Abstract
Salmonella enterica serotype Typhimurium requires a functional type III secretion system encoded by Salmonella pathogenicity island 1 (SPI1) to cause diarrhea. We investigated the role of genes encoding secreted target proteins of the SPI1-associated type III secretion system for enteropathogenicity in calves. Salmonella serotype Typhimurium strains having mutations in sptP, avrA, sspH1, or slrP induced fluid secretion in the bovine ligated ileal loop model at levels similar to that of the wild type. In contrast, mutations in sipA, sopA, sopB, sopD, or sopE2 significantly reduced fluid accumulation in bovine ligated ileal loops at 8 h postinfection. A strain carrying mutations in sipA, sopA, sopB, sopD, and sopE2 (sipA sopABDE2 mutant) caused the same level of fluid accumulation in bovine ligated ileal loops as a strain carrying a mutation in sipB, a SPI1 gene required for the translocation of effector proteins into host cells. A positive correlation was observed between the severity of histopathological lesions detected in the ileal mucosa and the levels of fluid accumulation induced by the different mutants. After oral infection of calves, the Salmonella serotype Typhimurium sipAsopABDE2 mutant caused only mild diarrhea and was more strongly attenuated than strains having only single mutations. These data demonstrate that SipA, SopA, SopB, SopD, and SopE2 are major virulence factors responsible for diarrhea during Salmonella serotype Typhimurium infection of calves.
Salmonella enterica serotype Typhimurium can infect a wide range of host species, including mice, cattle, and humans. In mice, Salmonella serotype Typhimurium causes a systemic disease which serves as an animal model to study typhoid fever, a disease caused by S. enterica serotype Typhi in humans. Murine typhoid is characterized by rapid bacterial multiplication in systemic organs and infiltration of mononuclear leukocytes in the intestinal mucosa which is not associated with diarrhea (36). Interestingly, mice exhibit a strikingly different response to Salmonella serotype Typhimurium infection than the human host. Salmonella serotype Typhimurium infection in humans commonly manifests as enterocolitis that is characterized by diarrhea and infiltration of polymorphonuclear leukocytes (PMN) in the intestinal mucosa (27, 41). The infection commonly remains localized in the intestine and mesenteric lymph nodes, while bacteremia is a rare complication (46). In addition to its frequent association with food-borne disease in humans, Salmonella serotype Typhimurium is a major cause of calf morbidity and mortality (14, 20, 33, 38, 40). The features of the diarrheal disease caused by Salmonella serotype Typhimurium in calves closely resembles the clinical and pathological features observed in humans infected with this pathogen (9, 39, 47, 56). Therefore, the calf is currently used as an animal model to study human enterocolitis caused by Salmonella serotype Typhimurium.
Mutations in genes located on Salmonella pathogenicity island 1 (SPI1), including prgH, hilA, and invH, result in strongly reduced virulence of Salmonella serotype Typhimurium during oral infection of calves (47, 52). PrgH and InvH are components of the invasion-associated type III secretion system whose expression is controlled by HilA (2, 3, 5). The main function of the invasion-associated type III secretion system is to translocate effector proteins into the cytosol of a host cell (11). In the first step, secreted target proteins are transported across inner and outer membranes of the bacterial cell by the type III secretion apparatus. A subset of these secreted proteins, including SipB (SspB), SipC (SspC), and SipD (SspD), form a translocation complex in the eukaryotic membrane, which is required for the delivery of other effector proteins into the cytoplasm (7, 10, 13, 16, 55). Nonpolar mutations in sipB, sipC, or sipD result in the same degree of attenuation of Salmonella serotype Typhimurium during oral infection of calves as deletion of genes (prgHIJK) encoding components of the invasion-associated type III secretion apparatus (48). Furthermore, inactivation of sipB in S. enterica serotype Dublin strongly reduces fluid secretion and inflammation measured at 12 h postinfection in bovine ligated ileal loops (13, 47).
