Abstract
A promoter-probe strategy was devised to identify genes specifically expressed by Legionella pneumophila during growth within the macrophage. Random fragments from the L. pneumophila chromosome were inserted upstream of a promoterless phage T4 td gene, and fragments that led to complementation of thymine auxotrophy during intracellular growth of the bacterium were identified. Two different selection strategies were employed to eliminate promoters that were also active during extracellular growth of the bacterium. Some of these genes were identified independently by using both of the selection strategies. The factors identified include orthologs of efflux-mediated resistance determinants and transporters, a transporter involved in protection from osmotic stress, a stress response GTP-binding protein, a response regulator, a sensor kinase, and two systems that increase the reducing potential of the bacterium, one of which encodes the L. pneumophila ortholog of ahpC. Five of the clones analyzed here were fusions to promoters that were closely linked to genes encoding three-component chemiosmotic efflux pumps that export heavy metals or toxic organic compounds. Analysis of ahpC gene expression indicates that levels increased at least sevenfold during intracellular growth of the bacterium. Inactivation of several of the genes at their chromosomal loci had no effect on the intracellular growth rate of L. pneumophila in cultured macrophages. This suggests that a number of genes with increased expression during intracellular growth may be part of redundant systems that allow survival and growth under the conditions encountered within host cells.
Pathogenic bacteria must be able to survive within host tissues in order to cause disease. It has long been appreciated that bacteria growing in intimate contact with the host express a unique complement of genes that collectively allow bacteria to prosper in this environment. To identify such genes, several studies have taken advantage of knowledge about the conditions found within the host to try to understand the bacterial response to the host-associated environment. For example, iron availability is extremely low in most sites of infection, and genes expressed in response to low iron concentrations have been identified in Shigella flexneri, Vibrio cholerae, and Pseudomonas spp. (29). Some of the genes expressed in response to these conditions have been shown to have effects on the virulence of the organism.
Unfortunately, not all of the conditions encountered by a microorganism during an infection are known, and this limits the number of loci that can be identified by using such an approach. Furthermore, a microorganism may be found in niches within a host that cannot be reproduced in culture. For this reason, a number of techniques have been developed to identify microbial genes that are expressed in response to growth within mammalian hosts or cultured cells (27). These strategies have involved using a variety of promoter trap vectors or subtractive techniques to identify transcripts expressed specifically in response to host cells. The analysis of such promoters has been confounded by the fact that many bacterial virulence factors known to be induced by contact with host tissues have not been isolated by using these strategies, presumably because they have significant expression outside of the host (26). Theoretically, nucleotide arrays could be used to identify bacterial mRNAs induced after contact with host cells, with the potential advantage of assaying the entire spectrum of genes encoded by the microorganism (18, 23). The disadvantage of this strategy is that it requires relatively high multiplicities of infection to obtain sufficient mRNA for analysis of gene expression, and this may not accurately mimic natural infections.
Legionella pneumophila is a gram-negative intracellular pathogen that is the causative agent of Legionnaires' disease (22). During disease, the bacteria enter alveolar macrophages, where they replicate, eventually causing pneumonia-like symptoms. This niche cannot easily be reproduced in any extracellular medium. Within the macrophage, the bacteria grow in an unusual compartment which appears to be surrounded by endoplasmic reticulum-derived membrane (56). Targeting to this site requires the products of the dot and icm genes, which encode a secretion system with high similarity to conjugative DNA transport (50, 61). Very little is known about how the Dot- and Icm-induced compartment develops or how the organism adapts to growth within this site.
The ability of L. pneumophila strains to grow within cultured macrophages correlates with their ability to cause disease in susceptible hosts (16, 39). The tissue culture model of L. pneumophila intracellular growth has been widely used to identify bacterial virulence factors and to test the virulence of environmental isolates (6, 8, 20, 31, 39, 51). It has been shown that the expression of as many as 35 proteins is increased during growth of the bacteria inside the macrophage (2). Several genes have been identified as being upregulated after contact with host cells, including gspA (general stress protein) (3), a pyrophosphatase gene (1), eml (4), and the helABC cluster, which appears to encode a three-component efflux-detoxifying system (32). In an effort to identify the nutritional and environmental challenges that the bacteria face during growth within the macrophage and to understand how the bacteria are able to grow at this site, we have developed a screen to identify the genes that are expressed by the bacterium during intracellular growth.
The screen is based on the observation that thymine auxotrophs of L. pneumophila rapidly undergo thymineless death in the macrophage (7, 36). By using a promoterless copy of a gene that complements the thymine auxotrophy, we were able to identify bacterial promoters that are specifically activated in response to the macrophage intracellular environment by using both positive and negative selection strategies. By using this strategy, we have identified genes whose products may comprise part of a larger cohort of proteins that allow adaptation to intracellular conditions.
MATERIALS AND METHODS
Growth of bacterial strains.
The bacterial strains used are described in Table 1. Escherichia coli strains were grown on L plates or in L broth supplemented with appropriate antibiotics as described. L. pneumophila Philadelphia 1 strains were all grown in ACES yeast extract (AYE) broth, on charcoal yeast extract (CYE) plates, or CYE plus thymidine (CYET), as described before (7). Both plates and broth were supplemented with 100 μg of thymidine and 20 μg of kanamycin sulfate per ml when appropriate. Antibiotic pressure was maintained unless otherwise noted. L. pneumophila used as a recipient in heterospecific matings was selected on CYE supplemented with 50 μg of streptomycin per ml or 5% sucrose, as appropriate. Bacterial strains were stored in 35% glycerol-65% broth at −70 or −20°C.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| L. pneumophila | ||
| Lp01 | Philadelphia 1, hsdR rpsL | 7 |
| Lp02 | Lp01, thyA | 7 |
| LpSR146 | Lp02, ahpC::kan | This work |
| LpSR237 | Lp02, stuC::pSR47 | This work |
| LpSR239 | Lp02, cadA1::pSR47 | This work |
| LpSR255 | Lp02, sflR::pSR47 | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | |
| MC1061 | ΔlacX74 galU galK rpsL thi | |
| SY547 | araD Δ(lac-pro) rpoB gyrA argE(Am) supE met thyA | |
| SY327λpir | araD Δ(pro-lac) argE(Am) rpoB gyrA recA56(λpir) | 34 |
| JM115 | SY327λpir F′ lacZM15 | 34 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km (λpir) | 35 |
| Plasmids | ||
| pBluescriptSK− | Stratagene, Inc. | |
| pMMB34ΔNotI | pMMB34, NotI site removed | This work |
| pTZtdΔi | Intron-free td gene of phage T4 | 14 |
| pMC1871 | lacZ gene with EcoRI site removed | 52 |
| pRS415 | trp-lac gene fusion | 62 |
| pRI261 | pMC1871 trp-lac | This work |
| pSR32 | Derivative of pMMB34 containing promoterless tdΔi gene | This work |
| pSR37 | Promoterless tdΔi-lacZ fusion | This work |
| pSR42 | pAY01/ahpC::Kanr | This work |
| pB30G, pA21E | pSR37/glrA, ahpC | This work |
| pA17A | pSR37obgL | This work |
| pA37F | pSR37/cadA1 | This work |
| pB3F, pB13F | pSR37/sflR | This work |
| pB17C, pB12H | PSR37/smlA | This work |
| pB30G-Δ5′ | Derivative of pB30G | This work |
| pB30G-Δ3′ | Derivative of pB30G | This work |
| pB30G-revΔ5′ | Derivative of pB30G | This work |
| pB30G-revΔ3′ | Derivative of pB30G | This work |
| pAY01 | Conditional replication, sucrose sensitivity | 64 |
| pSR47 | Conditional replication vector for integrative gene disruptions | 64 |
| pSR52 | Derivative of pSR37 lacking tdΔi gene | This work |
DNA manipulations.
