOH (33 mM final concentration) and incubating at 70°C for 10 min. HCl (33 mM) was added to each reaction mixture to neutralize the pH.
Reaction mixtures were purified with QIAGEN MinElute kits and eluted in 10-μl volumes, and 0.5 μl of 1 M NaHCO3 (pH 9.0) was added to adjust the pH for the coupling reactions. To couple the fluorescent dyes to cDNA, 1 μl freshly dissolved Cy3 or Cy5 dye (Amersham) was added to cDNA and incubated for 1 h in the dark, mixing every 15 min. Reactions were quenched by incubation with 1.4 M hydroxylamine for 15 min. Each experimental (Cy5-labeled) sample was mixed with an aliquot of reference RNA (labeled with Cy3), and mixed samples were purified with QIAGEN MinElute kits. The labeled samples were mixed with 10 μg salmon sperm DNA and 0.8 μg yeast tRNA, and the volume was adjusted to 14 μl. The samples were heated to 100°C for 5 min, spun down, mixed with 2× hybridization buffer (0.05% SDS, 5× SSC, 25% formamide final concentration) and hybridized to DNA on a microarray for at least 16 h at 42°C. Following hybridization, arrays were washed with 1× SSC-0.2% SDS for 5 min at 42°C, followed by a 5-min wash with 0.1× SSC-0.2% SDS at room temperature, and a final 5-min wash in 0.1× SSC at room temperature. Arrays were spun to remove extra liquid and dried with nitrogen gas.
Arrays were scanned and analyzed with GenePix 3.0 software (Axon Instruments, Inc.). The signal intensity for each spot (gene) [(Cy5/Cy3)g] was normalized to the total signal intensity [(Cy5/Cy3)t] on the array (essentially the sum of all the spots). Normalized ratios of experimental RNA abundance over reference RNA was obtained for each spot for which 80% of the pixels had intensities at least 1 standard deviation above background. In an average experiment, 96% of all genes gave such a signal. For genes of interest, we verified that the intensities of 80% of the pixels in the spot were at least 2 standard deviations above background. To compare two conditions, the normalized signals from condition A (e.g., cells treated with MC) were divided by the corresponding signals from condition B (cells not treated with MC): [(Cy5/Cy3)g/(Cy5/Cy3)t] under condition A/[(Cy5/Cy3)g/(Cy5/Cy3)t] under condition B.
Since all experiments were done at least in triplicate, we report the average ratio of ratios from all repetitions. Statistical analysis of microarrays (48) was used for all experiments. The input data contained the replicate ratios of experimental over reference samples in log2 format. Thus, for each gene, at least six values were input for calculation of significance—three from one condition and three from the other. For all experiments, an effect on a gene is considered statistically significant if there is less than 1% probability that this change occurred by chance (i.e., a false discovery rate of less than 1.0%).
RESULTS
Search for SOS operators.
Our strategy for identifying SOS genes was to search for SOS operators, or LexA binding sites, within promoter regions. The search was guided by the sequence requirements predicted from a thermodynamic analysis of LexA binding to recA operator mutants (Groban et al., submitted). The study showed that positions 2 to 5 (i.e., 5′-GAAC-3′) of the operator half site are particularly important for site-specific binding; two mismatches in these critical regions of either half site were shown to abolish site-specific binding. Using the Subtilist web server, we searched the B. subtilis genome for the abbreviated operator consensus sequence, 5′-GAACN4GTTC-3′, allowing for one mismatch in either of the two half sites and located within 300 bp upstream of a start codon. We further refined the search by eliminating candidates containing bases that destabilize binding enough to abolish operator binding (Fig. 1). For example, sequences lacking any of the AT base pairs were eliminated because these bases have been shown to be essential for operator binding (Groban et al., submitted). The search yielded at least one canonical SOS operator sequence upstream of 62 genes (listed in Table 1 and the top of Table 2), including the five previously characterized SOS genes.
TABLE 1.
B. subtilis genes with LexA binding sites in their putative promoter regions
| Genea | Operator sequenceb | Positionc | Kdd (nM) | Fold inductione |
|---|---|---|---|---|
| ybaK | aGAACATtTGTTCc | −126 (+15) | 6.5 | 2.3 |
| cwlD | 2.1 | |||
| ↑dinB | aGAACtcATGTTCG | −42 (−14) | 3.6 | 55 |
| ↓ydgG | CGAACATgaGTTCt | −114 (−63) | 3.6 | 1.4 |
| ydgH | 1.2 | |||
| ydiO | aGAACATtcGTTCt | −51 (+13) | 16 | 3.5 |
| ydiP | 3.6 | |||
| pcrA | aGAACgTATGTTtt | −22 (+8) | 22 | 2.0 |
| ligA | 2.0 | |||
| yerH | ND | |||
| yhaZ | aGAACgTAcaTTCc | −39 (−15) | 20 | 11 |
| yhaO | aGAACgTgcaTTCG | −50 (−27) | 29 | 3.5 |
| yhaN | 1.6 | |||
| yhaM | 2.0 | |||
| ↑yhjD | aGAACAaAcGTTCc | −21 (+14) | 7.7 | 16 |
| yhjC | 2.9 | |||
| yhjB | 3.8 | |||
| ↓yhjE | gGAACgTtTGTTCt | −119 (−60) | 7.7 | 1.2 |
| xkdA | aGAACAcAcGTTCG | −15 | 6.1 | 3.8 |
| ykvR | CGAACgTATGTTtG | −111 (−64) | 13 | 1.4 |
| recA | CGAAtATgcGTTCG | −73 (−44) | 4.6 | 9.4 |
| aprX | CGAACAaAcGTTCt | −166 (+9,−92) | 3.9 | 2.7 |
| lexA (1) | gGAAtgTtTGTTCG | −125 (−99) | 5.7 | 3.4 |
| lexA (2) | CGAACAaAcGTTtc | −88 (−62) | ||
| ↑lexA (3) | CGAACcTATGTTtG | −59 (−33) | ||
| ↓yneA (1) | CaAACATAgGTTCG | −49 (−44) | 5.7 | 37 |
| yneA (2) | gaAACgTtTGTTCG | −15 (−15) | ||
| yneA (3) | CGAACAaAcaTTCc | −12 (+23) | ||
| yneB | 44 | |||
| ynzC | 8.1 | |||
| parE | CaAACATAcGTTCt | −205 (+17) | 9.6 | 2.9 |
| parC | 3.0 | |||
| yozL (1) | CGAACtTtTGTTCt | −99 (−17) | 5.8 | 1.5 |
| ↑yozL (2) | gGAACgTtTGTTCt | −68 (+14) | ||
| yozK | 1.4 | |||
| yobH | 1.2 | |||
| ↓yozM (1) | aGAACAaAcGTTCc | −127 (−74) | 5.8 | 1.3 |
| yozM (2) | aGAACAaAaGTTCG | −97 (−44) | ||
| yorB | aGAACActTGTTCc | −62 | 12 | 1.7 |
| yolC (1) | aGAACAaAcGTTCt | −127 (−74) | 3.9 | 1.2 |
| ↑yolC (2) | aGAACAaAaGTTCG | −97 (−44) | ||
| ↓yolD (1) | CGAACtTtTGTTCt | −64 (−17) | 3.9 | 1.6 |
| yolD (2) | aGAACgTtTGTTCt | −34 (+14) | ||
| uvrX | 1.4 | |||
| hbs | gGAAtATtcGTTCG | −280 (−53) | 32 | −1.1 |
| ypuD | aGAACATAaaTTCG | −157 | 6.4 | 1.3 |
| yqjW (1) | CGAACATActTTCG | −43 (−15) | 4.1 | 3.5 |
| ↑yqjW (2) | CGAACATAaGTTCt | −15 (+14) | ||
| yqjX | 5.7 | |||
| yqjY | 1.8 | |||
| yqjZ | 1.9 | |||
| ↓yqzH (1) | aGAACtTATGTTCG | −138 (−114) | 4.1 | 1.7 |
| yqzH (2) | CGAAagTATGTTCG | −110 (−86) | ||
| ↑yqhB | CaAACtTtTGTTCt | −130 | 140 | 1.2 |
| ↓yqxL | aGAACAaAaGTTtG | −13 | 1.1 | |
| sda | aGAACgatTGTTCt | −113 (−60) | 10 | 1.7 |
| ruvA | CGAACATATGTTaa | −65 (−45) | 67 | 2.2 |
| ruvB | 2.2 | |||
| uvrC | aaAACAaAcGTTCG | −45 (−16) | 40 | 1.6 |
| dnaE | aGAACATtTGTTtc | −62 (−32) | 39 | 1.2 |
| uvrB | CGAACtTtaGTTCG | −79 (−42) | 4.1 | 11 |
| uvrA | 12 | |||
| dinC (1) | aGAACAagTGTTCt | −85 (−44) | 2.3 | 175 |
| dinC (2) | CGAACgTATGTTtG | −55 (−14) | ||
| vpr | CGAACgTATaTTCc | −177 (−55) | 6.8 | 1.1 |
| Half site consensus sequence | C28G56A66A66C58A34T40 |
Genes listed by map position. The first gene in operons is shown in boldface type; subsequent genes in putative multigene operons are shown indented. Arrows indicate divergently transcribed genes. The numbers in parentheses indicate different operators within the same promoter region.
