Abstract
When a mutation in an essential gene shows a temperature-sensitive phenotype, one usually assumes that the protein is inactive at nonpermissive temperature. DNA gyrase is an essential bacterial enzyme composed of two subunits, GyrA and GyrB. The gyrB652 mutation results from a single base change that substitutes a serine residue for arginine 436 (R436-S) in the GyrB protein. At 42°C, strains with the gyrB652 allele stop DNA replication, and at 37°C, such strains grow but have RecA-dependent SOS induction and show constitutive RecBCD-dependent DNA degradation. Surprisingly, the GyrB652 protein is not inactive at 42°C in vivo or in vitro and it doesn't directly produce breaks in chromosomal DNA. Rather, this mutant has a low kcat compared to wild-type GyrB subunit. With more than twice the normal mean number of supercoil domains, this gyrase hypomorph is prone to fork collapse and topological chaos near the terminus of DNA replication.
Temperature-sensitive (TS) mutants have been used to study complex biochemical pathways in prokaryotic and eukaryotic organisms alike. By comparing phenotypes of a TS mutant at permissive and restrictive temperatures, important insights can be obtained about the progression of intermediates through a biochemical pathway. For example, TS mutants guided early studies of DNA replication and provided the initial evidence for three different cellular polymerases (28). TS mutants also showed that replication involves numerous genes, and a large mutant collection guided the in vitro reconstruction of replication initiation and elongation (44, 72). In bacteriophages λ and Mu, TS repressors were found that undergo a change in protein conformation causing repressor-DNA complexes to dissociate at nonpermissive temperatures (24, 31, 69). Physiological studies after shifts from permissive to nonpermissive temperature revealed relevant promoters and important biochemical steps necessary for the onset of the lytic cycle in phage development (19, 30). Thus, it has become expected that when a gene confers a TS phenotype, the cause is a temperature-dependent change in the mutant protein's structure or catalytic activity.
DNA gyrase has a unique and essential role in prokaryotic replication and transcription. The enzyme catalyzes the introduction of negative supercoils into covalently closed DNA molecules at the expense of ATP binding and hydrolysis. The enzyme is essential in most prokaryotes and is a critical target for antimicrobial chemotherapy. The quinolone antibiotics are among the most potent antibacterial agents known, and they kill bacteria by turning DNA gyrase (and the closely related enzyme topoisomerase IV [Topo IV]) into a DNA-reactive cytotoxin (23).
Gyrase plays at least two essential roles in chromosome structure and function. First, it condenses DNA by converting relaxed DNA molecules into a dynamic, underwound, and plectonemically supercoiled state (12). DNA in living cells is compartmentalized into subchromosomal regions called domains, and the dynamic superhelical structure within a domain affects many processes like transcription, recombination, site-specific recombination, and transposition (12, 18, 33, 36). Second, to unwind the Watson and Crick strands during DNA synthesis, a cell must remove 400,000 positive coils in each chromosome-equivalent of DNA and resolve all catenane links between the replicated chromosomes. Gyrase-driven negative supercoils facilitate both the unwinding and the Topo IV-dependent unlinking of parental strands during bacterial DNA replication (54, 70).
We previously described TS mutants of gyrase with an increased number of superhelical domains. At the permissive temperature (30°C), mutations in gyrB were found to cause more than twice as many domains as wild-type (WT) strains over a 100-kb interval spanning the his and cob operons (64, 66). The gyrB652 mutation was originally isolated by Gari et al. (27), and it causes induction of the SOS response at 37°C and shows synthetic lethality in combination with recA or recB mutations. Here, we isolated GyrB652 gyrase and compared it to the WT enzyme. To our surprise, the GyrB652 gyrase was not TS in vitro or in vivo. The enzyme had a low turnover number (kcat) in vitro, and in vivo it caused a dramatic change in supercoil dynamics near the replication terminus.
MATERIALS AND METHODS
Strains and cloning.
Bacterial strains used in this work are listed in Table 1. Both the WT gyrB and the gyrB652 gene from Salmonella enterica serovar Typhimurium were cloned into pET21(a) in Escherichia coli DH5α. Then, plasmids were moved into E. coli BL121::λDE3 (67) for protein expression under control of the phage T7 RNA polymerase. Plasmid pRC06 contains the gyrB gene from WT Salmonella strain LT2, and pRC04 contains the gyrB652 gene with an A-C transversion that changes the Arg 436 codon to Ser.
TABLE 1.
Strains useda
| Strain | Genotype | Plasmid |
|---|---|---|
| NH2504 | cobT714::MudJr2 cobP712::Tn10dGn | pJBREScI |
| NH2585 | zeh-754::Tn10 | pUC19 |
| NH2589 | zib-6794::Tn10 gyrB652 | pUC19 |
| NH2598 | cobT714::MudJr2 cobP712::Tn10dGn zib-6794::Tn10 gyrB652 | pJBREScI |
| NH2689 | hsdS gal attB::DE3 | pRC04 |
| NH2691 | hsdS gal attB::DE3 | pRC06 |
| NH3432 | SMT1556::MudJr2 (Mud547) zij-8267::Tn10dGn | pJBREScI |
| NH3435 | SMT1556::MudJr2 (Mud547) zij-8267::Tn10dGn zib-6794::Tn10 gyrB652 | pJBREScI |
| NH2936 | zbj-8906::MudJr2 zbj-8907::Tn10dGn | pJBREScI |
| NH3440 | zbj-8906::MudJr2 zbj-8907::Tn10dGn zib-6794::Tn10 gyrB652 | pJBREScI |
| NH3500 | (STM1553 <FRT> STM1554) | pCP20 |
| NH3501 | (smpB <FRT> STM2689) | pCP20 |
| NH3502 | (STM3261 <FRT> STM3262) | pCP20 |
| NH3503 | (atpI <FRT> gidB) | pCP20 |
| NH3504 | (STM1553 <FRT res lacZYA gen res FRT> STM1554) | pJBREScI |
| NH3505 | (smpB <FRT res lacZYA gen res FRT> STM2689) | pJBREScI |
| NH3506 | (STM3261 <FRT res lacZYA gen res FRT> STM3262) | pJBREScI |
| NH3507 | (atpI <FRT res lacZYA gen res FRT> gidB) | pJBREScI |
| NH3508 | zib-6794::Tn10 gyrB652 (STM1553 <FRT res lacZYA gen res FRT> STM1554) | pJBREScI |
| NH3509 | zib-6794::Tn10 gyrB652 (smpB <FRT res lacZYA gen res FRT> STM2689) | pJBREScI |
| NH3510 | zib-6794::Tn10 gyrB652 (STM3261 <FRT res lacZYA gen res FRT> STM3262) | pJBREScI |
| NH3511 | zib-6794::Tn10 gyrB652 (atpI <FRT res lacZYA gen res FRT> gidB) | pJBREScI |
All strains are derivatives of S. enterica serovar Typhimurium strain LT2 except for NH2689 and NH2691, which are E. coli B derivatives. All strains were constructed for this study.
