Abstract
Formulae for calculating low-frequency twist-like and accordion-like modes of DNA molecules have been derived using a quasi-continuum model. The formulae can be employed in essentially all (viz. A, B, C, D, E, and Z) forms of DNA. Calculated results indicate that the experimentally observed low-frequency modes at 22 cm-1 for the A-form octanucleotide (d[CCCCGGGG]) and at 18 cm-1 for the B-form dodecanucleotide (d[CGCAA ATTTGCG]) may result from accordion-like motions, while those observed at 12 cm-1 and 15 cm-1 may result from combinations of twist-like oscillations excited in the intact segments of B- and A-DNA's, respectively. Frequency shifts in the low-frequency modes observed when DNA molecules undergo conformational changes among different forms are also discussed in terms of the current model.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooks B., Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. G., Erfurth S. C., Small E. W., Peticolas W. L. Conformationally dependent low-frequency motions of proteins by laser Raman spectroscopy. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1467–1469. doi: 10.1073/pnas.69.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C. Identification of low-frequency modes in protein molecules. Biochem J. 1983 Dec 1;215(3):465–469. doi: 10.1042/bj2150465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C. Low-frequency collective motion in biomacromolecules and its biological functions. Biophys Chem. 1988 May;30(1):3–48. doi: 10.1016/0301-4622(88)85002-6. [DOI] [PubMed] [Google Scholar]
- Chou K. C. Low-frequency motions in protein molecules. Beta-sheet and beta-barrel. Biophys J. 1985 Aug;48(2):289–297. doi: 10.1016/S0006-3495(85)83782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C. Low-frequency vibrations of DNA molecules. Biochem J. 1984 Jul 1;221(1):27–31. doi: 10.1042/bj2210027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Mao B. Collective motion in DNA and its role in drug intercalation. Biopolymers. 1988 Nov;27(11):1795–1815. doi: 10.1002/bip.360271109. [DOI] [PubMed] [Google Scholar]
- Chou K. C. Origin of low-frequency motions in biological macromolecules. A view of recent progress in the quasi-continuity model. Biophys Chem. 1986 Dec 15;25(2):105–116. doi: 10.1016/0301-4622(86)87001-6. [DOI] [PubMed] [Google Scholar]
- Eyster J. M., Prohofsky E. W. Lattice vibrational modes of poly(rU)-poly(rA). A coupled single-helical approach. Biopolymers. 1974 Dec;13(12):2527–2543. doi: 10.1002/bip.1974.360131210. [DOI] [PubMed] [Google Scholar]