Lowercase nucleotides and uppercase nucleotides indicate nonconsensus and consensus, respectively.
Location of the 3′ end relative to the ATG codon of the respective gene (and relative to the 3′ end of the −10 region of the canonical promoter sequence).
Apparent binding constant.
Fold induction following treatment with MC in wild-type cells; data also listed in Table 4.
TABLE 2.
Canonical SOS boxes within promoter regions that are not bound by LexA
| Gene | Sequencea | Positionb | No. of mismatches | Fold inductionc |
|---|---|---|---|---|
| flhO | aGAACgaAcGTTCc | −44 | 5 | −1.4 |
| yckD | CGAAtAatgGTTCG | −75 | 4 | 1.4 |
| yobQ | gGAACgcATGTTtt | −110 | 5 | 1.4 |
| yokF | aGAACAaAcaTTCt | −17 | 5 | 1.8 |
| yopS | gGAACgTgcGTTCt | −119 | 5 | 1.3 |
| yopT | aGAACgcAcGTTCc | −51 | 5 | 1.3 |
| yorL | aGAACtTgTGTTtt | −15 | 5 | 1.4 |
| yuiC | gGAACAatgaTTCG | −13 | 5 | 1.0 |
| Two 5′-GAAC-3′ mismatches | ||||
| azlB | CGAAtAaAaaTTCG | −57 | 4 | 1.2 |
| ctaA | aaAACAcATaTTCG | −70 | 4 | 1.2 |
| ctpA | CGAAtAagaaTTCG | −53 | 5 | 1.1 |
| icd | CaAACAaAaaTTCG | −200 | 4 | 1.3 |
| yprA | CaAACAaATaTTCG | −32 | 3 | 1.5 |
| yvsG | CaAACATAcaTTCt | −70 | 4 | 2.3 |
| Destabilizing mismatches | ||||
| appC | CGAACAaATtTTCa | −178 | 3 | −2.8 |
| divIC | CGAAacaATGTTtG | −146 | 4 | −1.2 |
| dltA | CGAAtAccgGTTCa | −39 | 5 | −1.2 |
| dnaX | CGAAacaAgGTTCa | −42 | 5 | 2.2d |
| hxlA | tGAACAataaTTCG | −33 | 5 | 1.4 |
| menE | CaAACATcaGTTCa | −127 | 4 | 1.1 |
| oxdC | CGAAaAgAaGTTtG | −184 | 4 | 1.3 |
| recO | gGAACgTATtTTCt | −151 | 4 | −1.3 |
| recQ | gGAACAgcgGTTCa | −46 | 5 | 1.1 |
| rocC | CaAACcaccGTTCa | −58 | 6 | 1.4 |
| sigE | CGAAaATgctTTCG | −90 | 4 | 1.2 |
| sigF | gGAACAacgaTTCG | −55 | 5 | 1.2 |
| xkdJ | tGAACAgcTGTTtG | −60 | 4 | 1.1 |
| ydjJ | CaAACATtcGTTCa | −87 | 4 | 1.2 |
| ycgM | tGAACgctgGTTtt | −60 | 7 | 3.1 |
| yqaL | tGAACtTccGTTtG | −70 | 5 | −1.2 |
| yraH | aGAAtcgcTGTTTt | −159 | 7 | 6.2 |
| yrdC | aGAACgcATtTTCc | −108 | 5 | 1.1 |
Lowercase and uppercase nucleotides indicate nonconsensus and consensus, respectively. Destabilizing mismatches are shown in boldface.
Location of the 3′ end relative to the ATG codon of the respective gene.
The dnaX gene is induced about twofold in both wild-type and recA mutant cells.
Identification of 54 genes with LexA binding sites within their putative promoter regions.
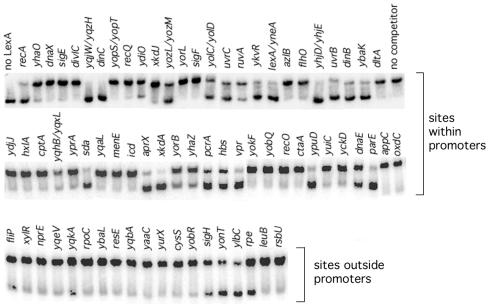
To determine whether LexA can bind specifically to the SOS operator candidates identified by our search, we tested the ability of DNA fragments containing the putative binding sites to compete with the recA operator in mobility shift assays. Figure 2 shows the results of mobility shift experiments in which LexA was incubated with recA promoter DNA and a 5- to 50-fold molar excess of competitor DNA. Using our assay conditions, a 50-fold molar excess of a DNA fragment that causes an easily detected 5% decrease in binding would correspond to a dissociation constant in the micromolar range, which is well beyond the range for specific binding (see below). DNA corresponding to the putative promoter regions of 34 operons comprising 54 genes, including the five known SOS genes, displaced LexA to various degrees from the recA promoter. Nine of these putative promoter fragments precede genes that are divergently transcribed: the yqjW and yqzH genes, the yolC and yolD genes, the yozL and yozM genes, the dinB and ydgG genes, the yhjD and yhjE genes, the yqhB and yqxL genes, and the lexA and yneA genes share LexA binding sites in their putative promoter regions. Twelve of the DNA fragments that bind LexA are upstream of putative operons containing two or more genes: ruvAB, uvrBA, parEC, ydiOP, yhjDCB, yhaONM, yqjWXYZ, yneAB-ynzC, pcrA-ligA, ybaK-cwlD, yolD-uvrX, and yozLK-yobH. The pcrA and ligA genes may be part of a four-gene operon, pcrB-pcrA-ligA-yerH, presumably transcribed from the pcrB promoter (38). However, the pcrA gene, which is homologous to the E. coli SOS gene uvrD (44), has an upstream SOS operator sequence immediately following the −10 region of a canonical σA promoter sequence.