Enzyme purification.
WT and mutant GyrB proteins were expressed and purified using the same protocol. Five milliliters of an overnight culture was used to inoculate 500 ml of Luria broth (LB) containing 50 μg/ml ampicillin. The culture was agitated at 30°C, and cell density was monitored hourly. When the culture reached an optical density at 650 nm of 0.85, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM, and incubation at 30°C was continued for 2 h. Cultures were chilled to 4°C, and the cells were harvested by centrifugation and resuspended in 60 ml of TGED buffer (50 mM Tris-HCl, pH 8.0, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT]). Lysis was induced by addition of 20×-concentrated lysis buffer which made the solution (at final concentration) 2 mM DTT, 20 mM EDTA, 100 mM KCl, 0.2% Brij 58, and 0.4 mg/ml egg white lysozyme. The mixture was placed in centrifugation tubes, incubated at 4°C for 30 min, and centrifuged at 35,000 rpm in a Beckman type 60 Ti ultracentrifuge rotor for 1 h at 4°C. Solid streptomycin sulfate was added to the supernatant at a final concentration of 4%, and the mixture was stirred at 4°C for 30 min. After centrifugation for 30 min in a Beckman J21 rotor at 7,000 rpm, solid ammonium sulfate was added to the supernatant at a final concentration of 50% saturation. After centrifugation in a Beckman J21 rotor at 7,000 rpm, the pellet was dissolved in 1× TGED buffer with 1 M NaCl. Dialysis was carried out overnight in TGED with 1 M NaCl.
Novobiocin was covalently coupled to CNBr-activated Sepharose 4B according to instructions supplied by Amersham Biotech. Ten milliliters of a dialyzed protein solution was applied to a 5-ml (bed volume) novobiocin-agarose column, and the applied protein solution was recirculated through on the column for 1 h. Washing with 15 ml of solution containing 2 M urea in 1× TGED eluted most of the protein bound to the resin. GyrB protein was then eluted in 6 M guanidine-HCl in 1× TGED. Fractions with the highest protein concentration were identified by Bio-Rad protein assays and dialyzed for 12 h against 1× TGED at a maximum protein concentration of 1 mg/ml, which allowed the refolding of the purified GyrB protein. The dialyzed solution was clarified by centrifugation, and the supernatant was assayed for supercoiling activity after addition of GyrA protein. Renatured GyrB protein was applied to a 5-ml DEAE column equilibrated with 1× TGED buffer. Active GyrB protein eluted in a 100-ml TGED gradient that varied linearly from 0 to 0.5 M NaCl. Fractions with the highest specific activity were identified using GyrA-complemented DNA supercoiling assays (35). Active fractions were pooled and applied to a 200-ml Sephacryl S-200 column equilibrated in 0.2 M KPO4, pH 7.4, 10% glycerol, 1 mM EDTA, and 5 mM DTT. Sephacryl fractions (20 ml) near the void volume were assayed and concentrated by dialysis into 50% glycerol containing 10 mM Tris-HCl, pH 7.4, 50 mM KPO4, 1 mM DTT, and 0.1 mM EDTA, which reduced the volume by half. Both the WT and GyrB652 subunits were stable at −20°C in this buffer.
Genetic methods.
PCR products were “sewn” into a 9-kb module containing a complete WT Lac operon (lacIZYA), the gentamicin resistance gene aacC1 [aminoglycoside-(3)-N-acetyltransferase] from Tn1696 flanked by a pair of res sites, and a pair of FLP recognition target (FRT) sites. The PCR product of the 9-kb module was cloned into plasmid pCR-XL-TOPO (Invitrogen) between nucleotides 336 and 337, generating the plasmid pPM1 (sequence available on request).
To introduce a Lac-Gen-res module into a chromosome, a FRT site was first inserted into a specific location in the Salmonella chromosome using techniques and reagents described by Datsenko and Wanner (14). Strains containing a single chromosomal FRT site and the pCP20 plasmid, which expresses FLP recombinase at 42°C, were grown to late log phase at 30°C and incubated at 42°C for 10 min to induce FLP expression. After chilling in ice water, cells were made competent by 30 s of centrifugation and resuspension in ice-cold water and were electroporated with plasmid pPM1. FLP recombinase catalyzes the excision of a FRT circle from pPM1 and its reintegration into the chromosomal FRT site. Each culture was infected with P22 at low multiplicity and incubated at 30°C in a shaking incubator until cell lysis (37). Strains NH3514 (LT2/pJBREScI) or NH3515 (gyrB652/pJBREScI) were transduced with the resulting P22 phage to generate strains NH3504 to -3511. Gentamicin-resistant transductants that stained blue on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates and were Kan sensitive were subsequently tested for the correct location of the Lac operon using primers in the bacterial chromosome and in the Lac-Gen-res module.
RESULTS
The GyrB652 protein is not TS for supercoiling.
Gari et al. isolated the Salmonella gyrB652 mutant and reported it to be constitutively induced for SOS functions and to show RecBCD-dependent DNA degradation (27). To examine the biochemical properties of this mutant, we purified Salmonella WT GyrB and GyrB652 proteins (see Materials and Methods). When mixed with either E. coli or Salmonella GyrA protein, the specific activity of WT S. enterica serovar Typhimurium GyrB-containing gyrase was comparable to purified WT E. coli gyrase— at 105 U/mg of purified protein. The specific activity of gyrase reconstituted with the Salmonella GyrB652 protein was 1/3 to 1/5 that of the WT protein reconstituted with either E. coli or Salmonella GyrA.
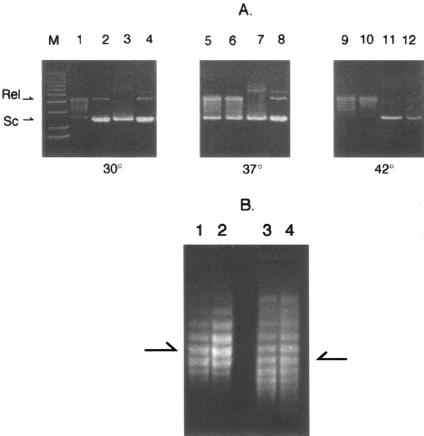
Gyrase containing the WT Salmonella GyrB or the GyrB652 protein was tested in supercoiling assays at three temperatures—30°C, which is permissive for growth; 37°C, which shows an SOS phenotype for GyrB652; and 42°C, which is nonpermissive for GyrB652. Supercoiling assays are not linear with respect to added enzyme because DNA intermediates become distributed in a family of bands with electrophoretic mobility between relaxed and fully supercoiled positions. A unit of gyrase has been defined by an endpoint dilution assay in which 1 unit of enzyme supercoils 0.2 μg of relaxed plasmid DNA to a highly supercoiled band after 30 min at 30°C, as measured by gel electrophoresis in an agarose gel lacking an intercalator (35). The activities of different gyrase preparations were compared using either 1 U or 4 U of mutant or WT enzyme incubated for half the normal time (15 min). Gyrase made with the WT GyrB subunit completely supercoiled the relaxed plasmid in 15 min at 30°C (Fig. 1A, lane 2), whereas the GyrB652 enzyme converted the substrate to a family of supercoiled bands that migrated between nicked and fully supercoiled forms (Fig. 1A, lane 1). At 37°C, the WT assay with 1 U of enzyme was slightly less active than the corresponding 30°C reaction (Fig. 1A, lane 6), but supercoiling was well matched to the GyrB652 enzyme (Fig. 1A, lane 5). At 42°C, WT enzyme was less active than at 37°C (Fig. 1A, lane 10), but the GyrB652 enzyme again showed activity comparable to WT enzyme (Fig. 1A, lane 9). Reactions containing 4 U of either the WT or GyrB652 gyrase completely supercoiled the relaxed plasmid at all temperatures (Fig. 1A, lanes 3, 4, 7, 8, 11, and 12). Thus the GyrB652 enzyme was no more temperature sensitive than WT gyrase as determined by an in vitro supercoiling assay.