Plasmid DNA was prepared from saturated overnight cultures of E. coli, generally DH5α or MC1061, grown in 2× YT and was routinely prepared by using the cetyltrimethylammonium bromide (CTAB) precipitation method (17). Unless otherwise stated, plasmid DNA was introduced into bacterial host strains by electroporation (7). Restriction enzymes, Vent DNA polymerase, T4 DNA polymerase, and calf intestinal phosphatase were obtained from New England Biolabs (Beverly, Mass.) and used according to the manufacturer's specifications unless otherwise indicated. Double-stranded DNA sequencing was performed by using the Sequenase kit from U.S. Biochemicals (Cleveland, Ohio).
Tissue culture.
U937 cells were obtained from the American Type Culture Collection, passaged once in RPMI plus 10% fetal bovine serum (FBS), and stored in 10% dimethyl sulfoxide in liquid nitrogen. Cells thawed rapidly at 37°C were pelleted to remove excess dimethyl sulfoxide and resuspended in RPMI supplemented with 10% FBS. Cells were maintained in this medium in suspension, passaged by 50-fold dilution into fresh medium, and discarded after approximately 14 passages. U937 cells were activated by treatment for 36 to 48 h with 10 ng of the phorbol ester 12-myristate 13-acetate (TPA) per ml in the same medium. The cells were resuspended in fresh medium without TPA, plated at approximately 106 cells/cm2 on tissue culture-treated plastic dishes, and used within 3 to 24 h. Growth curves of bacterial strains in monolayers of U937 cells were obtained as described previously (7).
Construction of pSR37.
Plasmid pSR37 was constructed in several steps. A promoterless copy of the tdΔi gene was amplified by PCR from plasmid pTZ tdDi by using primers 5312 (5′-CGGGATCCGCGGCCGCCAGTATATAAATGAAACAA) and 5753 (5′-CGAATTCGCTGACACCCGGGCGCAATAATAAAATTACACCGCC). The use of these primers adds a unique NotI site to the 5′ end of the gene and a SalI site just after the stop codon. The amplified fragment was digested with BamHI and EcoRI and inserted into a similarly digested pMMB34 derivative, pMMB34/NotIΔ. The resulting plasmid was digested with SalI, and a trp-lac fusion gene isolated from plasmid pRI261 was introduced into this site, yielding pSR37 (Fig. 1). The functional expression of the tdΔi gene was confirmed by complementation of the E. coli SY547 thyA mutant (data not shown).
FIG. 1.
Strategies used to identify L. pneumophila promoters induced during intracellular growth of the bacterium. A library of chromosomal fragments was introduced upstream of a promoterless tdΔi-lacZ fusion in plasmid pSR37. Lp02 (thyA) transformed with the library was then used to infect monolayers of U937 cells, and survivors were screened for a Thy− phenotype on bacteriologic medium (selection A). In selection B, transformants were plated directly on trimethoprim, and survivors were then used to infect monolayers of U937 cells. Strains surviving the selections were tested individually for the ability to form plaques on monolayers of U937 cells and for a strict Thy− phenotype on extracellular medium.
Construction of genomic library in pSR37.
Chromosomal DNA was partially digested with Sau3AI, and fragments of 1 to 2 kbp were gel purified. Synthetic oligonucleotides (5′-GATCCGAGCTCGC and 5′-GGCCGCGAGCTCG) were annealed together to form a NotI/SacI/Sau3AI adapter, and this adapter was ligated into the ends of NotI-digested pSR32. The vector with the adapters was gel purified, and the Sau3AI chromosomal fragments were ligated to the exposed Sau3AI ends. The inserts in this library were isolated by digestion with NotI and transferred to a similarly digested pSR37.
Identification of in vivo-activated promoters. (i) Selection A.
The pSR37 library was introduced into Lp02 (thyA) by electroporation, and 48 individual pools of approximately 104 transformants were plated on CYE supplemented with kanamycin. After 3 days, the colonies from each pool were swabbed off the plates, and 107 bacteria from each pool were used to infect 2 × 106 TPA-treated U937 cells in the presence of 100 μg of thymidine per ml. After 90 min, the monolayers were washed with sterile phosphate-buffered saline (PBS) to remove excess thymidine, fresh medium lacking thymidine was added, and the monolayers were returned to the incubator for 36 h. The monolayers were then washed again and lysed in 0.5 ml of sterile distilled H2O. The lysates were plated on CYET plus kanamycin. After 3 days the plates were overlaid with 3 ml of soft agar containing 600 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Eight individual colonies expressing relatively low levels of β-galactosidase (based on the results of the X-Gal overlay) were then picked from each pool, screened both for the ability to form plaques on monolayers of U937 cells as described previously (7) and for a Thy− phenotype on CYE plates lacking thymidine .
(ii) Selection B.
Forty-eight pools of transformants were plated directly on CYET plus kanamycin and trimethoprim. After 2 to 3 days, the colonies were swabbed off the plates and used to infect monolayers of U937 cells as described for selection A. After 2 days the monolayers were lysed, the lysate was plated on CYET plus kanamycin, and eight individual clones from each pool were tested for the ability to form plaques on monolayers of activated U937 cells and for a strict Thy− phenotype on CYE (the inability to form single colonies after 3 days on CYE).
RNA analysis.
Total RNA was isolated from broth-grown bacteria by a modification of the method of Chomczynski and Sacchi (13); 10 ml of a logarithmic-phase culture (A600 = 0.5 to 0.8) was pelleted and resuspended in 1 ml of diethyl pyrocarbonate-treated H2O. The cells were lysed in 10 ml of denaturing solution (13), 10 ml of warmed (65°C) H2O-equilibrated phenol was added, and the preparation was mixed and incubated at 65°C for 10 min. Then 2 ml of chloroform-isoamyl alcohol (24:1) was added, and the solutions were mixed and incubated on ice for 20 min. The aqueous layer was collected after centrifugation, and the extraction procedure was repeated at least once. The RNA was precipitated by the addition of an equal volume of isopropanol, and the pellet was washed in 70% ethanol. The samples were treated with RNase-free DNase (Boehringer, Ridgefield, Conn.) and applied to nylon membranes. Single-stranded radiolabeled RNA probes were made by using the Riboprobe kit (Promega, Madison, Wis.) on derivatives of the plasmid pBluescriptSK+ (Stratagene, La Jolla, Calif.).
β-Galactosidase assay of infected monolayers.
Bacteria from 1- to 2-day-old patches of the strains to be tested were suspended in RPMI plus 10% FBS and added to confluent monolayers of TPA-treated U937 cells in RPMI containing 10% FBS and 100 μg of thymidine per ml. The bacteria were centrifuged onto the monolayers (5 min, 1,000 rpm), and the medium was gently removed. At this point, the monolayer was lysed (for the time zero point) or fresh medium without thymidine was gently overlaid, and the dish was returned to the CO2 incubator.