A mutation in sipB may cause attenuation in calves, either because SipB directly engages targets in the host cell or because translocation of other effector proteins is prevented. The finding that SipB is an effector protein, which causes cell death in murine and bovine macrophages by binding to caspase 1, demonstrates that this protein can directly engage targets in the host cell (18, 34, 53). Furthermore, SipB-mediated macrophage cell death results in activation of the proinflammatory cytokine interleukin 1β (18), suggesting that this mechanism may contribute to eliciting an inflammatory response in the intestine. However, while inflammatory changes in bovine ligated ileal loops are observed as early as 1 h postinfection with Salmonella serotype Typhimurium, a significant increase in cell death detected by terminal deoxyribonucleotidyl transferase-dependent UTP nick end labeling in the bovine mucosa and lymphoid nodules is first observed 12 h postinfection (35). Thus, SipB-mediated macrophage cell death is unlikely to be required for triggering an early inflammatory response and fluid accumulation in the bovine ileum. These data raise the possibility that SipB-mediated translocation of effector proteins is the major virulence function of this protein during bovine enteritis pathogenesis.
In addition to proteins that act as translocases (SipBCD), SPI1 encodes three effector proteins, SipA (SspA), AvrA, and SptP (16, 19, 22-24). Furthermore, seven effector proteins, including SopA, SopB (SigD), SopD, SopE1, SopE2, SspH1, and SlrP, whose secretion is mediated by the SPI1 type III secretion system are encoded by genes which are located outside SPI1 (4, 13, 17, 21, 29, 43, 49, 54, 55). Much of the work on the role of SPI1 effector proteins during infection of calves has been performed with Salmonella serotype Dublin, a serotype that causes bacteremia rather than enterocolitis in humans (8). Inactivation of avrA does not reduce the ability of Salmonella serotype Dublin to induce fluid accumulation and PMN influx in bovine ligated ileal loops (37), suggesting that not all SPI1 effector proteins are required for enteropathogenicity in calves. The level of fluid secretion and inflammation elicited by a Salmonella serotype Dublin sopA, sopB, or sopD mutant is greater than that elicited by a Salmonella serotype Dublin sipB mutant. However, sopA, sopB, or sopD mutants cause significantly less fluid accumulation and PMN influx in bovine ligated ileal loops than the Salmonella serotype Dublin wild type causes (13, 21, 54). Furthermore, the enteropathogenicity of a Salmonella serotype Dublin sopB sopD double mutant is reduced compared to that of a sopB or sopD mutant, but it is still greater than that of a sipB mutant (21). These data suggest that several type III secreted effector proteins of Salmonella serotype Dublin are required for enteropathogenicity.
Although Salmonella serotype Dublin causes bacteremia rather than enterocolitis in humans (8), it likely employs virulence mechanisms to cause enterocolitis in calves that are similar to those used by Salmonella serotype Typhimurium. For instance, a mutation in sopB reduces the ability of Salmonella serotype Typhimurium to cause inflammation and fluid accumulation in bovine ligated ileal loops to a degree similar to that observed for Salmonella serotype Dublin (35). So far, the virulence of Salmonella serotype Dublin sopB, sopD, or sopA mutants has been characterized only in bovine ligated ileal loops. Oral infection studies with Salmonella serotype Typhimurium found that a sopB mutant appears to be fully virulent when calves are infected by this route (47). The lack of attenuation of a Salmonella serotype Typhimurium sopB mutant during oral infection is likely due to the fact that several type III secreted effector proteins act in concert during the pathogenesis of bovine enteritis. However, it is currently not known how many type III secreted effector proteins are involved in eliciting fluid secretion and inflammation in bovine ligated ileal loops. If the main role of SipB during infection is the translocation of effector proteins involved in enteropathogenicity (i.e., SopB and other proteins), it should be possible to reduce the virulence of Salmonella serotype Typhimurium to that of a sipB mutant by inactivating the genes encoding these effector proteins. To test this prediction, we determined which genes encoding targets for the SPI1 type III secretion system are required for fluid accumulation in bovine ligated ileal loops. Furthermore, a Salmonella serotype Typhimurium strain lacking all effector proteins involved in fluid secretion was constructed and characterized in ligated ileal loops and during oral infections of calves.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella serotype Typhimurium and Escherichia coli strains used in this study are listed in Table 1. Bacteria were cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar plates. If appropriate, antibiotics were added at the following concentrations: kanamycin, 100 mg/liter; chloramphenicol, 30 mg/liter; ampicillin, 100 mg/liter; nalidixic acid, 50 mg/liter; and tetracycline, 20 mg/liter.