FIG. 1.
Temperature effects on DNA supercoiling in vitro or in vivo. A. Supercoiling assays were assembled with relaxed pMP1000 plasmid DNA and either 1 U (lanes 1, 2, 5, 6, 9, and 10) or 4 U (lanes 3, 4, 7, 8, 11, and 12) of DNA gyrase containing GyrB652 subunits (odd-numbered lanes) or WT GyrB (even-numbered lanes). After 15 min of incubation at 30°C (lanes 1 to 4), 37°C (lanes 5 to 8), or 42°C (lanes 9 to 12), reactions were terminated by addition of SDS and the products were run on a 1% agarose gel. Arrows at the side indicate the position of the relaxed substrate (Rel) and supercoiled product (Sc). Intermediates in the supercoiling reaction run between these two positions. B. Plasmid DNAs (pUC19) isolated from exponential cultures of strain NH2585 (WT; lanes 1 and 2) or NH2589 (GyrB652; lanes 3 and 4) were run in an agarose gel containing Tris-borate-EDTA and 25 μM chloroquine phosphate. In lanes 1 and 3, cells were incubated continuously at 30°C, while in lanes 2 and 4, cells were shifted to 42°C for an hour before harvest. The arrows indicate the centers of the topoisomer distribution.
Supercoiling reactions in vitro might differ from in vivo reactions in critical ways. For example, at the restrictive temperature in vivo a mutant enzyme could be refolded by molecular chaperones or degraded by cellular proteases. Thus, the in vivo supercoiling phenotype for a TS GyrB652 strain might not be apparent in vitro. To test gyrase activity in vivo, the supercoil density of the plasmid pUC19 was measured at permissive and nonpermissive temperatures. Plasmid supercoiling is exquisitely sensitive to change in gyrase activity (32, 46, 57). For example, inhibition of gyrase with novobiocin or a fluoroquinolone leads to a loss in plasmid linking number within minutes of drug addition (53, 75). To test in vivo supercoiling, a WT and a GyrB652 mutant strain was grown to mid-log phase (optical density at 65 nm, 0.6) at 30°C. A portion of each culture was shifted to 42°C and incubated for 1 h. Plasmid DNA was purified from both the 30°C and 42°C cultures, and aliquots of plasmid DNA were loaded onto an agarose gel containing 25 μM chloroquine phosphate, which partially unwinds DNA to reveal the distribution of topoisomers. For Salmonella strain NH2585 (WT), the average superhelical density of plasmids isolated from cells held at 30°C was similar to plasmids from cells incubated at 42°C (Fig. 1B, lanes 1 and 2). For strain NH2589 (gyrB652), the 30°C pUC91 plasmid topoisomer distribution differs from that for WT cells grown at 30°C in two ways (Fig. 1B, lane 3). First, the distribution is broader. Second, the median of the distribution is shifted one or two topoisomers to the less-supercoiled level (toward the gel bottom in this system). These results agree with supercoil distributions published previously (64), where we found a slightly less supercoiled plasmid in the gyrB652 mutant at 30°C. When the NH2585 strain is incubated for 1 h at the nonpermissive temperature of 42°C, the pUC19 plasmid supercoil distribution remained the same (Fig. 1B, lane 4). Treatment of either NH2589 (WT) or NH2585 (gyrB652) with an inhibitory but nonlethal dose of the gyrase inhibitor novobiocin caused a shift to a less-supercoiled band position (data not shown). Thus, the GyrB652 subunit does not cause a significant temperature-dependent loss of plasmid supercoiling at 42°C in vivo.
The GyrB652 subunit has low catalytic activity.
The in vitro and in vivo supercoiling results (Fig. 1) lead to a dilemma. The mutant enzyme does not behave as a classic TS protein; it shows no loss of activity at the nonpermissive temperature. Nonetheless, it has a striking TS phenotype. The measurable difference in plasmid supercoil density when the mutant is compared to WT was not influenced by temperature and there was no evidence of temperature-dependent gyrase-induced DNA cleavage in vitro or in vivo (26, 27; R. Chen, unpublished data). These observations required a new hypothesis. If the difference in the specific activity of the WT GyrB and GyrB652 preparations is real (i.e., not due to a much larger fraction of misfolded or otherwise inactive protein in the GyrB652 preparation), then low catalytic efficiency, kcat, might cause the SOS response and RecBCD-dependent degradation of DNA.
To test the hypothesis, an assay was required that is not biased by the possible presence of inactive enzyme. Supercoiling assays do not distinguish between slow and partially inactive proteins because the reaction is somewhat distributive. Measuring a supercoil rate per enzyme molecule requires knowing the number of active enzymes, but cleavage assays meet the requirement. A high-affinity binding site for gyrase in bacteriophage Mu (the strong gyrase site [SGS]) is bound and cleaved efficiently after addition of fluoroquinolones in vitro and in vivo (50, 60). The SGS is highly preferred as a gyrase binding site, and only one gyrase tetramer can bind to the SGS. During strand transfer the complex passes through a covalent intermediate wherein the DNA backbone is linked to tyrosine-122 of the GyrA subunit (11, 43). In the absence of drugs, covalent gyrase-DNA complexes are a fleeting species because biochemical equilibrium strongly favors a noncovalent form of enzyme-bound DNA (34). Enoxacin shifts the equilibrium so that gyrase makes an efficient double-strand break following the addition of denaturing agents like sodium dodecyl sulfate (SDS) (60). Because gyrase does not turn over in cleavage assays, DNA breakage of preformed gyrase-DNA complexes monitors only the active protein bound to a single site on DNA.