At various times after infection, representative monolayers were lysed and assayed for β-galactosidase activity. The monolayer to be assayed, having 2 × 106 U937 cells, was lysed in 0.5 ml of sterile H2O, and an aliquot of this lysate was reserved for quantitation of CFU by plating on CYET. The remainder of the lysate was pelleted and resuspended in 0.5 ml of PBS. β-Galactosidase activity of this suspension was measured by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a chromogenic substrate, and the amount of activity was normalized to the number of CFU, as determined by plating (33). Units reported are 108 ΔA420/CFU/min, where A420 is corrected for the level of ONPG hydrolysis by an uninfected monolayer. All samples were analyzed in triplicate, and values are represented as means ± standard deviations.
Inactivation of the ahpC gene.
The insert in plasmid pB30G was isolated from the plasmid by digestion with NotI and introduced into the NotI site of pBluescriptSK− (Stratagene). A 1.1-kbp BamHI fragment containing the kanamycin resistance gene from Tn903 (37) was introduced into the BclI site internal to the ahpC gene. The resulting plasmid was digested with SacI, liberating a 2.7-kbp fragment containing the entire insert from the original pB30G plasmid as well as the kanr gene, which was then inserted into the SacI site in plasmid pAY01, generating plasmid pSR42. Plasmid pSR42 was introduced into SM10λpir, which was used as a conjugative donor to introduce the plasmid into Lp02 (7). Exconjugants containing a single crossover in which pSR42 was integrated into the chromosomal ahpC gene were selected by plating on CYET supplemented with kanamycin and streptomycin. In a second step, to select for loss of the plasmid, the recipient strains were plated on CYET supplemented with kanamycin and sucrose. Strains which survived these selections were screened by genomic Southern blot for generation of an L. pneumophila ahpC::kan strain lacking an intact ahpC gene.
Inactivation of stuC, cadA1, and sflR genes of L. pneumophila.
PCR-amplified plasmid inserts from plasmids pA41B, pA37F, and pB3F were generated with vector-specific primers. Internal gene fragments of the genes therein (stuC, 537-bp DraI-DraI fragment; cadA1, 599-bp SspI-SspI fragment; and sflR, 323-bp EcoRI-BspEI fragment) were introduced into the polylinker of plasmid pSR47, and the resulting plasmids were introduced into Lp02 by using E. coli SY327 λpir/pRK600, a streptomycin-sensitive strain that carries the transfer functions necessary for mobilizing pSR47. L. pneumophila recipients were selected by plating on CYET plus kanamycin (to select for the plasmid) and streptomycin (to select against the donor). Kanamycin-resistant strains were colony purified, and integration events were confirmed by genomic Southern blot analysis.
Sequence analysis.
L. pneumophila sequences were compared to previously identified genes by using the Basic Local Alignment Search Tool (BLAST) as described by Altschul et al. (5) against the NCBI protein databases. In cases where a limited L. pneumophila genomic sequence was obtained in this work, an additional sequence was obtained from the unfinished microbial genome web server at NCBI, submitted by the Columbia University Genome Center (CUGC). Mapping of clones relative to one another was based on the contigs presented at the CUGC website (http://genome3.cpmc.columbia.edu/≈legion/index.html). When numbers are used to refer to open reading frames (ORFs) (such as orf360), the numbers correspond to those given by the CUGC and are used when there are no clear orthologs. These ORF numbers were temporary designations of CUGC and may no longer be referred to as such by the Center. ORF numbers presented here, however, were submitted to GenBank and can be accessed by using the gene designations in Table 2. In the case of orf1419.5, the ORF was not designated as such by CUGC and is located between orf1419 and orf1420.
TABLE 2.
L. pneumophila genes identified and their relationship to previously identified genes
| Plasmid(s) | Related genea | Organism | L. pneumophila gene (GenBank accession no.) | Predicted property or function | Similarityb | GenBank accession no. of homolog |
|---|---|---|---|---|---|---|
| pB30G, pA21E | ahpC | Mouse | ahpC (AF480906) | Oxidative and nitrosative stress response | 4e−50 | AF032722 |
| ydhD | E. coli | glrA | Glutaredoxin-like protein | 2e−18 | L01622 | |
| pA41B | tutC | Thauera aromatica | stuC (AF480911) | Response regulator | 9e−87 | U57900 |
| pA17A | obg | B. subtilis | obgL (AF480915) | G protein; stress response | 5e−81 | Z99118 |
| pA45G | hydD | Pyrococcus abyssi | hydD (AF480908) | Cytochrome c hydrogenase | 4e−58 | AJ248285 |
| pB3F, pB13F | floR | E. coli | sflR (AF480908) | Florfenicol efflux/resistance | 4e−34 | AAG16656 |
| pB17C, pB12H | milA | L. pneumophila | smlA (AF480909) | Intracellular growth | 2e−29 | AF153695 |
| narK | B. subtilis | Nitrite extrusion | 1e−11 | Z99123 | ||
| pB49A | cya | Anabaena sp. | ladC (AF480916) | Adenylate cyclase | 2e−38 | D89623 |
| pB33A | citA/proP | E. coli | tphB (AF480910) | Proline/betaine transporter; osmotic stress-induced gene | 3e−51 | M83089 |
| pA37F | Hypothetical protein | Streptomyces coelicolor | orf1419.5 (AF480912) | Conserved hypothetical; contiguous to cadA1 (Cd efflux family) | 8e−24 | AL109962 |
| pB1H | phoR | B. subtilis | shkA (AF480907) | Sensor kinase | 7e−20 | M23549 |
| pA39H | S. enterica serovar Typhimurium | plpA (AF480912) | Probable lipoprotein; contiguous to Cu efflux family (copA2) | 5e−04 | AE008711 | |
| pB47G | None | orf360 (AF480914) | Contiguous to Na+/Ca+ exchanger family (nceA) | NA | NA | |
| pA39C | Coxiella burnetti cryptic plasmid | orf505 (AF480913) | Probable membrane protein; contiguous to Mg2+/Co2+ transporter family (corA) and As efflux ortholog (lrsB) | 6e−12 | X75356 | |
| pA15B | CC329 | Caulobacter crescentus | orf853 (AF480917) | Zn2+ metalloprotein, M20/M25/M40 family | e−100 | AAK25201 |
| pB47A, pA35A | purA (wrong orientation) | E. coli | purA (AF480918) | Adenylosuccinate synthetase | e−160 | AP002568 |
| pA44H | fimV (wrong orientation) | P. aeruginosa | fimV (AF499441) | Pilus biosynthesis | 7e−36 | U93274 |
Genes shown in this column are the best characterized of the closely related genes identified by homology search. In some cases there are more closely related ORFs whose functions have not been characterized.
Expect values are from a filtered BLAST search. NA, not available.
Nucleotide sequence accession numbers.
All sequences have been submitted to GenBank under the accession numbers shown in Table 2.
RESULTS
Identification of promoters induced during growth within the macrophage.
The probe strategy to identify intracellularly induced promoters took advantage of the observation that thymine auxotrophs of L. pneumophila are unable to grow in macrophages (36) and that this auxotrophy can be complemented by the phage T4 tdΔi gene encoding thymidylate synthetase (7). Plasmid pSR37 (Fig, 1), a derivative of the broad-host-range plasmid pMMB34 (19), contains a promoterless copy of the tdΔi gene (14) followed by the gene encoding β-galactosidase (lacZ) to allow easy assay of gene expression levels. This plasmid was unable to complement the thymine auxotrophy of Lp02 either on laboratory medium (CYE) or during infection of monolayers of U937 cells, indicating that there was little or no plasmid-derived expression of the tdΔi gene in pSR37 (Fig. 2B). There was detectable basal β-galactosidase activity, however, as the colonies that arose on medium in the presence of thymidine were a pale blue color in the presence of the chromogenic substrate X-Gal.