- Genzel L., Keilmann F., Martin T. P., Winterling G., Yacoby Y., Fröhlich H., Makinen M. W. Low-frequency Raman spectra of lysozyme. Biopolymers. 1976 Jan;15(1):219–225. doi: 10.1002/bip.1976.360150115. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Ito K., Shimanouchi T. Vibrational frequencies and modes of alpha-helix. Biopolymers. 1970;9(4):383–399. doi: 10.1002/bip.1970.360090402. [DOI] [PubMed] [Google Scholar]
- Lamba O. P., Wang A. H., Thomas G. J., Jr Low-frequency dynamics and Raman scattering of crystals, of B-, A-, and Z-DNA, and fibers of C-DNA. Biopolymers. 1989 Feb;28(2):667–678. doi: 10.1002/bip.360280210. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R. M., Srinivasan A. R., Olson W. K., McCammon J. A. Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers. 1984 Jun;23(6):1099–1112. doi: 10.1002/bip.360230610. [DOI] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- Mao B., McCammon J. A. Theoretical study of hinge bending in L-arabinose-binding protein. Internal energy and free energy changes. J Biol Chem. 1983 Oct 25;258(20):12543–12547. [PubMed] [Google Scholar]
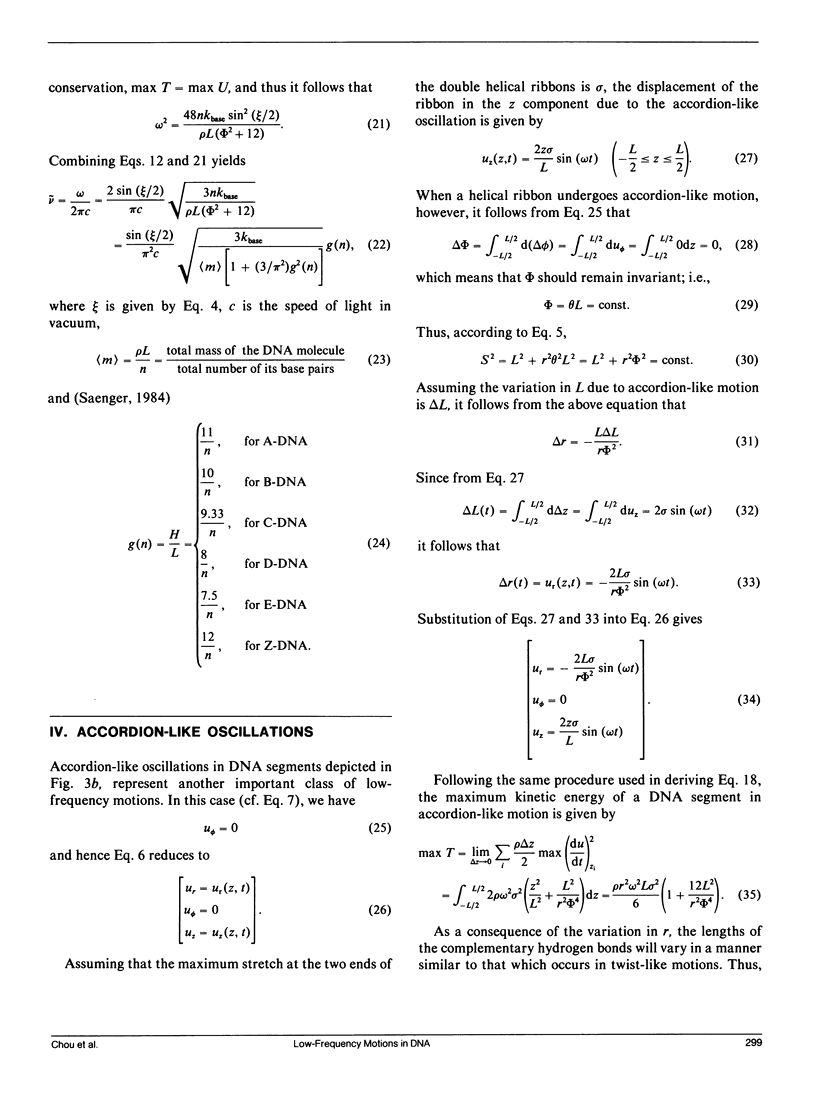
- Miller K. J. Interactions of molecules with nucleic acids. I. An algorithm to generate nucleic acid structures with an application to the B-DNA structure and a counterclockwise helix. Biopolymers. 1979 Apr;18(4):959–980. doi: 10.1002/bip.1979.360180415. [DOI] [PubMed] [Google Scholar]
- Painter P. C., Mosher L. E., Rhoads C. Low-frequency modes in the Raman spectra of proteins. Biopolymers. 1982 Jul;21(7):1469–1472. doi: 10.1002/bip.360210715. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suezaki Y., Go N. Breathing mode of conformational fluctuations in globular proteins. Int J Pept Protein Res. 1975;7(4):333–334. doi: 10.1111/j.1399-3011.1975.tb02448.x. [DOI] [PubMed] [Google Scholar]
- Tidor B., Irikura K. K., Brooks B. R., Karplus M. Dynamics of DNA oligomers. J Biomol Struct Dyn. 1983 Oct;1(1):231–252. doi: 10.1080/07391102.1983.10507437. [DOI] [PubMed] [Google Scholar]
- Urabe H., Tominaga Y. Low-lying collective modes of DNA double helix by Raman spectroscopy. Biopolymers. 1982 Dec;21(12):2477–2481. doi: 10.1002/bip.360211212. [DOI] [PubMed] [Google Scholar]
- Wittlin A, Genzel L, Kremer F, Häseler S, Poglitsch A, Rupprecht A. Far-infrared spectroscopy on oriented films of dry and hydrated DNA. Phys Rev A Gen Phys. 1986 Jul;34(1):493–500. doi: 10.1103/physreva.34.493. [DOI] [PubMed] [Google Scholar]