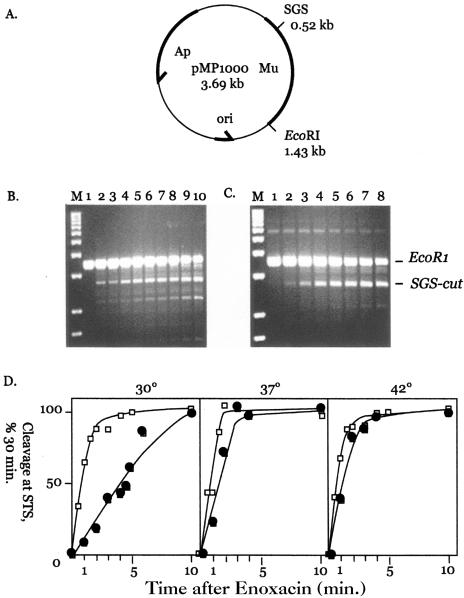
To measure the transition from a bound but uncleaved complex to cleavage complex, WT GyrB- and GyrB652-reconstituted gyrase was incubated for an hour with plasmid pMP1000, which has a 1-kb fragment with the Mu SGS cloned into a pBR322-derived plasmid (50). Digestion with EcoRI generates a 3.69-kb linear molecule, but after gyrase cleavage at the SGS, two bands appear: a 2.78-kb fragment and a small rapidly migrating 0.91-kb band (not seen in Fig. 2). SDS was added at different time intervals after enoxacin addition to measure the kinetics of cleavage. This experiment eliminates the contribution of inactive GyrB subunits because they don't bind the SGS or cleave DNA.
FIG. 2.
A. Physical map of plasmid pMP1000. The positions of the phage Mu DNA (bold line) and the SGS are indicated along with the position of a single EcoRI restriction site. B. Kinetics of WT gyrase cleavage in the presence of enoxacin. Plasmid pMP1000 was incubated with EcoRI and reconstituted WT gyrase as indicated at 37°C for 1 h. After the addition of enoxacin, aliquots were withdrawn and quenched with SDS and proteinase K at different times. The marker in lane M is a 1-kb DNA ladder; gyrase cleavage at the SGS produces a 2.7-kb band. Incubation with enoxacin was as follows: 0 min, lane 1; 0.5 min, lane 2; 1 min, lane 3; 1.5 min, lane 4; 2 min, lane 5; 3 min, lane 6; 4 min, lane 7; 5 min, lane 8; 10 min, lane 9; and 30 min, lane 10. C. Kinetics of GyrB652 gyrase cleavage in the presence of enoxacin. pMP1000 DNA was incubated as described above with GyrB652 gyrase. Enoxacin was added and incubation continued at 37°C for the following times: 0 min, lane 1; 1 min, lane 2; 2 min, lane 3; 3 min, lane 4; 4 min, lane 5; 5 min, lane 6; 10 min, lane 7; and 30 min, lane 8. The positions of EcoRI-linearized pMP1000 and gyrase-cleaved product are marked as EcoRI and SGS-cut at the right edge of panel C. D. Temperature effects on the kinetics of enoxacin-dependent cleavage. Cleavage reactions were carried out using WT gyrase (open squares) and GyrB652 gyrase (filled squares) at 30°C (left panel), 37°C (central panel), and 42°C (right panel). Cleavage levels were quantitated using Bio-Rad Quantity One video imaging software, and the level is given as a percentage of product formed after 30 min of incubation with enoxacin (see panels B and C). The estimated time to achieve half-maximal cleavage was about 1 min at 30°C, 45 s at 37°C, and 30 s at 42°C for WT gyrase and 5 min at 30°C, 1.6 min at 37°C, and 1 min at 42°C for GyrB652 gyrase.
DNA cleavage was slower for GyrB652 gyrase than for the WT gyrase (Fig. 2B, C, and D). At 30°C, the time required to reach half-maximal cleavage of the SGS was 1 min for the wild type and 3.5 min for the GyrB652 gyrase (Fig. 2D). Measurable differences were also found at 37°C and 42°C: wild-type and mutant enzymes reach half-maximal cleavage in 45 s versus 1.5 min at 37°C and 30 s versus 1 min at 42°C, respectively. The Q10 was observed with both forms of gyrase; the cleavage rates increased about twofold as assay temperatures rose from 30°C to 42°C. Nonetheless, gyrase made with the GyrB652 subunit cleaved DNA more slowly than WT at all temperatures tested. These data support a kinetic explanation for the Gyr652 TS phenotype rather than enzyme inactivation at nonpermissive temperature.
Strains with the gyrB652 allele lose supercoil dynamics near the dif site.
If gyrase is active at 42°C, why does a GyrB652 strain stop growth and DNA synthesis at 42°C rather than just slowing down? The terminus of replication has unique topological roles in bacterial chromosome replication and segregation. Bidirectional DNA synthesis in both E. coli and Salmonella leads the two forks that initiate replication at oriC to converge on the terminus. Twin domains of supercoiling predict that the terminus could experience a loss of negative supercoiling or might even become positively supercoiled during fork convergence (25, 73). In addition, dimeric chromosomes that arise from reciprocal recombination must be resolved to monomers by site-specific recombination at dif prior to cell division (45, 52).
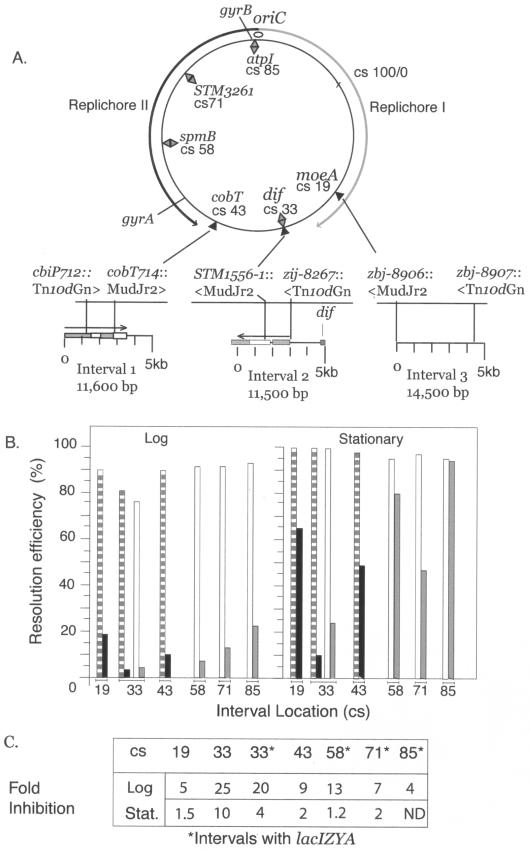
To study supercoiling at dif, transposons with associated res sites were isolated near dif. The resolvase system uses movement of plectonemic negative supercoils to form a recombination synapse—a three-node superhelix that pairs two 120-bp resolution sites (res sites) (65). Resolution reactions were studied at three different locations, the his-cob interval, which is 468 kb from dif (37), an interval within 3 kb of the dif site, and a third interval in the opposite replichore 600 kb from dif near moeA (Table 1 and Fig. 3A). Each interval is defined by two transposons with a single res site (37). In strains with WT gyrase, resolution was efficient at the cob, dif, and moeA regions of the chromosome. Over 80% of the cells in exponentially growing populations generated a deletion after a 10-min induction of γδ resolvase expression (Fig. 3B, left panel). When resolution was tested in stationary-phase cells, essentially all chromosomes were recombinant (Fig. 3B, right panel). Thus, supercoil behavior in WT cells was similar at cob, dif, and moeA (Fig. 3A).