FIG. 2.
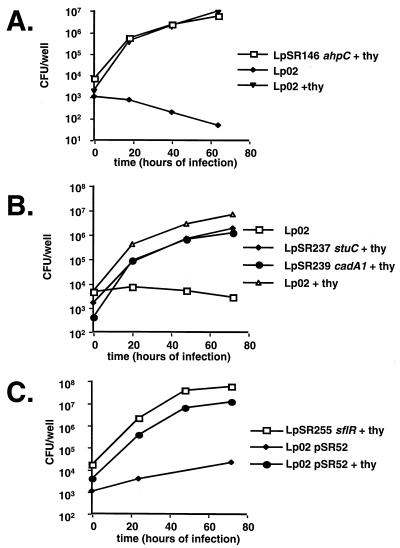
Growth of L. pneumophila in macrophages is not affected by inactivation of genes identified in the screen. Monolayers of U937 cells were infected with strains as indicated, and thymidine was added as noted. At various times after infection, representative monolayers were lysed, and the bacterial titers were determined. (A) Insertional inactivation of the ahpC gene on the L. pneumophila chromosome has no effect on the ability of the stain to grow in U937 cells. Strains tested were Lp02 (thyA) grown in the absence of thymidine (diamonds), Lp02 grown in the presence of thymidine (triangles), and LpSR146 (thyAahpC::kan) grown in the presence of thymidine (squares). (B) Inactivation of stuC or cadA1 has no effect on intracellular growth. Strains tested were Lp02 (thyA) grown in the absence of thymidine (squares), Lp02 grown in the presence of thymidine (triangles), LpSR237 (thyA stuC) grown in the presence of thymidine (diamonds), and LpSR239 (thyA cadA1) grown in the presence of thymidine (circles). (C) Inactivation of sflR has no effect on intracellular growth. Strains tested were Lp02 (thyA)/pSR52 grown in the absence of thymidine (diamonds), Lp02/pSR52 grown in the presence of thymidine (circles), and LpSR255 (thyA sflR) grown in the presence of thymidine (squares). Two independent isolates of each new strain were tested in duplicate.
A library of chromosomal fragments was cloned into the unique restriction site upstream of the tdΔi gene, and the resulting collection of plasmids was introduced into the L. pneumophila Thy− strain Lp02. Transformants were then screened for the presence of promoters that allowed both complementation of the Thy− defect within cultured cells and retention of the Thy− phenotype on laboratory medium. Two selection strategies were used on each of 48 pools of transformants, as described below and illustrated in Fig. 1.
In selection A, monolayers of U937 cells were infected directly with pools of transformants. Strains recovered from the monolayers after a 2-day incubation were plated, and colonies that arose were overlaid with β-galactosidase indicator medium. Individual colonies with low levels of β-galactosidase activity, based on having no color or faint blue color, were screened for the ability to form plaques on monolayers of U937 cells and for a strict Thy− phenotype on CYE plates. In the second selection strategy (selection B), 48 pools of transformants were plated first on trimethoprim (10 μg/ml), which inhibits the growth of Thy+ strains. Following the trimethoprim selection, survivors were used to infect monolayers of U937 cells. After 2 days, the U937 cells were lysed, and the surviving bacteria were plated nonselectively. Eight individuals surviving each enrichment procedure were isolated from each pool for further analysis. Each of the 768 strains identified was then tested individually for the ability to form plaques on monolayers of U937 cells and to confirm a strict thymine requirement during extracellular growth (7). Finally, a total of 96 individuals (one from each of 48 pools by using both strategies) were chosen for more detailed characterization. Plasmids isolated through selection A and selection B have been assigned names beginning with pA and pB, respectively.
The inserts were amplified by PCR. Dot blot DNA hybridization combined with restriction fragment analysis indicated that several of the inserts had been isolated multiple times independently, in some cases by using different selection strategies (data not shown). Plasmids from these strains were isolated and introduced into fresh background strains to confirm that the phenotype was plasmid dependent.
The inserts from a number of plasmids were partially or completely sequenced, including all those that were isolated multiple times from independent selections. Several of the plasmids contained partial or complete ORFs with similarities to previously identified genes from a number of organisms. The results are summarized in Table 2 for all clones in which there was sufficient sequence information from the inserts and from the unfinished L. pneumophila genome sequencing project (Columbia University Genome Center) to identify the gene regulated by the cloned sequence. With the exception of one gene, which had been identified by a signature-tagged mutagenesis approach (stuC) (41), all of the L. pneumophila genes are newly described here.
ORFs with similarities to a heavy metal transporter, redox proteins, a member of the response regulator class of signal transduction proteins, a sensor histidine kinase, an ortholog of the E. coli adenylate cyclase, and an ORF that encodes a membrane transporter or efflux-mediated resistance determinant were among the genes identified in the screen. Also identified was the L. pneumophila ortholog of ahpC, a gene family which has recently been implicated in resistance to both reactive oxygen intermediates (44) and reactive nitrogen intermediates (12). One of the plasmids contained a gene (obg) encoding a GTP-binding protein found in a number of organisms (38, 49, 60). In Bacillus subtilis, the Obg protein appears to regulate the master sigma factor that controls stress-dependent transcription (49). Two of the inserts contained genes (the apparent purA gene and a gene highly similar to Pseudomonas aeruginosa fimV) in the wrong orientation relative to the direction of transcription of td and will not be discussed further (Table 2).
To rule out the possibility that increased copy number of insert sequences conferred on the strains bearing the selected plasmids the ability to grow in the intracellular environment, a frameshift mutation was introduced into the tdΔi gene of several of the plasmids. Plasmids pB13F, pB47A, pB30G, and pA41B were digested at a restriction site within the tdΔi gene. Fill-in and religation of these plasmids resulted in a frameshift mutation in and subsequent inactivation of the tdΔi gene. In all cases, this modification resulted in the loss of ability to complement the Thy− phenotype intracellularly (data not shown). This confirms that expression of the tdΔi gene per se confers on the selected strains the ability to grow in cultured cells and that growth is not due to a cryptic thymine prototrophy conferred by sequences within the inserts.
Genomic analysis of the identified clones.
There were four noteworthy features of the sequences identified by these procedures. First, several of the genes are associated with protecting against conditions of stress in other organisms. Second, several of the inserts regulate ORFs that were orthologs of detoxification efflux systems or were closely linked to such ORFs on the genome. Third, fusions were identified that were physically linked to each other in the genome. Finally, one of the clones contained a presumed regulatory intragenic region that is repeated in several places in the L. pneumophila genome.
(i) Stress response loci.
In addition to the role of obg in regulating the general stress response in B. subtilis, several of the identified loci have been shown in other organisms to be important for protecting against stress insults or regulated in response to stress (Table 2). These genes include ahpC, which is a stress response gene in Salmonella enterica serovar Typhimurium and Mycobacterium tuberculosis; efflux transporters (sflR and smlA), which are expressed in response to a variety of stress conditions in B. subtilis; and tphB, which is similar to genes encoding proline/glycine betaine transport systems (40). Glycine/betaine uptake has been demonstrated to play a central role in protecting against hyperosmotic stress in a number of bacterial species (15). L. pneumophila has at least two paralogs of this uptake system (50).
(ii) Linkage to efflux system.