FIG. 2.
Binding of B. subtilis LexA to potential SOS promoters. Mobility shift assays were conducted with purified LexA, radiolabeled recA promoter DNA (5 to 10 nM), and a 5- to 50-fold molar excess of the indicated promoter DNA as described in Materials and Methods. The lower and upper bands correspond to unbound and LexA-bound recA promoter DNA, respectively. Lanes with no LexA protein or competitor DNA added are indicated.
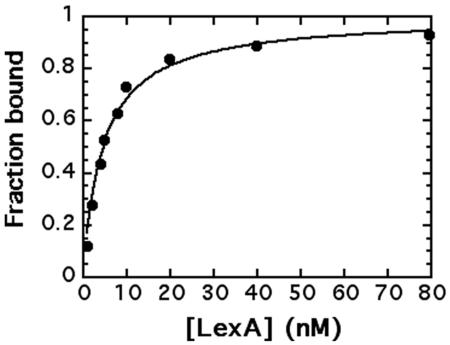
The apparent binding constant, Kd, was recently determined for the recA, dinB, dinC, and uvrB operators by titrating promoter fragments with purified LexA in mobility shift assays (Groban et al., submitted). In these experiments, LexA is treated as the ligand and DNA is the acceptor. Figure 3 shows a binding curve for such an experiment where the fraction of the recA operator fragment bound was quantified from mobility shift gels by densitometry. Consistent with previous determinations, the Kd for the recA promoter was determined by curve fitting the ligand binding equation, fraction bound = [LexA]unbound/Kd + [LexA]unbound, to be 4.6 nM. Apparent binding constants were similarly determined for each of the promoter fragments that displaced LexA from the recA operator (Table 1).
FIG. 3.
Binding of B. subtilis LexA to the recA promoter. Graphical analyses of mobility shift titration of 32P-labeled recA promoter (10 nM) incubated with increasing concentrations of LexA as described in Materials and Methods.
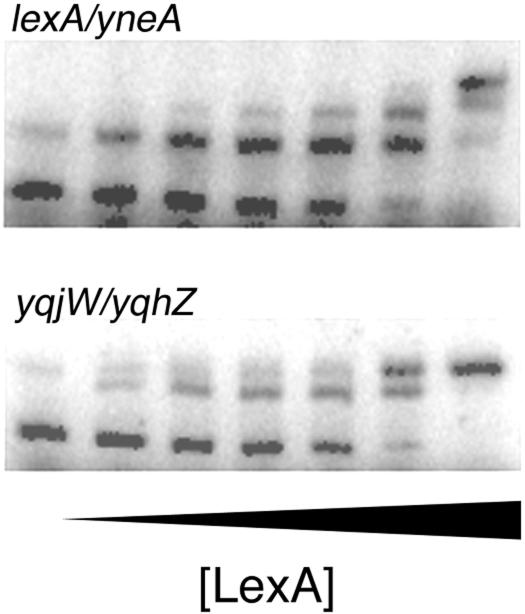
Five of the promoter regions that bind LexA, including the two previously identified SOS genes, lexA and dinC, have multiple binding sites. The binding of LexA to both dinC sites has been reported (27, 31) and binding of LexA to two of the three sites of lexA has been reported (15, 53), but binding to the entire lexA promoter region (which is also the yneA promoter region) has not been demonstrated previously. A mobility shift titration of the lexA and yneA promoter region with purified LexA shows three distinct complexes (Fig. 4). The other putative SOS genes with multiple sites are the yqjW-yqzH pair, the yolC-yolD pair, and the yozL-yozM pair. Mobility shift titrations with the corresponding promoter fragments indicate that LexA binds to both sites in each promoter region as shown for the yqjW or yqzH promoter fragment (Fig. 4). The apparent Kd values for these genes were determined as the concentration of LexA that binds to one half of the total DNA sites.
FIG. 4.
Binding of B. subtilis LexA to the lexA and yqjW promoters. Mobility shift assays were conducted with purified LexA (0 to 96 nM) and radiolabeled lexA (12 nM) or yqjW (12 nM) promoter DNA as described in Materials and Methods.
Seven of the promoter regions identified in our operator search did not compete with the recA promoter for LexA binding. The corresponding genes and their canonical operator sequences are listed at the top of Table 2; two of the genes—yopS and yopT—are divergently transcribed and share the same operator-like sequence. We further tested the ability of LexA to bind to these sites by titrating the radiolabeled promoter fragments directly with LexA. In every case we did not detect any shift at LexA concentrations under 400 nM. LexA concentrations above 400 nM produce a diffuse supershifted band, which we attribute to nonspecific LexA binding because a similar supershift is observed at comparably high LexA concentrations with any DNA fragments and the supershifted band can be eliminated by the addition of excess nonspecific DNA.
B. subtilis SOS operator consensus sequence.
Among the operator sequences listed in Table 1, there are 33 distinct sites or 66 half sites. Alignment of these distinct half site sequences gives the consensus operator sequence, 5′-CGAACATATGTTCG-3′ (bottom of Table 1), which expands the previously determined consensus sequence by four internal base pairs. This sequence has also been shown to be the thermodynamically preferred sequence for LexA binding (Groban et al., submitted). Although sequences outside the operator may contribute to binding and all mismatches are not equal, there is a correlation between binding affinity and the number of mismatches relative to the consensus sequence. The SOS operators with the highest LexA affinity generally differ from the expanded consensus sequence by 2 or 3 base pairs, unless they are adjacent to another site that could contribute to binding through cooperative interactions. Lower affinity binding sites have four or five mismatches, as do the sequences that are not bound by LexA. There are no sequences in the entire genome with less than two mismatches relative to the expanded consensus sequence.
Identification of six additional LexA binding sites in the B. subtilis genome.
To search for other LexA binding sites that may be located elsewhere in the B. subtilis chromosome, we searched the entire genome for the expanded consensus sequence allowing for up to five nondestabilizing mismatches. In this search we also allowed for one mismatch in both of the GAAC sections to test the possibility that we missed some binding sites in our initial search (which allowed for only one GAAC mismatch). In addition to the sequences shown in Table 1, we found 18 canonical sites. Six of these sites (listed in the middle of Table 2) are located within putative promoter regions; they were not identified in our initial search because they all contain one mismatch in both of the 5′-GAAC-3′ sites. Consistent with the prediction, none of these sites displaced LexA from the recA operator when added in 50-fold molar excess over the recA site. Of the remaining 12 canonical sites located outside putative promoter regions, only 4 of these sites, located upstream of the yonT gene and inside the fliP, ylbC, and yqkA genes, bind specifically to LexA (Table 3).
TABLE 3.