FIG.3.
A. Genetic and physical map of the S. enterica serovar Typhimurium chromosome. Intervals analyzed with Tn3 resolution assays are shown for seven map positions on the standard 100-CS Salmonella map. Replichore I goes from oriC clockwise, while replichore II goes counterclockwise. The map coordinates for intervals defined by transposon insertions are indicated by black triangles, and inserted Lac modules are indicated by diamonds. B. The data for resolution efficiencies shown in Table 2 for three transposon-defined intervals at CS 43, CS 33, and CS 19 are shown as striped bars for WT gyrase and black bars for GyrB652. Resolution efficiencies for deletion of Lac intervals are indicated by white bars for WT gyrase and gray bars for GyrB652. C. Relative to WT gyrase, the reduction in recombination efficiency for GyrB652 derivatives is shown for all chromosomal positions.
However, gyrB652 derivatives had a markedly different profile (Fig. 3B). In exponential-phase cells, resolution near dif was reduced 25-fold compared to WT cells. This reduction in resolution efficiency was much greater than that observed for intervals of comparable size at the cob operon (ninefold), which is about 450 kb clockwise on the standard map, or the interval near moeA, which is 600 kb counterclockwise of dif (fivefold) (Fig. 3B; Table 2). In stationary phase, GyrB652 strains at the cob and moeA regions had only 1.5- and 2-fold reductions relative to WT, but a 10-fold reduction in resolution was still seen near dif (Fig. 3B, right panel).
TABLE 2.
Effect of gyrB652 mutation on resolution in different chromosome regionsa
| Strain | Region | gyrB | Deletions in exponential growth (%) | Deletions in stationary growth (%) |
|---|---|---|---|---|
| NH2504 | CS 43 | WT | 90 | 97 |
| NH2598 | CS 43 | gyrB652 | 10 | 49 |
| NH3432 | CS 33 | WT | 82 | 99 |
| NH3435 | CS 33 | gyrB652 | 3 | 10 |
| NH2936 | CS 19 | WT | 90 | 99 |
| NH3440 | CS 19 | gyrB652 | 18 | 65 |
| NH3504 | CS 33 | WT | 75 ± 5 | 97 ± 3 |
| NH3508 | CS 33 | gyrB652 | 3.9 ± 0.2 | 23 ± 1 |
| NH3505 | CS 58 | WT | 92 ± 2 | 95 ± 1 |
| NH3509 | CS 58 | gyrB652 | 7 ± 2 | 80 ± 5 |
| NH3506 | CS 71 | WT | 91 ± 2 | 98 ± 1 |
| NH3510 | CS 71 | gyrB652 | 13 ± 1 | 46 ± 4 |
| NH3507 | CS 85 | WT | 93 ± 4 | 94 ± 6 |
| NH3511 | CS 85 | gyrB652 | 22 ± 3 | 93 ± 3 |
Resolution assays were carried out as described previously (37, 64). Briefly, cells grown at 30°C to mid-log phase were shifted to 42°C for 10 min to induce expression of Tn3 resolvase from pJBREScI. Cells were diluted into LB, incubated at 30°C in LB for an hour to allow chromosomal segregation, and plated on selective medium containing X-Gal. Recombinant colonies appearing on plates incubated at 30°C were scored after 3 to 5 days. The recombination numbers reported here represent average values from three or more independent cultures. Variation between replicas was less than 15% of the reported values.
Resolution assays at three chromosomal locations showed differences in supercoiling caused by the GyrB652 mutant. These assays monitor the loss of the MudJr2 element, which includes a lacZ gene and a Kan resistance gene. However, each interval is slightly different in size and there might be supercoil influences associated with the bacterial genes altered by transposon insertions or caused by the resulting deletion. To eliminate these complications, a copy of the complete E. coli lac operon, flanked by directly repeated res sites, was introduced into four intergenic locations of the Salmonella chromosome (Fig. 4; see Materials and Methods). Resolution of the 9-kb Lac-Gen module was tested at four positions in the replichore II segment of the Salmonella chromosome. These positions were near centisome (CS) 33 (which is within 3 kb of dif), CS 58, CS 71, and CS 85 (which is near oriC) (Fig. 3A). By measuring deletion of the same interval at multiple locations, we eliminated minor variations in comparisons that are due to different segment lengths and no Salmonella gene was disrupted. Data from these four intervals confirmed and extended our conclusions from the three transposon-defined intervals at CS 43, CS 33, and CS 19. First, resolution of all intervals was 80% or greater in WT cells when cells were tested in log phase, and it was essentially 100% in stationary cultures (Fig. 3B). As shown by others working with both E. coli and Salmonella, supercoil structure in WT cells is similar in all regions of the genome (48, 51). However, in the GyrB652 mutant, exponential cells exhibited a gradient of supercoil disruption (Fig. 3B). The region least affected was near oriC (22% resolution efficiency), and the zone with the most severe loss of supercoil structure was near dif (4% resolution; Fig. 3B, left panel). In stationary-phase cultures, resolution near oriC was unaltered by GyrB652 (97%), whereas the resolution for the dif site remained low (22%; Fig. 3B, right panel). When these results are expressed as an n-fold reduction in recombination relative to strains with WT gyrase, the interval near dif showed 20- to 25-fold decreases for resolution in log phase, whereas the oriC region showed a modest fourfold decrease (Fig. 3C).
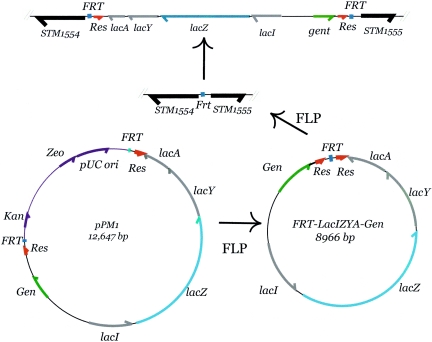
FIG. 4.
Introduction of a Lac-Gent module by FLP recombination. A FRT site, which includes a 34-bp FRT core sequence, was introduced into the S. enterica serovar Typhimurium genome using the bacteriophage λ red recombination system (14, 74). Strains containing a single FRT site and the plasmid pCP20 (not shown), which contains a thermoinducible FLP recombinase, were induced for FLP expression and immediately transformed by electroporation with the 12-kb plasmid pPM1 (shown at bottom left). FLP recombination releases a 9.2-kb FRT-generated circle (bottom right) which can recombine into the chromosomal FRT site to generate a gentamicin-resistant strain with a lac operon flanked by res and FRT sites (top).