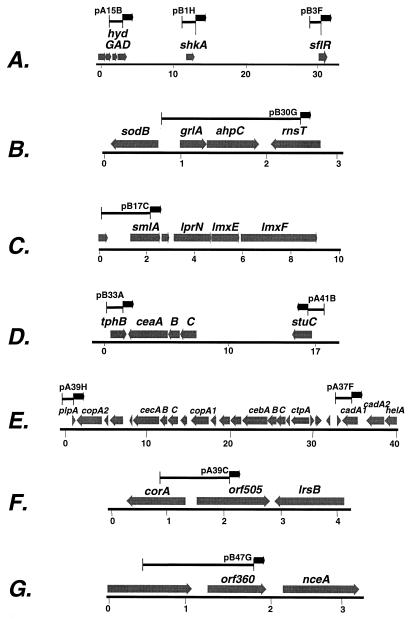
Two of the 14 identified sequences regulated ORFs (sflR and smlA [Table 2]) similar to others encoding solute efflux systems. sflR is an ortholog of a variety of antibiotic efflux systems (Table 2), whereas smlA is similar to nitrite efflux pumps and to milA, a gene previously identified as being required for maximal growth of L. pneumophila in macrophages (21). Five of the fusions were located contiguous to ORFs encoding orthologs of chemiosmotic efflux systems involved in heavy metal or organic compound detoxification (Fig. 3C, D, and E; called cea, ceb, and cec for chemiosmotic efflux system a, etc.; helA; and lprNlmxEF). Two of these plasmids contained DNA located in a 40-kb genomic island that appears primarily dedicated to encoding detoxification efflux systems (Fig. 3E) (see Discussion). Finally, two fusions are contiguous to other proteins involved in metal transport. One was located between an ortholog of a Co2+/Mg2+ transporter and an arsenite efflux channel (Fig. 3F, orf505), while the other was directly upstream of a Ca2+/Na+ exchanger family member (Fig. 3G, orf360).
FIG. 3.
Positions of inserts in clones that survived the enrichment relative to nearby loci. Each map displays contiguous DNA sequences from the unfinished L. pneumophila genome sequence, as determined by the Columbia University Genome Center (http://genome3.cpmc.columbia.edu/≈legion/index.html). Numbers refer to distances (in kilobases). Gray arrows show the predicted ORFs and the directions of translation. Black bars are the regions of the genome cloned in each plasmid that survived the selections. Black arrows refer to direction of translation of the tdΔi gene in the fusions. Numbers above the bars refer to plasmids harboring fusions. Gene names are based on similarities to database entries. (A) Three fusions are fused to ORFs on a 30-kb contiguous region of the genome. Displayed are hydGAD (similar to cytochrome c hydrogenases with identical operon structure), shkA (sensor histidine kinase ortholog), and sflR (similar to floR efflux pump). Other ORFs in this region are not displayed. (B) A genomic cassette encoding factors that protect against reactive nitrogen intermediate and reactive oxygen intermediate killing mechanisms. The sodB gene, encoding the Fe-dependent superoxide dismutase (48), and ahpC are divergently transcribed. (C) The smlA gene is directly upstream of a region encoding a three-component chemiosmotic efflux system. The lprNlmxElmxF cassette shows highest similarity to systems involved in efflux of antibiotics, such as the P. aeruginosa oprNmexEF operon (GenBank accession no. X99514) (25). (D) The tphB gene (similar to proline/glycine betaine transporter proP) and stuC (response regulator) are linked to ORFs encoding a putative chemiosmotic efflux system (ceaCBA; P. aeruginosa accession no. Y14018). Other ORFs in this region are not displayed. (E) Two macrophage-expressed fusions are derived from a genomic island encoding efflux proteins involved in metal homeostasis. Two small, conserved hypothetical ORFs found in several microbial genomes are linked to helA, previously shown to have enhanced expression after incubation with macrophages (32), and are within a 40-kb region encoding a large number of efflux systems. Shown are cadA1 and cadA2 (similar to Pseudomonas putida cadA, cadmium efflux ATPase; accession no. AF333961); helA, cebCBA, and cecCBA (chemiosmotic efflux systems); ctpA (similar to human Ca+-exporting ATPases; accession no. AF181120); and copA1 and copA2 (similar to copA, Cu efflux ATPase; E. coli accession no. Q47452). (F) A macrophage-expressed gene is divergently transcribed from an ORF encoding a presumed Co2−/Mg2− transporter (corA). Shown is orf505 located contiguous to corA (similar to S. enterica serovar Typhimurium accession no. AF250878) and lrsB, an ortholog of the membrane component of arsenite efflux, arsB (Sulfolobus solfataricus: accession no. AE006653). (G) A macrophage-regulated sequence is directly upstream of an ortholog of an Na+/Ca2+ exchanger (nceA; E. coli accession no. U18997).
(iii) Linkage of the cloned sequences to each other.
Three of the inserts had ORFs that were within 30 kb of each other (Fig. 3A, hydD, shkA, and sflR, ORFs most closely related to cytochrome c hydrogenase, sensor histidine kinases, and antibiotic efflux pumps, respectively). Two other pairs of inserts were also closely linked to each other (Fig. 3D, tphB and stuC; Fig. 3E, plpA and orf1419.5). One of these latter pairs of fusions was found to be tightly linked to the helA gene (Fig. 3E), which had been previously demonstrated to be induced during intracellular growth of L. pneumophila (32).
(iv) Potential regulatory site upstream of obgL.
The obgL regulatory elements contained in plasmid pA17A (Table 2) include an intragenic sequence found at least three other times in the genome. Two of the nucleotide blocks showed identity over a region of almost 70 bases (Fig. 4) and were located either upstream of an operon that encodes subunits of the flagellar apparatus (fliM; Fig. 4) (28) or upstream of an ORF similar to tehB, which encodes tellurite resistance (58). It is noteworthy that genes similar to those encoding tellurite resistance proteins are induced in B. subtilis in response to stress conditions (40), and as stated above, several macrophage-induced genes are in regions of the chromosome rich in genes encoding detoxification systems. The final patch, which is remarkable for its identity over a 150-nucleotide region, is upstream of an ORF apparently unique to L. pneumophila (Fig. 4, Lpn 001794).
FIG. 4.
Intragenic region upstream of obgL is repeated on the L. pneumophila chromosome. Displayed are four nucleotide sequences of extensive identity found in intragenic regions. Each sequence is found directly upstream of the ORFs noted. ORF numbers refer to designations given by the Columbia University Genome Center (http://genome3.cpmc.columbia.edu/≈legion/index.html) at the time of submission. Where a gene name is given, the ORFs correspond to the genes with closest similarities. fliM, flagellin biosynthesis; tehB, similar to tellurite resistance locus.
Several of the genes identified are not required for intracellular growth.
As the genes in this study were identified based on their expression during intracellular growth of the bacterium, it was not known whether intracellular growth of the bacterium required expression of these genes. In fact, the above analysis suggested that there may be considerable functional redundancy among the factors identified. To test this possibility, several of the genes were inactivated at their chromosomal loci, and the resulting strains were tested for the ability to grow in monolayers of phorbol ester-treated U937 cells. The L. pneumophila gene with homology to ahpC was inactivated at its chromosomal locus by insertion of a drug resistance marker, generating strain LpSR146.