Canonical SOS boxes located outside putative promoter regions
| Gene | Operator sequencea | Positionb | No. of mismatches | Kd (nM)c |
|---|---|---|---|---|
| LexA binding sites | ||||
| fliP | aaAACgTAaGTTCG | Inside (+54) | 4 | >200 |
| rpe | CGAACATgTtTTCG | Inside (+369) | 2 | >200 |
| sigH | CGAAacTtTGTTCG | Inside (+127) | 3 | >200 |
| ylbC | aGAACATAgGTTCc | Inside (+46) | 3 | 9.2 |
| yonT | CGAACATAaGTTtt | −320 | 3 | 16 |
| yqkA | gaAACtTgTGTTCG | Inside (+535) | 4 | >200 |
| Sites that do not bind LexA | ||||
| cysS | gaAACATtcGTTCc | Inside | 5 | |
| leuB | tGAAaATATGTTCG | Inside | 2 | |
| nprE | aGAACATATtTTCc | Inside | 3 | |
| resE | gGAAttTATGTTtG | Inside | 4 | |
| rpoC | CGAACgcATtTTCG | Inside | 3 | |
| rsbU | gGAACtTtaGTTCc | Inside | 5 | |
| xylR | gGAACAatcGTTCt | Inside | 5 | |
| yaaC | gaAACATtTaTTCG | Inside | 4 | |
| ybaL | aGAAtATgTGTTtG | Inside | 4 | |
| yobR | CaAACAcATaTTCG | Inside | 3 | |
| yqbA | CGAAagTATGTTCa | Inside | 3 | |
| yqeV | aGAAttTATGTTCt | Inside | 4 | |
| yurX | CGAACATAaGTTaa | Inside | 3 |
Nonconsensus nucleotides are shown in lowercase type, consensus nucleotides are shown in uppercase, and destabilizing mismatches are shown in boldface type.
Location of the 3′ end relative to the ATG codon of the respective gene.
Apparent binding constant.
We also tested the possibility that we missed some binding sites by eliminating candidates with the destabilizing bases indicated in Fig. 1. In fact, the binding site upstream of the ruvA gene has two destabilizing mismatches in the first two positions of one half site (although these are the only mismatches). Despite the unfavorable mismatches, we included it in our initial search because its E. coli counterpart is an SOS gene. Although LexA binds to this site, its affinity is significantly lower than for the other sites that differ from the expanded consensus sequence by only two bases. To identify other potential sites, we searched the genome for canonical sequences that have no more than one mismatch in the two GAAC regions but contain destabilizing bases in positions other than the four essential AT base pairs. The search yielded the sites listed in Tables 2 and 3. Only two sites, inside the rpe and sigH genes, with two and three mismatches, respectively, relative to the expanded consensus sequence, bind LexA (albeit very weakly). None of the other sequences competed with the recA operator for LexA binding in mobility shift assays. Thus, our search identified only 39 LexA binding sites in the entire genome, and 33 of them are located in promoter regions.
Microarray analysis of DNA damage-inducible genes.
According to the SOS system model, any genes with LexA binding sites overlapping their promoters in a way that inhibits RNA polymerase binding should be induced by DNA damage. We used genomic microarrays to identify genes that are induced by mitomycin C and UV radiation in a RecA+ strain, but not in a recA null strain. UV radiation and MC are known inducers of the SOS response that generate the inducing signal differently (29, 42), and our microarray analysis shows that they cause substantially different changes in overall gene expression (data not shown). By definition, an SOS gene should be induced by both treatments but only in cells containing a functional RecA protein.
Microarray experiments were done, in triplicate, on samples from RecA+ cells and recA null cells treated with either MC or UV radiation as described in Materials and Methods. During the 60-minute treatment time, there was no visible effect on growth. We found 37 genes that met the SOS induction criteria; that is, they were induced by both MC and UV in wild-type cells, but not in a recA mutant (Table 4). Thirty-three of these genes, corresponding to 18 operons, are preceded by LexA binding sites (Table 1). All but four genes (yqjY, yqjZ, uvrC, and yhaN) in Table 4 exhibited statistically significant induction, corresponding to a 99% confidence level, by both treatments. Although induction of the yqjY, yqjZ, uvrC, and yhaN genes did not meet the 99% confidence level, we presume they are transcribed from promoters containing LexA binding sites, and they show slight, but reproducible, RecA-dependent induction by both treatments. The uvrC gene has a LexA binding site overlapping its promoter, and the other three genes are apparently part of damage-inducible operons in which the upstream genes are induced. The lower induction levels for the yqjY, yqjZ, and yhaN genes are consistent with the reduced induction we observed for the downstream genes in all the putative operons containing more than two genes. For example, ynzC and yhjB induction levels by both treatments are about 20% of the yneA and yhjD levels, respectively. The induction levels of yhaN and yqjY or yqjZ by both treatments are about 50% of the yhaO and yqjW levels, respectively.
TABLE 4.
B. subtilis genes induced by UV and MC in wild-type cells, but not in recA null mutants
| Genea | Fold inductionb
|
|||||
|---|---|---|---|---|---|---|
| MC (wt) | MC (recA) | UV 30′ (wt) | UV 30′ (recA) | UV 60′ (wt) | UV 60′ (recA) | |
| ybaK | 2.3 | −1.1 | 1.3 | −1.3 | 2.6 | 1.5 |
| cwlD | 2.1 | 1.0 | 1.3 | −1.1 | 2.6 | 1.5 |
| dinB | 55 | 1.0 | 30 | 1.0 | 43 | 3.5 |
| ydiO | 3.5 | 1.7 | 2.8 | 1.2 | 2.5 | 1.6 |
| ydiP | 3.6 | 1.8 | 3.1 | 1.3 | 2.8 | 1.7 |
| pcrA | 2.0 | 1.5 | 2.0 | 1.4 | 1.6 | 1.1 |
| ligA | 2.0 | 1.3 | 2.3 | 1.4 | 1.9 | 1.3 |
| yhaZ | 11 | −1.2 | 11 | 1.0 | 11 | 2.3 |
| yhaO | 3.9 | 1.0 | 3.0 | −3.2 | 3.2 | −1.2 |
| yhaN | 1.6 | −1.1 | 1.5 | −1.2 | 1.3 | 1.5 |
| yhaM | 2.0 | 1.0 | 1.7 | 1.0 | 1.9 | 1.2 |
| yhjD | 16 | 1.0 | 12 | 1.1 | 7.7 | 2.0 |
| yhjC | 2.9 | 1.0 | 1.5 | −1.1 | 1.6 | 1.2 |
| yhjB | 3.8 | 1.1 | 1.9 | 1.0 | 1.8 | 1.3 |
| xkdA | 3.8 | 1.0 | 3.1 | −1.2 | 2.6 | 1.2 |
| recA | 9.4 | 1.2 | 9.1 | 1.0 | 6.7 | 1.7 |
| aprX | 2.7 | 1.1 | 1.6 | −1.2 | 1.9 | 1.2 |
| ymaCc | 4.0 | 1.4 | 3.6 | 1.1 | 2.5 | 1.6 |
| ymaDc | 2.5 | 1.5 | 1.8 | 1.2 | 1.4 | 1.6 |
| lexA | 3.4 | −1.6 | 4.1 | 1.0 | 4.4 | 1.6 |
| yneA | 37 | 1.0 | 32 | 1.1 | 25 | 2.8 |
| yneB | 44 | 1.5 | 9.7 | −1.5 | 12 | 2.8 |
| ynzC | 8.1 | 1.2 | 5.6 | −1.1 | 4.3 | 1.9 |
| parE | 2.9 | 1.5 | 2.0 | 1.1 | 1.9 | 1.0 |
| parC | 3.0 | 1.5 | 2.5 | 1.2 | 2.1 | 1.3 |
| yqjW | 3.5 | 1.0 | 2.2 | −1.1 | 2.8 | 1.5 |
| yqjX | 5.7 | 1.1 | 4.9 | −1.1 | 4.3 | 1.8 |
| yqjY | 1.8 | −1.2 | 1.4 | 1.0 | 1.5 | 1.1 |
| yqjZ | 1.9 | −1.2 | 1.3 | −1.2 | 1.5 | −1.1 |
| ruvA | 2.2 | 1.1 | 2.2 | 1.1 | 2.1 | 1.5 |
| ruvB | 2.2 | 1.1 | 2.0 | 1.1 | 2.2 | 1.5 |
| uvrC | 1.6 | 1.1 | 1.7 | 1.1 | 1.6 | 1.0 |
| yvsGc | 2.3 | 1.1 | 2.5 | 1.5 | 2.0 | 1.7 |
| uvrB | 11 | 1.0 | 7.8 | 1.0 | 6.8 | 1.8 |
| uvrA | 12 | 1.0 | 8.8 | 1.0 | 8.3 | 2.0 |
| dinC | 175 | 1.2 | 39 | −1.2 | 54 | 4.8 |
| licAc | 3.3 | 1.2 | 3.6 | −1.3 | 4.6 | 1.7 |
Genes listed by map position. The first gene in operons is shown in boldface type; subsequent genes in putative multigene operons are shown indented.