DISCUSSION
Gyrase formed with the GyrB652 subunit is not a TS enzyme in vitro or in vivo (Fig. 1 and 2). Rather, GyrB652 gyrase has a poor kcat relative to the WT gyrase at 30°C, 37°C, and 42°C (Fig. 1 and 2). The gyrB652 mutation resides in a region of the protein that interacts with the GyrA subunit (8, 42, 56). Thus, the mutation may make gyrase less efficient by impeding interactions between the A and B subunits. The two- to fourfold effect of the gyrB652 mutation on the enoxacin-dependent shift to a cleavable complex (Fig. 2) matches the specific activity that is three- to fivefold lower than for WT GyrB in supercoiling assays conducted at 30°C. Defining the precise loss in catalytic efficiency is important but difficult. Single-molecule assays (15) for negative supercoiling are not yet available (N. R. Cozzarelli, personal communication). Nonetheless, contacts between protein subunits can clearly modulate enzyme efficiency. For example, protein-protein contacts between DNA-bound RecA protomers may explain the differences in recombinational efficiency of the E. coli and Pseudomonas aeruginosa RecA proteins (4).
Since the TS response is not a failure of the enzyme to work at 42°C, why do cells stop growth? As a central enzyme in DNA metabolism, the catalytic rate of gyrase could be required to operate within specific limits to accommodate DNA compaction and cell division timing mechanisms. We suggest that three factors could contribute to replication problems in gyrB hypomorphic mutants. First, even at permissive temperature (30°C) the number of supercoil domains per genome equivalent of DNA in a GyrB652 strain increases to over 1,000 from the 400 domains found in WT cells (17, 55, 66). Although our assay restricts us to studies at 30°C because of the TS repressor control on resolvase expression, others have shown that the number of DNA genome equivalents per cell at the fastest growth rates reaches a median between 2 and 4 (62). Under these conditions, unless gyrase expression increases, domains will exceed the number of cellular gyrase molecules (about 1,000/cell) by severalfold. Second, during chromosomal replication, oriC is physically separated from the terminus during formation of sister nucleoids (5, 20, 49, 68). Work from several labs shows that the physical separation of ori and the terminus is a highly reproducible part of a bacterial cell cycle (5, 20, 71). Third, the chromosomes in GyrB652 cells show a gradient of supercoil disruption in which the region closest to oriC is nearest to normal WT and the dif region is the most disorganized (Fig. 3B to C). Because of a supercoil-sensing mechanism that drives the gyrB promoter (46, 53), gyrB's position near oriC may preclude it sensing a loss of supercoiling at dif.
Another factor may involve Topo IV (a tetramer of ParC and ParE proteins), which relaxes both positive and negative supercoils in addition to decatenating the replicated sister chromosomes (13, 16, 22). Topo IV contributes to the supercoil balance in vivo (75), and it becomes concentrated near the terminus of replication (21), possibly occupying a high-affinity site near dif (38). Topo IV relaxation activity near dif would exacerbate the loss of gyrase-dependent supercoiling in a GyrB652 mutant.
Dichotomous chaos and the terminus.
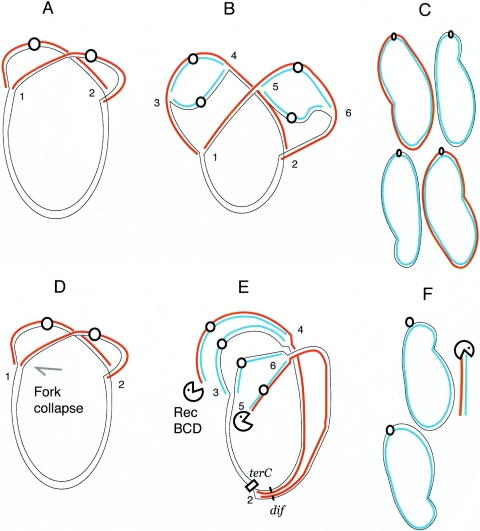
In addition to discovering the complex GyrB652 phenotype, Gari et al. found that cell growth is required for gyrase mutants to trigger an SOS response (26). A genetic selection for cells that survive recA- or recBCD-gyrA208 synthetic lethality revealed TS mutants in tRNA aminoacyl synthetases. When protein synthesis was blocked soon after temperature up-shift, cells were protected from synthetic lethality and SOS induction. Garil et al. (26) proposed that chromosome breaks were caused by unbalanced growth and suggested a link to thymine-less death that occurs in both prokaryotes and eukaryotes (3). We propose that the factors listed above contribute to unbalanced growth during dichotomous growth, which is the poly-fork DNA replication pattern that accompanies growth of E. coli and Salmonella in rich medium at a high temperature (Fig. 5A to C). As growth rate increases, cells reinitiate replication before the initial round is complete. Initiation at oriC (Fig. 5A) leads to bidirectional synthesis with two replisomes (labeled 1 and 2 in Fig. 5A) proceeding in opposite directions (7). When replication proceeds to a specified distance from the origin, initiation occurs again at each replicated oriC. This adds four new replication forks (3 to 6 in Fig. 5B), and at cell division in WT cells, two daughters inherit a complete chromosome and partially replicated chromosomes (data not shown). The products of two rounds of initiation are four chromosomes with strands of the original template, strands from the first initiation, and strands from the second initiation (Fig. 5C).
FIG. 5.
Model for topological chaos at dif and SOS and Rec-less degradation in a GyrB652 mutant. A. In strains with WT gyrase, initiation at oriC (0) leads to semiconservative (red strands) and bidirectional replication using two forks (labeled 1and 2). B. At 42°C in rich medium, rapid cell growth allows a second round of replication initiates at each oriC leading to four more forks (labeled 3, 4, 5, and 6) and new blue strands of DNA. C. As replication forks 1and 2 complete synthesis at the terminus, cell division proceeds with daughter cells inheriting two complete and two partially replicated chromosomes (not shown). When the second round is complete, four genome equivalents are produced with two black/blue strands and two red/blue strands. D. Strains with a GyrB652 gyrase have well organized origin regions and poorly organized termini. Collapse of replication fork 1 stalls replichore II while fork movement through replichore I continues unabated. E. Rapid cell growth triggers reinitiation at both oriCs. If forks 3 and 5 overtake stalled fork 1, double-strand ends expose red/blue strands to RecBCD degradation. The residual elements of forks 3 and 5 (black/blue strands) can restart one fork while forks 4 and 6 proceed to the terminus. In a RecA mutant, rescue of DNA ends by strand invasion is impaired, which leads to degradation of the partial chromosomal replicas with red/blue strands. F. The outcome is two genome equivalents of DNA having both an original template (black) and one blue strand derived from the second initiation.