Inactivation of the ahpC gene had no effect on the ability of the strain to grow in cultured cells (Fig. 2A). In addition, the L. pneumophila genes with similarity to antibiotic efflux systems (sflR; Fig. 3A), to Cd2+ efflux systems (cadA1; Fig. 3E), and to response regulators (stuC; Fig. 3D) were inactivated at their chromosomal loci by insertion of a nonreplicating plasmid containing internal gene fragments. The resulting strains were able to grow in U937 cells as well as the parental strain, Lp02 (Fig. 2B and 2C). Identical results were obtained when mouse bone marrow-derived macrophages were challenged with the mutants (data not shown; technique described in reference 56). Previously, inactivation of stuC had been reported to have no effect on intracellular growth within U937 cells or in human monocytes, although it was found to depress growth within amoebae (41). We found that this mutant grew with normal kinetics in the amoeba Dictyostelium discoideum (data not shown; technique described in reference 54), so the previously reported growth defect may be dependent on the strain of amoeba used as the host.
As predicted, we were unable to inactivate the gene encoding obgL (data not shown), an ORF that is highly similar to the essential obg genes in other organisms (30, 38, 63).
The presence of the lacZ gene in the vector allowed measurement of β-galactosidase expression from the inserts. In order to measure expression levels during intracellular growth, monolayers were infected with individual strains, and the levels of β-galactosidase activity were measured at several time points following U937 cell infection. For several of the fusions, it was difficult to detect an increase in lacZ expression during intracellular growth, possibly due to the presence of a promoter internal to the tdΔi gene in the vector that masked increased expression from upstream regulatory signals (Fig. 5). Therefore, to determine the source of transcription that allows intracellular growth of the identified strains in the absence of thymidine, we undertook a detailed analysis of one of the clones which showed a particularly complex structure.
FIG. 5.
Transcriptional analysis of gene regulation by the insert in plasmids pB30G and pA21E. Derivatives of the plasmid, as drawn in the chart, were constructed and analyzed for β-galactosidase activity and tdΔi RNA levels in extracellular medium, as well as for the ability to grow in U937 cells or on plates without thymidine supplementation. The data indicate that transcriptional activity contained upstream of ahpC, as well as a negative regulatory element downstream of the gene, ensured that strains carrying the original plasmids would survive the selection. RNA isolation was performed and β-galactosidase activities (in Miller units) were determined in mid-logarithmic phase on bacteria grown in AYE medium. ++, plaque size of L. pneumophila strains in the absence of thymidine on U937 cell lawns that is indistinguishable from that of the LP02 control in the presence of thymidine; +, plaque size in the absence of thymidine is smaller than that of the LP02 control grown on U937 cell lawns in the presence of thymidine; −, no ability to plaque on U937 lawns. ND, not determined; t, transcription terminator.
Analysis of tdΔi regulation by the pB30G insert.
The clone pB30G encoding ahpC, which was isolated by both selection strategies (Fig. 3B) (GenBank accession number L46863), was chosen for further analysis because it contained several ORFs. There are genes found in both orientations within the insert, and we wanted to understand which allowed growth in an intracellular environment in the absence of thymine. Furthermore, genes in the ahpC family have been implicated as being important for survival in the presence of a variety of stress conditions that pathogens may face within macrophages (10, 12). The insert in this plasmid contains two ORFs oriented in the same direction as tdΔi. Upstream and in the same orientation as ahpC is an ORF predicted to encode a protein of high sequence similarity to several glutaredoxins and thioredoxins (grlA) (43, 45). Downstream of ahpC, extending from the tdΔi fusion joint in the opposite orientation, is a third partial ORF encoding RNase T (Fig. 3B, rnsT). This ORF is missing its 5′ end as well as any upstream regulatory elements.
Analysis of fragments harboring these ORFs indicated that a promoter upstream of ahpC controlled expression of tdΔi, and a transcription terminator between ahpC and rnsT was required for the clone to exhibit regulated complementation of the Thy− auxotrophy. Portions of the insert were deleted or inverted in vitro, and strains carrying these constructs were tested for growth on bacteriological medium and in U937 cells cultured in the absence of thymidine. Consistent with the selection strategy, strains carrying the full-length insert (pB30G) grew in macrophages, whereas thymine supplementation was required during extracellular growth (Fig. 5). Interestingly, reconstitution of this exact phenotype required the full-length insert. For instance, direct fusion of ahpC to tdΔi (as in pB30G-Δ3′, Fig. 5) resulted in a Thy+ phenotype both during intracellular growth and on extracellular medium. On the other hand, an insert containing only the sequence downstream of ahpC (pB30G-Δ5′) conferred no complementation of the Thy− phenotype under any condition tested (Fig. 5).
Therefore, a promoter upstream of ahpC is responsible for transcription of the tdΔi gene in pB30G. Furthermore, a transcription terminator must be present downstream of ahpC (denoted t, Fig. 5), since the presence of this additional DNA sequence is required for thymidine auxotrophy on bacteriological medium. Presumably, the full-length insert allows growth within U937 cells because the promoter upstream of ahpC is so highly activated intracellularly that the resulting transcript runs through the terminator into the tdΔi gene. The ability of strains harboring the pB30G clone to survive the original selection on trimethroprim plates, on the other hand, was probably dependent on the presence of this terminator, because direct fusion of ahpC to tdΔi gave sufficiently high levels of expression on bacteriological medium to confer a Thy+ phenotype.
Transcriptional fusion with ahpC is regulated by association with animal cells.
The analyses of β-galactosidase and mRNA levels were consistent with both the complementation data and the presence of a transcription terminator in the parental clone (Fig. 5). Direct fusion of ahpC to tdΔi-lacZ (pB30G-Δ3′) resulted in β-galactosidase levels in bacteriological medium that were 10-fold higher than that observed for the original plasmid having the putative transcription terminator between ahpC and tdΔi-lacZ (pB30G). Furthermore, strains carrying the ahpC gene directly adjacent to the tdΔi gene (plasmid pB30G-Δ3′) showed high tdΔi RNA levels relative to strains carrying the originally isolated plasmid, consistent with the Thy+ phenotype of this deletion derivative on laboratory medium (Fig. 5).
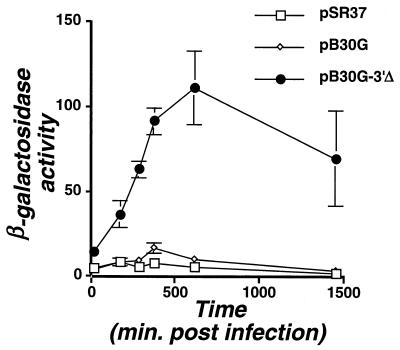
To test the model that a promoter upstream of ahpC was upregulated during intracellular growth, the strain harboring a direct fusion of ahpC to tdΔi-lacZ (pB30G-Δ3′; Fig. 5) was analyzed. For this fusion, the level of expression increased approximately sevenfold during growth in cultured cells relative to bacteriological medium (Fig. 6), with expression rising within 2 to 4 h after infection and peaking at approximately 10 h. This contrasted with a strain harboring the parental p30G clone, which showed clear upregulation of tdΔi in response to intracellular growth (Table 2), but showed weak induction of the lacZ fusion after introduction of the bacteria into U937 cells (Fig. 6). Apparently, the presence of the tdΔi gene upstream of lacZ interfered with the ability to detect increased expression of such a weak transcript. Control strains with inserts in the opposite orientation relative to lacZ also showed no increased expression in the intracellular environment. Therefore, only the fragment placed in the proper orientation, with the reporter fused within the coding region of ahpC, showed activated expression in response to interaction with cultured cells, verifying that the selection procedure allowed identification of intracellularly activated promoters.
FIG. 6.