Induction levels were determined by microarray analyses as described in Materials and Methods and are given as fold induction in wild-type cells (wt) or recA null mutants relative to untreated cells. UV treatment was performed for 30 min (30′) or 60 min (60′).
This gene does not have a LexA binding site within its promoter region.
Several features of the microarray data are worth noting. (i) There is a wide range of induction levels, from 175-fold for dinC induction by MC to less than twofold, with most genes induced between two- to fourfold. (ii) Induction by MC is typically greater than that by UV treatment, consistent with earlier studies of RecA induction (28). (iii) A low level of induction is observed at 60 min after UV treatment in the absence of RecA. (iv) In several cases there is variable induction for genes within the same putative operon; as mentioned above, there is a general decrease in induction level as distance from the promoter increases for yneA-yneB-ynzC, yhjDCB, yhaONM, and yqjWXYZ (although the yqjX gene is induced more than 50% higher by both MC and UV than yqjW). For the other putative operons-uvrBA, ruvAB, ydiOP, parEC, pcrA-ligA, and ybaK-cwlD-the level of induction for both genes is similar. The pcrA-ligA operon is unusual in that the pcrB and yerH genes, which bracket the pcrA-ligA genes, are not induced by either treatment.
Four of the genes listed in Table 4—licA, ymaC, ymaD, and yvsG—do not have LexA binding sites. Of these, licA and the ymaCD operon have no upstream sequences resembling an SOS operator and LexA does not bind specifically to their promoter regions. The yvsG promoter contains a canonical SOS operator sequence (Table 2), but LexA does not bind specifically to DNA containing this sequence. As with the other genes listed in Table 2, no binding was detected at LexA concentrations below 400 nM and a supershift was observed at higher LexA concentrations (data not shown).
The 20 genes listed in Table 5 contain LexA binding sites in their putative promoter regions, but they did not meet our induction criteria. In every case there was no statistically significant RecA-dependent induction following one or both of the treatments. Many of them show low RecA-dependent induction but not enough to support including them in our list of SOS genes. We also found five genes that showed statistically significant RecA-dependent induction by MC, but not by UV treatment (Table 6). Of these, the yraH gene and the ycgMNO operon have canonical SOS operator sites in their promoter regions (Table 2). Both sites have seven mismatches relative to the expanded consensus, and LexA does not bind to either site. The yolC gene, which has a LexA binding site overlapping its putative promoter, is the only gene that showed RecA-dependent induction by UV, but not by MC (Table 5).
TABLE 5.
B. subtilis genes that contain LexA binding sites within their promoter regions, but are not significantly induced by MC or UV in wild-type cells
| Genea | Fold inductionb
|
|||||
|---|---|---|---|---|---|---|
| MC (wt) | MC (recA) | UV 30′ (wt) | UV 30′ (recA) | UV 60′ (wt) | UV 60′ (recA) | |
| ydgG | 1.4 | −1.1 | −1.1 | −1.2 | 1.3 | −1.1 |
| ydgH | 1.2 | 1.0 | −1.1 | −1.2 | 1.1 | 1.0 |
| yhjE | 1.2 | 1.0 | 1.1 | 1.1 | 1.0 | 1.0 |
| ykvR | 1.4 | 1.3 | 1.2 | −1.1 | 1.0 | 1.0 |
| yozL | 1.5 | −1.1 | 1.3 | −1.3 | −1.0 | 1.2 |
| yozK | 1.4 | 1.1 | 1.1 | −1.2 | 1.6 | 1.1 |
| yobH | 1.2 | −1.3 | 1.0 | −1.1 | 1.7 | 1.1 |
| yozM | 1.3 | −1.2 | −1.1 | −1.1 | −1.2 | −1.5 |
| yorB | 1.7 | −1.3 | 1.4 | −1.0 | 1.9 | −1.0 |
| yolC | 1.2 | 1.0 | −1.1 | −1.1 | 2.5 | −1.2 |
| yolD | 1.6 | −1.1 | 1.1 | −1.2 | 1.1 | 1.1 |
| uvrX | 1.4 | −1.5 | 1.1 | −1.1 | −1.1 | 1.1 |
| hbs | −1.1 | −1.4 | −1.0 | −1.3 | −1.9 | −1.2 |
| ypuD | 1.3 | 1.4 | 1.1 | 1.1 | 1.1 | 1.3 |
| yqzH | 1.7 | ND | 1.2 | ND | ND | 1.3 |
| yqhB | 1.2 | 1.1 | 1.3 | −1.1 | −1.0 | 1.1 |
| yqxL | 1.1 | −1.0 | −1.0 | 1.1 | 1.4 | 1.2 |
| sda | 1.7 | 1.5 | 1.7 | −1.2 | 1.2 | 2.2 |
| dnaE | 1.2 | 1.0 | 1.7 | 1.4 | 1.2 | 1.1 |
| vpr | 1.1 | −1.1 | 1.3 | −1.2 | −1.3 | −1.2 |
Genes listed by map position. The first gene in operons is shown in boldface type; subsequent genes in putative multigene operons are shown indented.
Induction levels were determined by microarray analyses as described in Materials and Methods and are given as fold induction in wild-type cells (wt) or recA mutants relative to untreated cells. UV treatment was performed for 30 min (30′) or 60 min (60′). ND, no data.
TABLE 6.
B. subtilis genes that exhibit RecA-dependent induction by MC, but not by UV
| Gene | Fold inductiona
|
|||||
|---|---|---|---|---|---|---|
| MC (wt) | MC (recAΔ) | UV 30′ (wt) | UV 30′ (recA) | UV 60′ (wt) | UV 60′ (recAΔ) | |
| ycgM | 3.1 | −1.2 | −1.1 | −1.2 | 1.2 | 1.0 |
| ycgN | 2.5 | −1.2 | −1.0 | −1.1 | 1.1 | 1.1 |
| ycgO | 1.9 | −1.1 | 1.0 | 1.1 | 1.4 | 1.1 |
| yobU | 25 | 5.6 | 15 | 14 | 4.9 | 11 |
| yraH | 6.2 | 2.2 | 1.8 | 1.9 | 1.3 | 1.9 |
Induction levels were determined by microarray analyses as described in Materials and Methods and are given as fold induction in wild-type cells (wt) or recAΔ cells relative to untreated cells. UV treatment was performed for 30 min (30′) or 60 min (60′).