All barriers to the DnaB helicase must be eliminated to allow unimpeded fork movement at the normal rate of >800 bp/s. We hypothesize that DNA replication in cells carrying the sluggish hypomorphic gyrase bogs down due to poor supercoil condensation of DNA near the terminus. If a fork collapses while cell growth continues (10, 47), there is a critical time for restarting before new forks overtake the stalled fork (Fig. 5). When a second fork runs into a stalled fork, it exposes a double-stranded end (see Bidnenko et al. [6]). This won't happen nearly as much if DNA initiation is blocked or delayed by slow protein synthesis, and chromosomal termini can gain time to condense and resolve DNA at low fork density. The condition of a GyrB652 mutant mimics the killing of cells with quinolones, which also requires growth and protein synthesis for achieving a lethal effect (39).
The terminal chaos hypothesis is consistent with several recent observations. In an E. coli gyrase hypomorph, Jeong et al. found a large disruption in gene expression near dif, and their chromosome immunoprecipitation assays indicated gyrase depletion near the terminus (41). Moreover, several genetic conditions that increase oriC initiation also trigger fork failure. One example is the overproduction of DnaA, which leads to overinitiation, fork collapse (29), and breakage of chromosomal DNA with loss of cell viability (61). Terminal chaos can explain the sickness of SeqA mutants, which must be grown in minimal medium at a low temperature because they lack the mechanism by which SeqA inhibits premature reinitiation by blocking access of DnaA protein to oriC (9, 63). In addition to gyrase, SeqA, and DnaA, phenotypes of growth rate toxicity are found with mutants of priA (58, 59), mukB (1), diaA (40), and parC and parE (2). If resolution near dif is specifically impaired in several of these mutants, then defining a comprehensive set of genes that change domain structure at high growth rates would be an important step toward completing our understanding of DNA fork termination and chromosome segregation.
Acknowledgments
This work was supported by National Institutes of Health grant GM-33143-20 and MCB 0110675 from the National Science Foundation.
We thank N. Bossi, N. Figueroa, and M. Schmid for the gift of mutant strains.
REFERENCES
- 1.Adachi, S., and S. Hiraga. 2003. Mutants suppressing novobiocin hypersensitivity of a mukB null mutation. J. Bacteriol. 185:3690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, D. E., E. M. Shekhtman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 4.Bakhlanova, I. V., T. Ogawa, and V. A. Lanzov. 2001. Recombinogenic activity of chimeric recA genes (Pseudomonas aeruginosa/Escherichia coli): a search for RecA protein regions responsible for this activity. Genetics 159:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121:899-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidnenko, V., S. D. Ehrlich, and B. Michel. 2002. Replication fork collapse at replication terminator sequences. EMBO J. 21:3898-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breier, A. M., H.-U. G. Weier, and N. R. Cozzarelli. 2005. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. USA 102:3942-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabral, J. H. M., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Siddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 11.Cozzarelli, N. R. 1980. DNA gyrase and the supercoiling of DNA. Science 207:953-960. [DOI] [PubMed] [Google Scholar]
- 12.Cozzarelli, N. R., and J. C. Wang. 1990. DNA topology and its biological effects. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 13.Crisona, N. J., T. R. Strick, D. Bensimon, V. Croquette, and N. R. Cozzarelli. 2000. Preferential relaxation of positively supercoiled DNA by Escherichia coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14:2881-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using polymerase chain reaction products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport, R. J., J. L. Wuite, R. Landick, and C. Bustamante. 2000. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science 287:2497-2500. [DOI] [PubMed] [Google Scholar]
- 16.Deibler, R. W., S. Rahmati, and E. L. Zechiedrich. 2001. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 15:748-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, S., R. A. Stein, and N. P. Higgins. Submitted for publication.
- 18.Deng, S., R. A. Stein, and N. P. Higgins. 2004. Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome. Proc. Natl. Acad. Sci. USA 101:3398-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echols, H., and G. Guarneros. 1983. Control of integration and excision, p. 75-92. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Elmore, S., M. Muller, N. Vischer, T. Odijk, and C. Woldringh. 2005. Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J. Struct. Biol. 136:53-66. [DOI] [PubMed] [Google Scholar]
- 21.Espeli, O., C. Levine, H. Hassing, and K. J. Marians. 2003. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell 11:189-201. [DOI] [PubMed] [Google Scholar]
- 22.Espeli, O., and K. J. Marians. 2004. Untangling intracellular DNA topology. Mol. Microbiol. 52:925-931. [DOI] [PubMed] [Google Scholar]
- 23.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:21429-21432. [DOI] [PubMed] [Google Scholar]
- 24.Gaitanaris, G. A., A. G. Papavassiliou, P. Rubock, S. J. Silverstein, and M. E. Gottesman. 1990. Renaturation of denatured l repressor requires heat shock proteins. Cell 61:1013-1020. [DOI] [PubMed] [Google Scholar]
- 25.Gamper, H. B., and J. E. Hearst. 1982. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell 29:81-90. [DOI] [PubMed] [Google Scholar]
- 26.Gari, E., L. Bossi, and N. Figueroa-Bossi. 2001. Growth-dependent DNA breakage and cell death in a gyrase mutant of Salmonella. Genetics 159:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gari, E., N. Figueroa-Bossi, A.-B. Blanc-Potard, F. Spirito, M. B. Schmid, and L. Bossi. 1996. A class of gyrase mutants of Salmonella typhimurium show quinolone-like lethality and require Rec functions for viability. Mol. Microbiol. 21:111-122. [DOI] [PubMed] [Google Scholar]
- 28.Gefter, M. L., Y. Hirota, T. Kornberg, J. A. Wechsler, and C. Barnoux. 1971. Analysis of DNA polymerases II and 3 in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc. Natl. Acad. Sci. USA 68:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grigorian, A. V., R. B. Lustig, E. C. Guzman, J. M. Mahaffy, and J. W. Zyskind. 2003. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 185:630-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gussin, G. N., A. D. Johnson, C. O. Pabo, and R. T. Sauer. 1983. Repressor and cro protein: structure, function and role in lysogenization, p. 93-121. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Hecht, M. H., H. C. M. Nelson, and R. T. Sauer. 1983. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc. Natl. Acad. Sci. USA 80:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed] [Google Scholar]
- 33.Higgins, N. P. 1999. DNA supercoiling and its consequences for chromosome structure and function, p. 189-202. In R. L. Charlebois (ed.), Organization of the prokaryotic genome, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 34.Higgins, N. P., and N. R. Cozzarelli. 1982. The binding of gyrase to DNA: analysis by retention to nitrocellulose filters. Nucleic Acids Res. 10:6833-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins, N. P., C. L. Peebles, A. Sugino, and N. R. Cozzarelli. 1978. Purification of the subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc. Natl. Acad. Sci. USA 75:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins, N. P., and A. Vologodskii. 2004. Topological behavior of plasmid DNA, p. 181-201. In G. Phillips and B. Funnell (ed.), Plasmid biology. ASM Press, Washington, D.C.