Expression of L. pneumophila ahpC is increased during intracellular growth. Monolayers of U937 cells were infected with strains carrying pB30G or derivatives thereof in medium containing thymidine. At various times after infection, representative monolayers were lysed and assayed for β-galactosidase levels. Strains assayed were Lp02/pSR37 (no insert; squares); Lp02/pB30G (intact insert containing promoter and terminator; diamonds); and Lp02/pB30G-3′Δ (insert lacking terminator; circles). Note that units reported are 108 ΔA420/CFU/min, where A420 is corrected for the level of ONPG hydrolysis by an uninfected monolayer. This contrasts with Fig. 5, in which activity is measured as Miller units (33).
DISCUSSION
There is ample evidence that pathogenic microorganisms express a unique complement of genes when they interact with host tissues. A strategy to identify L. pneumophila genes induced in response to intimate contact with host cells was designed based on complementation of an auxotrophy that abolishes the ability of L. pneumophila to grow in the intracellular environment. This procedure allowed the identification of promoters that were expressed in response to growth in the intracellular environment. By using this selection, we have identified several novel genes and described the regulation of one of these clones in detail. No genes previously identified as being required for growth within mammalian macrophages were isolated by using the strategies described here. This result may have been predicted, because the mutations that cause the greatest defects in intracellular growth are in the dot and icm genes, all of which are expressed during growth in bacteriological medium (65). Other genes required for intracellular growth may be similarly expressed prior to contact with host cells.
The largest class of genes identified here consisted of a number of putative efflux-mediated resistance determinants or ORFs closely linked to efflux systems. The substrates for these systems in other species include heavy metals, antibiotics, and toxic organic compounds. Although we have not identified the substrates for these efflux pumps, the large number of these systems found in the genome suggests that there is an extremely broad range of small toxic substrates that L. pneumophila can effectively expel or that there exists considerable functional redundancy among the various efflux systems. Consistent with the latter proposition, an insertion mutation in one of multiple orthologs of the Staphylococcus aureus cadA gene, which is an ATP-dependent Cd2+ efflux pump, has no effect on intracellular growth (Fig. 2B). Similarly, previous work demonstrated that the helCBA locus, another ortholog of chemiosmotic metal efflux systems (Fig. 3E), could be mutated with no striking effects on either resistance to heavy metals or intracellular growth (32).
The close proximity of the A37F and A39H sequences to the hel locus, which is also expressed at increased levels in response to intracellular conditions (32), suggests that this region of the genome contains several genes that are transcriptionally activated by contact with host cells. This region of the genome is remarkable for its abundance of efflux pumps (Fig. 3E). Located nearby are three chemiosmotic efflux systems (helCBA, ceaCBA, and cebCBA), and five ORFs similar to ATP-dependent Cd2+, Cu2+, and Ca2+ efflux pumps (cadA1, cadA2, copA1, copA2, and ctpA). This is almost certainly a genomic island, presumably derived from an extrachromosomal element that is dedicated to detoxification and maintenance of proper metal ion balance within the bacterial cell. The importance of efflux in intracellular growth is further confirmed by the fact that mutations in the L. pneumophila milA gene, whose product may also be involved in ion extrusion, cause defects in intracellular growth (21; S. Vanrheenen and R. Isberg, data not shown). milA has several paralogs in the genome, including one identified by using the selections described here (smlA, Table 2).
The apparent importance of these systems is surprising, given that there are no well-characterized antimicrobial tactics within phagocytic cells that bathe bacteria in the small-molecule substrates of these efflux systems. In fact, the best-characterized relevant antimicrobial system elicited by macrophages appears to be that mediated by the NRAMP-1 protein, which removes divalent cations from the lumen of the phagosome (24) and presumably depletes the phagosome of metal ions. Perhaps in response to intraphagosomal conditions that cause cation starvation, scavenging transporters that sequester potentially toxic metal ions are activated in L. pneumophila, causing an overabundance of metal ions within the bacterial cytoplasm. To counteract this imbalance, efflux systems may be activated to maintain metal ion homeostasis.
A second possibility is that reactive oxygen and nitrogen intermediates or their detoxified products may be funneled out of the cell by these efflux systems. For instance, the closest characterized ortholog of smlA is narK (Table 2), which promotes nitrite efflux (46). Nitrite can be generated by bacterial metabolism of reactive nitrogen intermediates and is toxic under aerobic growth conditions (55). Efflux of this and similar solutes may be required for successful intraphagosomal replication.
We have shown that the expression of the ahpC gene in L. pneumophila is increased at least sevenfold during association of the bacterium with cultured cells. The increased expression of ahpC by L. pneumophila during infection may occur in response to the oxidative burst produced by macrophages during phagocytosis or due to continued exposure to reactive nitrogen intermediates during the growth cycle of the bacterium. ahpC genes from other species have been shown to confer protection from oxidative stress as part of a global response to an oxidizing environment (57). The ability of strains in which the ahpC gene is inactivated to grow in the intracellular environment indicates that this gene may be part of a redundant system for the prevention and repair of oxidative damage. In fact, in organisms as diverse as E. coli and mammals, there are multiple genes encoding members of this family. In addition, overexpression of ahpC increases the survival rate of M. tuberculosis strains that have lesions in the gene encoding catalase (53). These observations support the notion of redundant systems in the detoxification of reactive intermediates.
Recent work has demonstrated that the AhpC protein confers resistance to reactive nitrogen intermediates in both eukaryotic and prokaryotic cells (12) and that the protein has a peroxynitrite reductase activity that confers this resistance (10). Reactive nitrogen intermediates are generated in a number of cellular processes. Most notably, the inducible form of nitric oxide synthase is used to generate second messengers (nitric oxide) and to combat microbial infection in lymphoid cells (9). Nitric oxide produced by lung macrophages has been shown to help prevent mycobacterial replication in vitro (11), and it appears to act together with oxidative mechanisms to limit microbial replication (59).
It is interesting that the genes in the L. pneumophila ahpC region are dedicated to protecting against such dual assaults encountered within the host. The gene for the essential Fe-dependent superoxide dismutase (sodB) (48) and the grlA and ahpC genes are divergently transcribed in a compact cassette (Fig. 3B). In this small region of the genome, L. pneumophila appears to coordinate protection against both reactive oxygen and reactive nitrogen intermediate-dependent killing mechanisms and promotes maintenance of reducing potential within the bacterium. This is particularly intriguing in light of the proposed synergy of reactive oxygen intermediates and reactive nitrogen intermediates in protecting against the replication of pathogens within mice (59).
In the system described in this report, the basal expression of the tdΔi gene must be extremely low for the strains to survive the initial negative selection. Even very low levels of expression of the tdΔi gene resulted in a Thy+ phenotype, which would be lethal during selection on trimethoprim in bacteriologic medium. In contrast, most known stress response genes, such as those protecting against reactive oxygen intermediates, are expressed at significant levels in the absence of the inducing stress. It is possible that the negative selection used in the experiments presented here selected against the majority of L. pneumophila genes with increased expression in the macrophage intracellular environment.
As we have shown, strains harboring a direct fusion of the ahpC promoter to the tdΔi gene would not have survived the selection, since the basal expression of ahpC was sufficient to result in a Thy+ phenotype during growth on laboratory medium. The rigorous negative selection of this and similar strategies enriches most strongly for inserts that are transcriptionally silent in an extracellular environment. The ahpC gene-containing insert survived the selection because, in addition to a promoter, the fragment contained a terminator downstream from the ahpC gene. Transcription was attenuated sufficiently in this construction to allow survival on trimethoprim, but there was also a significant increase in expression in the presence of the cultured cells, allowing transcription to read through this element into the tdΔi gene. Perhaps inclusion of a terminator in the promoter-probe vector might allow a less stringent selection to be performed, allowing isolation of genes that are expressed at significant levels under noninducing conditions.