DISCUSSION
We have shown that the B. subtilis LexA protein binds to the putative promoter regions of 54 B. subtilis genes in vitro and that 33 of these genes, organized in 18 putative operons, are induced by both MC and UV radiation in a RecA+ strain, but not in recA null cells. That is, there are at least 33 genes in the B. subtilis SOS regulon. We also identified four genes without LexA binding sites that show RecA-dependent induction by both treatments. These genes may be considered secondary SOS genes; they could be induced by a LexA-regulated gene product or regulated by another protein that responds to RecA activation. Of the 33 primary SOS genes identified here (Fig. 5), 8 of them correspond to E. coli genes whose roles in SOS regulation, excision repair, and recombinational repair have been well characterized (8). The most highly conserved of these are ruvB, uvrA, uvrB, and recA, which share 60%, 59%, 58%, and 56% amino acid identity, respectively, between the two distantly related species. Conserved to a lesser degree are ruvA, uvrC, pcrA (uvrD), and lexA, which have 41%, 39%, 38%, and 33% amino acid identity, respectively, with their E. coli counterparts. Functional conservation between the two bacteria has been reported for the products of the recA (30), lexA (31, 54), ruvAB (3), pcrA (38, 39), and uvrC (23) genes (although E. coli uvrC is not SOS regulated).
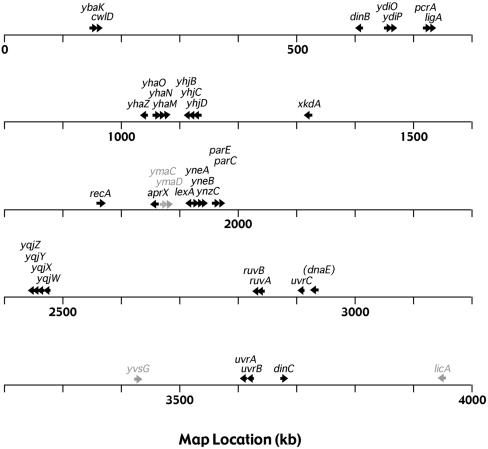
FIG. 5.
Genetic map locations of B. subtilis SOS genes. Primary (black) and secondary (gray) SOS genes are indicated, with arrows depicting the direction of transcription.
Excision repair.
The E. coli UvrABC exinuclease catalyzes the excision of a variety of bulky DNA lesions leaving a short gap that is presumably filled in by DNA polymerase I (41). The high level of conservation of the uvrA and uvrB genes together with the ability of the B. subtilis UvrC protein to substitute for its E. coli counterpart (23) indicates that a B. subtilis UvrABC exinuclease exists with activities like the E. coli enzyme. The E. coli UvrD protein is a DNA helicase involved in excision repair, mismatch repair, and the dismantling of RecA nucleoprotein filaments (2, 13, 50). Its B. subtilis homolog, PcrA, is an essential DNA helicase that suppresses the UV sensitivity of an E. coli uvrD mutant and functions in rolling-circle replication (38).
Although the excision repair proteins have been highly conserved in E. coli and B. subtilis, there are differences in the organization and regulation of the corresponding uvr genes. Unlike in E. coli, where the uvrA and uvrB genes are separated on the chromosome, the B. subtilis genes are contiguous and presumably transcribed from the same promoter; correspondingly, they are induced to about the same degree following DNA damage—about 8- to 12-fold, which is about twice the induction level of the E. coli genes (37). Unlike in E. coli, the B. subtilis uvrC gene has a LexA binding site and is marginally induced by MC and UV. The E. coli uvrD gene is induced five- to sevenfold by DNA damage (37) compared with about twofold for pcrA; the uvrD gene is also not part of a multigene operon like its B. subtilis counterpart. The inducible gene following pcrA in that putative operon, ligA, codes for DNA ligase, which shares 49% amino acid identity with E. coli DNA ligase; the E. coli gene has not been shown to be a damage-inducible gene.
Recombinational repair.
The main proteins involved in recombinational repair are RecA and the Ruv proteins. The contiguous B. subtilis ruvA and ruvB genes apparently comprise an operon as they do in E. coli. The RuvA and RuvB proteins have also been highly conserved in the two bacteria, and genetic evidence indicates a similar enzymatic role (3). Prior to resolution of the Holliday junction in homologous recombination (and recombinational repair), the E. coli RuvA and RuvB proteins are thought to act together, presumably with RecA, to promote ATP-dependent branch migration (36). In E. coli the Holliday junction is cleaved by the RuvC protein, which is not SOS regulated (51, 52). There is no RuvC homolog in B. subtilis; however, the B. subtilis RecU protein, which is found in gram-positive, but not gram-negative, bacteria has a similar Holliday junction resolvase activity (1).
The sequences of other SOS genes suggest possible involvements in recombinational repair. The product of the yneB gene, part of the yneA operon, has significant homology with several Bacillus species site-specific recombinases of the resolvase family. The product of the ynzC gene, also part of the yneA operon, shares 31% identity with part of the B. subtilis GyrB protein. The putative parEC operon codes for the subunits of topoisomerase IV, a type II topoisomerase that is essential for segregation of replicated chromosomes in B. subtilis (19); the products of the parC and parE genes are homologous with the GyrA and GyrB subunits of DNA gyrase.
Error-prone repair (translesion DNA synthesis).
The yqjW gene product shares 26% identity with E. coli UmuC protein and the N-terminal two-thirds of YgjW is 33% identical with E. coli DinP (also called DinB). The two E. coli proteins belong to the Y superfamily of DNA polymerases, which can replicate over various DNA lesions (11, 21). The better characterized is the product of the umuC gene, part of the damage-inducible umuDC operon, whose products catalyze translesion DNA synthesis during the SOS response (i.e., error-prone repair) (46). The UmuD protein is cleaved by activated RecA to produce UmuD′, which associates as a dimer with UmuC to form the functional UmuCD′2 enzyme. No homolog of E. coli UmuD exists in B. subtilis.
Deletion of the B. subtilis yqjW gene decreases UV-induced mutagenesis (45), suggesting that the product of the yqjW gene and/or downstream members of the putative operon are involved in error-prone repair. yqjW is the first gene in a putative operon also containing the yqjX, yqjY, and yqjZ genes. The functions of the yqjX, yqjY, and yqjZ gene products are unknown, and none of them have any homology with E. coli UmuD. The yqjX gene product shares limited homology with the products of the B. subtilis yolD and yozL genes, which both have SOS boxes but did not meet our induction criteria. (It is noteworthy that the product of the uvrX gene, which follows yolD in a putative operon, is homologous with YqjW and other Y-family DNA polymerases.) The yqjY gene codes for a protein with a GCN5-related N-acetyltransferase (GNAT) domain that is homologous with other Bacillus acetyltransferases as well as the product of the E. coli yfiQ gene. The yqjZ gene product shares 59% identity with a conserved bacterial protein involved in polyketide biosynthesis, and it is homologous with the C-terminal domains of P. aeruginosa CTP synthase and E. coli DNA photolyase (17).