- 37.Higgins, N. P., X. Yang, Q. Fu, and J. R. Roth. 1996. Surveying a supercoil domain by using the γδ resolution system in Salmonella typhimurium. J. Bacteriol. 178:2825-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hojgaard, A., H. Szerlong, C. Tabor, and P. Kuempel. 1999. Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol. Microbiol. 33:1027-1036. [DOI] [PubMed] [Google Scholar]
- 39.Hooper, D. C. 1993. Quinolone mode of action—new aspects. Drugs 45:8-14. [DOI] [PubMed] [Google Scholar]
- 40.Ishida, T., N. Akimitsu, T. Kashioka, M. Hatano, T. Kubota, Y. Ogata, K. Sekimizu, and T. Katayama. 2004. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 279:45546-45555. [DOI] [PubMed] [Google Scholar]
- 41.Jeong, K. S., J. Ahn, and A. B. Khodursky. 2004. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 5:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kampranis, S. C., A. D. Bates, and A. Maxwell. 1999. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. USA 96:8414-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kampranis, S. C., and A. Maxwell. 1998. The DNA gyrase-quinolone complex. ATP hydrolysis and the mechanism of DNA cleavage. J. Biol. Chem. 273:22615-22626. [DOI] [PubMed] [Google Scholar]
- 44.Kornberg, A., and T. Baker. 1991. DNA replication, 2nd ed. W. H. Freeman & Co., New York, N.Y.
- 45.Louarn, J. M., P. Kuempel, and F. Cronet. 2005. The terminus region of the E. coli chromosome or all's well that ends well, p. 251-273. In N. P. Higgins (ed.), The bacterial chromosome. ASM Press, Washington, D.C.
- 46.Menzel, R., and M. Gellert. 1983. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34:105-113. [DOI] [PubMed] [Google Scholar]
- 47.Michel, B., G. Grompone, M.-J. Flores, and V. Bidnenko. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 101:12783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, W. G., and R. W. Simons. 1993. Chromosomal supercoiling in Escherichia coli. Mol. Microbiol. 10:675-684. [DOI] [PubMed] [Google Scholar]
- 49.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14:212-223. [PMC free article] [PubMed] [Google Scholar]
- 50.Pato, M., and M. Banerjee. 1996. The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. Mol. Microbiol. 22:283-292. [DOI] [PubMed] [Google Scholar]
- 51.Pavitt, G. D., and C. F. Higgins. 1993. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol. Microbiol. 10:685-696. [DOI] [PubMed] [Google Scholar]
- 52.Perals, K., F. Cornet, Y. Merlet, and J.-M. Louarn. 2000. Functional polarization of the Escherichia coli chromosome terminus. The dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarities. Mol. Microbiol. 36:33-43. [DOI] [PubMed] [Google Scholar]
- 53.Peter, B. J., J. Arsuaga, A. M. Breier, A. B. Khodursky, P. O. Brown, and N. R. Cozzarelli. 2004. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 5:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postow, L., N. J. Crisona, B. J. Peter, C. D. Hardy, and N. R. Cozzarelli. 2001. Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. USA 98:8219-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Postow, L., C. D. Hardy, J. Arsuaga, and N. R. Cozzarelli. 2004. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18:1766-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reece, R., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 57.Richardson, S. M. H., C. F. Higgins, and D. M. J. Lilley. 1984. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J. 3:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandler, S. 1996. Overlapping functions for recF and priA in cell viability and UV-inducible SOS expression are distinguished by dnaC809 in Escherichia coli K-12. Mol. Microbiol. 19:871-880. [DOI] [PubMed] [Google Scholar]
- 59.Sandler, S., H. Samra, and A. Clark. 1996. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics 143:5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheirer, K. E., and N. P. Higgins. 1997. The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J. Biol. Chem. 272:27202-27209. [DOI] [PubMed] [Google Scholar]
- 61.Simmons, L. A., A. M. Breier, N. R. Cozzarelli, and J. M. Kaguni. 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51:349-358. [DOI] [PubMed] [Google Scholar]
- 62.Skarstad, K., R. Bernander, and E. Boye. 1995. Analysis of DNA replication in vivo by flow cytometry. Methods Enzymol. 262:604-613. [DOI] [PubMed] [Google Scholar]
- 63.Skarstad, K., N. Torheim, S. Wold, R. Lurz, W. Messer, S. Fossum, and T. Bach. 2001. The Escherichia coli SeqA protein binds specifically to two sites in fully and hemimethylated oriC and has the capacity to inhibit DNA replication and affect chromosome topology. Biochimie 83:49-51. [DOI] [PubMed] [Google Scholar]
- 64.Staczek, P., and N. P. Higgins. 1998. DNA gyrase and topoisomerase IV modulate chromosome domain size in vivo. Mol. Microbiol. 29:1435-1448. [DOI] [PubMed] [Google Scholar]
- 65.Stark, M. W., and M. R. Boocock. 1995. Topological selectivity in site-specific recombination, p. 179. In D. Sherratt (ed.), Mobile genetic elements, vol. 58. IRL Press, Oxford, England. [Google Scholar]
- 66.Stein, R., S. Deng, and N. P. Higgins. 2005. Measuring chromosome dynamics on different timescales using resolvases with varying half-lives. Mol. Microbiol. 56:1049-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 68.Sunako, Y., T. Onogi, and S. Hiraga. 2001. Sister chromosome cohesion of Escherichia coli. Mol. Microbiol. 42:1233-1241. [DOI] [PubMed] [Google Scholar]
- 69.Vogel, J. L., Z. J. Li, M. M. Howe, A. Toussaint, and N. P. Higgins. 1991. Temperature-sensitive mutations in the bacteriophage Mu c repressor locate a 63-amino-acid DNA-binding domain. J. Bacteriol. 173:6568-6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vologodskii, A. V., W. Zhang, V. V. Rybenkov, A. A. Podtelezhnikov, D. Subramanian, J. D. Griffith, and N. R. Cozzarelli. 2001. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl. Acad. Sci. USA 98:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, X., C. Possoz, and D. J. Sherratt. 2005. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 19:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wechsler, J. A., V. Nusslein, B. Otto, A. Klein, F. Bonhoeffer, R. Herrmann, L. Gloger, and H. Schaller. 1973. Isolation and characterization of thermosensitive Escherichia coli mutants defective in deoxyribonucleic acid replication. J. Bacteriol. 113:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, H.-Y., S. Shyy, J. C. Wang, and L. F. Liu. 1988. Transcription generates positively and negatively supercoiled domains in the template. Cell 53:433-440. [DOI] [PubMed] [Google Scholar]
- 74.Yu, D., J. A. Sawitzke, H. Ellis, and D. L. Court. 2003. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc. Natl. Acad. Sci. USA 100:7099-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zechiedrich, E. L., B. K. Arkady, S. Bachellier, D. Chen, D. M. Lilley, and N. R. Cozzarelli. 2000. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275:8103-8113. [DOI] [PubMed] [Google Scholar]