The selection strategy described here allowed identification of promoters induced under specific conditions, with the great advantage that the biochemical details of the inducing environment were not known. Furthermore, because this strategy required neither the introduction of chromosomal mutations nor impairment of intracellular growth, genes encoding functionally redundant factors could be identified. In fact, functional redundancy may be a common feature of proteins that are involved in supporting the pathogenesis of L. pneumophila, since the mutations that have the most drastic effects on intracellular growth appear to be limited to those located in the dot and icm genes (6, 42, 47). Further analysis of the loci identified here and the use of strains having multiple lesions in related genes should allow detailed investigation of proteins necessary to support the lifestyle of L. pneumophila during its stay within the macrophage.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (HHMI), training grant 5T32GM07310 from the NIGMS, and Program Project Award grant P30DK34928 from the NIDDK. R.R.I is an investigator of HHMI.
We thank Guillaume Dumènil, Isabelle Derre, Zhao-Qing Luo, and Susan VanRheenen for review of the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Abu Kwaik, Y. 1998. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect. Immun. 66:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., L. Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. [DOI] [PubMed] [Google Scholar]
- 4.Abu Kwaik, Y., and L. L. Pederson. 1996. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol. Microbiol. 21:543-556. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 8.Bezanson, G., R. Fernandez, D. Haldane, S. Burbridge, and T. Marrie. 1994. Virulence of patient and water isolates of Legionella pneumophila in guinea pigs and mouse L929 cells varies with bacterial genotype. Can. J. Microbiol. 40:426-431. [DOI] [PubMed] [Google Scholar]
- 9.Bogdan, C., M. Rollinghoff, and A. Diefenbach. 2000. The role of nitric oxide in innate immunity. Immunol. Rev. 173:17-26. [DOI] [PubMed] [Google Scholar]
- 10.Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211-215. [DOI] [PubMed] [Google Scholar]
- 11.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., Q. W. Xie, and C. Nathan. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1:795-805. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Chu, F. K., G. F. Maley, F. Maley, and M. Belfort. 1984. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc. Natl. Acad. Sci. USA 81:3049-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culham, D. E., A. Lu, M. Jishage, K. A. Krogfelt, A. Ishihama, and J. M. Wood. 2001. The osmotic stress response and virulence in pyelonephritis isolates of Escherichia coli: contributions of RpoS, ProP, ProU and other systems. Microbiology 147:1657-1670. [DOI] [PubMed] [Google Scholar]
- 16.Davis, G. S., W. C. Winn, Jr., D. W. Gump, and H. N. Beaty. 1983. The kinetics of early inflammatory events during experimental pneumonia due to Legionella pneumophila in guinea pigs. J. Infect. Dis. 148:823-835. [DOI] [PubMed] [Google Scholar]
- 17.Del Sal, G., G. Manfioletti, and C. Schneider. 1989. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. BioTechniques 7:514-520. [PubMed] [Google Scholar]
- 18.Diehn, M., and D. A. Relman. 2001. Comparing functional genomic datasets: lessons from DNA microarray analyses of host-pathogen interactions. Curr. Opin. Microbiol. 4:95-101. [DOI] [PubMed] [Google Scholar]
- 19.Frey, J., M. Bagdasarian, D. Feiss, F. C. Franklin, and J. Deshusses. 1983. Stable cosmid vectors that enable the introduction of cloned fragments into a wide range of gram-negative bacteria. Gene 24:299-308. [DOI] [PubMed] [Google Scholar]
- 20.Gao, L. Y., O. S. Harb, and Y. A. Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harb, O. S., and Y. Abu Kwaik. 2000. Characterization of a macrophage-specific infectivity locus (milA) of Legionella pneumophila. Infect. Immun. 68:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, M. A., and S. Silverstein. 1980. Legionnaire's disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. van't Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabado, N., A. Jankowski, S. Dougaparsad, V. Picard, S. Grinstein, and P. Gros. 2000. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192:1237-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. H., and A. Camilli. 2000. Novel approaches to monitor bacterial gene expression in infected tissue and host. Curr. Opin. Microbiol. 3:97-101. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, R. K. Henderson, D. S. King, L. Huang, S. Kustu, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 29.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddock, J., A. Bhatt, M. Koch, and J. Skidmore. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 179:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marra, A., M. A. Horwitz, and H. A. Shuman. 1990. The HL-60 model for the interaction of human macrophages with the Legionnaires' disease bacterium. J. Immunol. 144:2738-2744. [PubMed] [Google Scholar]
- 32.McClain, M. S., M. C. Hurley, J. K. Brieland, and N. C. Engleberg. 1996. The Legionella pneumophila hel locus encodes intracellularly induced homologs of heavy-metal ion transporters of Alcaligenes spp. Infect. Immun. 64:1532-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 34.Miller, V. L., and J. J. Mekalanos. 1985. Genetic analysis of the cholera toxin-positive regulatory gene toxR. J. Bacteriol. 163:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mintz, C. S., J. X. Chen, and H. A. Shuman. 1988. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect. Immun. 56:1449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn 903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, S., and K. Ochi. 1998. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30:107-119. [DOI] [PubMed] [Google Scholar]
- 39.Pearlman, E., A. H. Jiwa, N. C. Engleberg, and B. I. Eisenstein. 1988. Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb. Pathog. 5:87-95. [DOI] [PubMed] [Google Scholar]
- 40.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polesky, A. H., J. T. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuven, N. B., E. V. Koonin, K. E. Rudd, and M. P. Deutscher. 1995. The gene for the longest known Escherichia coli protein is a member of helicase superfamily II. J. Bacteriol. 177:5393-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Manzaneque, M. T., J. Ros, E. Cabiscol, A. Sorribas, and E. Herrero. 1999. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8180-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe, J. J., T. Ubbink-Kok, D. Molenaar, W. N. Konings, and A. J. Driessen. 1994. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol. Microbiol. 12:579-586. [DOI] [PubMed] [Google Scholar]
- 47.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadosky, A. B., J. W. Wilson, H. M. Steinman, and H. A. Shuman. 1994. The iron superoxide dismutase of Legionella pneumophila is essential for viability. J. Bacteriol. 176:3790-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor sigma B. J. Bacteriol. 181:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shapira, S. K., J. Chou, F. V. Richaud, and M. J. Casadaban. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene 25:71-82. [DOI] [PubMed] [Google Scholar]
- 53.Sherman, D. R., K. Mdluli, M. J. Hickey, C. E. Barry III, and C. K. Stover. 1999. AhpC, oxidative stress and drug resistance in Mycobacterium tuberculosis. Biofactors 10:211-217. [DOI] [PubMed] [Google Scholar]
- 54.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tartaglia, L. A., G. Storz, and B. N. Ames. 1989. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210:709-719. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, D. E., Y. Hou, R. J. Turner, and J. H. Weiner. 1994. Location of a potassium tellurite resistance operon (tehA tehB) within the terminus of Escherichia coli K-12. J. Bacteriol. 176:2740-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ischiropoulos, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vidwans, S. J., K. Ireton, and A. D. Grossman. 1995. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:3308-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 62.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn 10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 63.Welsh, K. M., K. A. Trach, C. Folger, and J. A. Hoch. 1994. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 176:7161-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, Y., and R. R. Isberg. 1993. Cellular internalization in the absence of invasin expression is promoted by the Yersinia pseudotuberculosis yadA product. Infect. Immun. 61:3907-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]