A role for DnaE in error-prone repair has been suggested by its ability to bypass certain DNA lesions and by the loss of UV-induced mutagenesis when DnaE is depleted (22). Although we did not find significant induction of the dnaE gene in our microarray analyses, it probably is an SOS gene; it has an upstream LexA binding site, and it was recently shown to be induced about threefold by both MC and nalidixic acid (22). The corresponding protein is an essential DNA polymerase that lacks 3′→5′ proofreading exonuclease activity.
Filamentation.
Inhibition of cell division, or filamentation, is a property that has long been associated with the SOS response. In E. coli the product of the sulA gene inhibits cell division by binding to the major component of the cell division machinery, FtsZ (6, 34). There is no sulA homolog in B. subtilis, but there is evidence that the B. subtilis yneA gene product plays a similar role in the inhibition of B. subtilis cell division. Studies of SOS-induced filamentation in yneA and lexA mutants suggest that the YneA protein suppresses cell division by inhibiting FtsZ ring formation (20). yneA is the first gene of the yneAB-ynzC operon.
Other putative DNA repair functions.
Some of the SOS genes code for proteins homologous with DNA repair or modification enzymes. The yhaZ gene codes for a protein that shares 40 to 50% identity with DNA alkylation repair enzymes of a variety of bacterial species. The ydiO and ydiP gene products have some homology with each other and are similar to cytosine-specific methyltransferases from a variety of bacteria; recent evidence suggests that these proteins are responsible for the modification of BsuM restriction sites (35). The yhaO gene codes for a protein that is homologous with DNA repair endonucleases, the C-terminal end of the yhaN gene product has a DNA repair-associated ATPase domain, and yhaM has a metal-dependent phosphohydrolase domain.
Of the remaining SOS gene products, only three—AprX, CwlD, and LicA—have known activities, but their roles in the SOS response are not clear. The aprX gene codes for a subtilisin-like protease that appears to be a member of a new family of proteases (49). The gene has two putative σA promoters, but transcriptional analysis showed that only the downstream promoter is used and only during stationary phase (47). The aprX SOS box overlaps the upstream promoter sequence, which could explain why no transcription was observed from this promoter. Our induction results are consistent with repression of both promoters during exponential growth; DNA damage would induce transcription from the upstream promoter through LexA cleavage, and the downstream promoter may be induced by another mechanism during stationary phase.
The cwlD gene codes for N-acetylmuramoyl-l-alanine amidase, an enzyme that hydrolyzes a linkage in the cell wall (10). It is located immediately downstream of the ybaK gene, and together the two constitute an operon; the function of the ybaK gene product is unknown. The licA gene, the third gene of the licCBAH operon codes for an enzyme IIA component of the lichenan phosphotransferase system (47). It is not clear why licA is induced by DNA damage and not the other genes in the operon, or how the phosphotransferase system could be involved in the SOS response.
Are there more SOS genes? We have identified 33 primary SOS genes, but we do not rule out the possibility that other genes listed in Table 1 are also part of the B. subtilis SOS regulon. Indeed, our inability to detect significant dnaE induction suggests that we may have missed other inducible genes using our microarray conditions. However, assuming we have identified all of the LexA binding sites, we can put an upper limit of 54 on the number of primary SOS genes in YB886 cells. Although it is possible that our search missed some LexA binding sites, the results from our less stringent search of the entire genome argues that we probably did not. Besides the genes regulated by LexA, the induction of prophage genes has long been associated with the SOS response. Because the strains we used in this study are noninducible for prophage PBSX and have been cured of prophage SPβ, we did not detect the induction of the corresponding bacteriophage genes.
A recent study of the B. subtilis oxidative stress response suggests that the SOS regulon may overlap with other regulons comprising the B. subtilis adaptational network. Macroarray analysis of B. subtilis cells exposed to hydrogen peroxide showed the induction of several SOS operons: recA, lexA, uvrBA, uvrC, dinB, dinC, yhaONM, yhaZ, yneAB-ynzC, and the secondary SOS genes ymaCD (33). However, in a similar study of B. subtilis cells exposed to the same concentration of hydrogen peroxide and analyzed using microarrays, no SOS gene induction was reported; however, there was significant down-regulation of prophage PBSX genes, contrary to what would be expected if the SOS response were activated (18). Both analyses showed significant induction of the perR and sigB regulons by hydrogen peroxide, and the macroarray analysis also showed a stringent response. Neither the perR nor sigB regulon is induced by MC or UV, and there is no stringent response following either treatment (A. I. Goranov, E. Kuester-Schoeck, J. D. Wang, and A. D. Grossman, unpublished results). Thus, there is no conclusive evidence for overlap between the oxidative stress response and the SOS response.
Differential binding and expression of B. subtilis SOS genes.
The binding constants for LexA binding to B. subtilis SOS operators range from 2 nM to over 100 nM, similar to the range reported for E. coli (43). There is also a wide range of induction levels, ranging from about 2-fold to 175-fold, although the very high induction levels for dinB and dinC are probably due, in part, to low basal levels of expression. Genes that are needed during normal growth, such as recA and lexA, have higher basal levels of expression and lower induction levels. Nevertheless, there is some correlation between binding affinity and induction level. As expected, genes with higher affinity binding sites are generally induced to a greater extent, but the induction level also depends on the strength of the promoter, the position of the operator relative to the promoter, stability of the mRNA, and interactions with other molecules. Our results indicate that the position of the operator may be particularly important. For example, the operators for the dinB and dinC genes, which have high-affinity binding sites and show the greatest induction, are both located in the same position relative to the putative promoters. In general, the operators of genes that showed the greatest level of induction are located between 14 and 45 base pairs upstream of the 3′ end of the −10 region of the promoter; genes with operators downstream of the −10 region were moderately induced; and genes with operators more than 50 base pairs upstream of the −10 region were not significantly induced regardless of binding affinity.
In summary, we have demonstrated a very effective and generally applicable approach for identifying specific DNA binding sites that regulate genes scattered throughout the genome. Using the results from a thermodynamic analysis of LexA binding to recA operator mutants, we identified 40 potential SOS boxes and we showed that 33 of them are bound specifically by LexA. A less stringent search did not reveal any additional sites within promoter regions, although we found six sites located outside promoters. Without the operator binding study as a guide, a search for a 14-mer allowing for up to five mismatches relative to the SOS operator consensus sequence yields over 18,000 sequences in the B. subtilis genome.
Acknowledgments
This work was supported by NSF grants MCB-9601398 and MCB-0135899 to C.M.L. and NIH grant GM41934 to A.D.G.
REFERENCES
- 1.Ayora, S., B. Carrasco, E. Doncel, R. Lurz, and J. C. Alonso. 2004. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl. Acad. Sci. USA 101:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron, P. R., S. R. Kushner, and L. Grossman. 1985. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc. Natl. Acad. Sci. USA 82:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco, B., M. C. Cozar, R. Lurz, J. C. Alonso, and S. Ayora. 2004. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J. Bacteriol. 186:5557-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1991. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J. Bacteriol. 173:1696-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1993. Elucidation of regulatory elements that control damage induction and competence induction of the Bacillus subtilis SOS system. J. Bacteriol. 175:5907-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordell, S. C., E. J. Robinson, and J. Lowe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg, E. C., G. C. Walker, and W. Seide. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 9.Gillespie, K., and R. E. Yasbin. 1987. Chromosomal locations of three Bacillus subtilis din genes. J. Bacteriol. 169:3372-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilmore, M. E., D. Bandyopadhyay, A. M. Dean, S. D. Linnstaedt, and D. L. Popham. 2004. Production of muramic delta-lactam in Bacillus subtilis spore peptidoglycan. J. Bacteriol. 186:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 12.Granger-Schnarr, M., M. Schnarr, and C. A. van Sluis. 1986. In vitro study of the interaction of the LexA repressor and the UvrC protein with a uvrC regulatory region. FEBS Lett. 198:61-65. [DOI] [PubMed] [Google Scholar]
- 13.Guarne, A., S. Ramon-Maiques, E. M. Wolff, R. Ghirlando, X. Hu, J. H. Miller, and W. Yang. 2004. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 23:4134-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadden, C. T. 1979. Gap-filling repair synthesis induced by ultraviolet light in a Bacillus subtilis Uvr− mutant. J. Bacteriol. 139:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haijema, B. J., D. van Sindren, K. W. Winterling, J. Kooistra, G. Venema, and L. W. Hamoen. 1996. Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol. Microbiol. 22:75-85. [DOI] [PubMed] [Google Scholar]
- 16.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for bacillus. John Wiley and Sons, Ltd., Chichester, England.
- 17.Heelis, P. F., S. T. Kim, T. Okamura, and A. Sancar. 1993. The photo repair of pyrimidine dimers by DNA photolyase and model systems. J. Photochem. Photobiol. B 17:219-228. [DOI] [PubMed] [Google Scholar]
- 18.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, W. M., J. L. Libbey, P. van der Hoeven, and S. X. Yu. 1998. Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc. Natl. Acad. Sci. USA 95:4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai, Y., S. Moriya, and N. Ogasawara. 2003. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol. Microbiol. 47:1113-1122. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Chatelier, E., O. J. Becherel, E. d'Alencon, D. Canceill, S. D. Ehrlich, R. P. Fuchs, and L. Janniere. 2004. Involvement of DnaE, the second replicative DNA polymerase from Bacillus subtilis, in DNA mutagenesis. J. Biol. Chem. 279:1757-1767. [DOI] [PubMed] [Google Scholar]
- 23.Lin, J. J., and A. Sancar. 1990. Reconstitution of nucleotide excision nuclease with UvrA and UvrB proteins from Escherichia coli and UvrC protein from Bacillus subtilis. J. Biol. Chem. 265:21337-21341. [PubMed] [Google Scholar]
- 24.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 81:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love, P. E., M. J. Lyle, and R. E. Yasbin. 1985. DNA-damage-inducible (din) loci are transcriptionally activated in competent Bacillus subtilis. Proc. Natl. Acad. Sci. USA 82:6201-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love, P. E., and R. E. Yasbin. 1984. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J. Bacteriol. 160:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovett, C. M., K. C. Cho, and T. M. O'Gara. 1993. Purification of an SOS repressor from Bacillus subtilis. J. Bacteriol. 175:6842-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovett, C. M., P. E. Love, R. E. Yasbin, and J. W. Roberts. 1988. SOS-like induction in Bacillus subtilis: induction of the RecA protein analog and a damage-inducible operon by DNA damage in Rec+ and DNA repair-deficient strains. J. Bacteriol. 170:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovett, C. M., T. M. O'Gara, and J. N. Woodruff. 1994. Analysis of the SOS inducing signal in Bacillus subtilis using Escherichia coli LexA as a probe. J. Bacteriol. 176:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovett, C. M., and J. W. Roberts. 1985. Purification of a RecA protein analogue from Bacillus subtilis. J. Biol. Chem. 260:3305-3313. [PubMed] [Google Scholar]
- 31.Miller, M. C., J. B. Resnick, B. T. Smith, and C. M. Lovett. 1996. The Bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein. J. Biol. Chem. 271:33502-33508. [PubMed] [Google Scholar]
- 32.Moolenaar, G. F., C. A. van Sluis, C. Backendorf, and P. van de Putte. 1987. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res. 15:4273-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohshima, H., S. Matsuoka, K. Asai, and Y. Sadaie. 2002. Molecular organization of intrinsic restriction and modification genes BsuM of Bacillus subtilis Marburg. J. Bacteriol. 184:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons, C. A., A. Stasiak, R. J. Bennett, and S. C. West. 1995. Structure of a multisubunit complex that promotes DNA branch migration. Nature 374:375-378. [DOI] [PubMed] [Google Scholar]
- 37.Peterson, K. R., and D. W. Mount. 1987. Differential repression of SOS genes by unstable lexA41 (tsl-1) protein causes a “split-phenotype” in Escherichia coli K-12. J. Mol. Biol. 193:27-40. [DOI] [PubMed] [Google Scholar]
- 38.Petit, M., E. Dervyn, M. Rose, K. Entian, S. McGovern, S. D. Erlich, and C. Bruand. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 29:261-273. [DOI] [PubMed] [Google Scholar]
- 39.Petit, M. A., and D. Ehrlich. 2002. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 21:3137-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raymond-Denise, A., and N. Guillen. 1991. Identification of dinR, a DNA damage-inducible regulator gene of Bacillus subtilis. J. Bacteriol. 173:7084-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancar, A., and G. B. Sancar. 1988. DNA repair enzymes. Annu. Rev. Biochem. 57:29-67. [DOI] [PubMed] [Google Scholar]
- 42.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 43.Schnarr, M., P. Oertel-Buchheit, M. Kazmaier, and M. Granger-Schnarr. 1991. DNA binding properties of the LexA repressor. Biochimie 73:423-431. [DOI] [PubMed] [Google Scholar]
- 44.Siegel, E. C. 1983. The Escherichia coli uvrD gene is inducible by DNA damage. Mol. Gen. Genet. 191:397-400. [DOI] [PubMed] [Google Scholar]
- 45.Sung, H. M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 47.Tobisch, S., P. Glaser, S. Kruger, and M. Hecker. 1997. Identification and characterization of a new beta-glucoside utilization system in Bacillus subtilis. J. Bacteriol. 179:496-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valbuzzi, A., E. Ferrari, and A. M. Albertini. 1999. A novel member of the subtilisin-like protease family from Bacillus subtilis. Microbiology 145:3121-3127. [DOI] [PubMed] [Google Scholar]
- 50.Veaute, X., S. Delmas, M. Selva, J. Jeusset, E. Le Cam, I. Matic, F. Fabre, and M. A. Petit. 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]
- 52.West, S. C. 1996. The RuvABC proteins and Holliday junction processing in Escherichia coli. J. Bacteriol. 178:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winterling, K. W., A. S. Levine, R. E. Yasbin, and R. Woodgate. 1997. Characterization of DinR, the Bacillus subtilis SOS repressor. J. Bacteriol. 179:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witkin, E. M. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev. 40:869-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasbin, R. E. 1977. DNA repair in Bacillus subtilis. I. The presence of an inducible system. Mol. Gen. Genet. 153:211-218. [PubMed] [Google Scholar]
- 57.Yasbin, R. E. 1977. DNA repair in Bacillus subtilis. II. Activation of the inducible system in competent bacteria. Mol. Gen. Genet. 153:219-225. [PubMed] [Google Scholar]
- 58.Yasbin, R. E., D. L. Cheo, and K. W. Bayles. 1992. Inducible DNA repair and differentiation in Bacillus subtilis: interactions between global regulons. Mol. Microbiol. 6:1263-1270. [DOI] [PubMed] [Google Scholar]
- 59.Yasbin, R. E., P. I. Fields, and B. J. Anderson. 1980. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene 12:155-159. [DOI] [PubMed] [Google Scholar]