TABLE 1.
Salmonella serotype Typhimurium strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Salmonella serotype Typhimurium strains | ||
| ATCC 14028 | ATCCb | |
| IR715 | ATCC 14028 wild type, nalr | 44 |
| CAS152 | ATCC 14028 phoN::Cm sipB | 49 |
| M119 | ATCC 14028 sopE::tetr | Mirold et al.c |
| JLR137 | ATCC 14028 phoN::Cm glpD::Tn5-lacZY | Miller laboratory |
| STN39 | ATCC 14028 nalrslrP::mini-Tn5Km2 | 49 |
| ZA9 | ATCC 14028 nalrsopE2::tetr | This study |
| ZA10 | ATCC 14028 nalr ΔsipA (Δ16-606/685) | This study |
| ZA11 | ATCC 14028 nalr ΔsptP (Δ30-478/544) | This study |
| ZA13 | ATCC 14028 nalr ΔavrA (Δ19-213/300) | This study |
| ZA14 | ATCC 14028 nalr ΔsspH1 (Δ53-354/702) | This study |
| BA1567 | ATCC 14028 sopB::mudJ | 1 |
| ZA15 | ATCC 14028 nalrsopB::mudJ | 15 |
| ZA16 | ATCC 14028 nalrsopBE2 | This study |
| ZA17 | ATCC 14028 nalrsopD | This study |
| ZA18 | ATCC 14028 nalrsopBED | This study |
| ZA19 | ATCC 14028 nalr ΔsopA (Δ38-734/783) | This study |
| ZA20 | ATCC 14028 nalr ΔsopABDE2 | This study |
| ZA21 | ATCC 14028 nalr ΔsipAΔsopAsopBDE2 | This study |
| ZA26 | ATCC 14028 nal ΔsipAsopB | This study |
| Plasmids | ||
| pCR2.1 | Ampr KanrlacZα | Invitrogen |
| pEP185.2 | Cmr | 25 |
| pRDH10 | CmrsacB | 26 |
| pWSK29 | Ampr | 51 |
| pZA10 | pRDH10 carrying the flanking regions of sipA | This study |
| pZA11 | pRDH10 carrying the flanking regions of sptP | This study |
| pZA13 | pRDH10 carrying the flanking regions of avrA | This study |
| pZA14 | pRDH10 carrying the flanking regions of sspH1 | This study |
| pZA17 | pEP185.2 carrying sopD fragment (bp 101 to 621) | This study |
| pZA19 | pRDH10 carrying the flanking region of sopA | This study |
| pSipA | pWSK29 carrying the sipA gene | This study |
The numbers in parentheses are the deleted codons/total number of codons of avrA, sipA, sopA, sptP, and sspH1.
ATCC, American Type Culture Collection.
Mirold et al., submitted.
Construction of mutants.
Mutant ZA9 (sopE2) was created by phage P22-mediated transduction by using Salmonella serotype Typhimurium strains M119 (S. Mirold, W Rabsch, H. Tschäpe, and W.-D. Hardt, submitted for publication) and IR715 (44) as the donor and the recipient, respectively. Inactivation of sopE2 in ZA9 was confirmed by Southern hybridization. Briefly, a DNA fragment containing nucleotides 3 to 702 of the sopE2 open reading frame was amplified by PCR by using primers SopE1 and SopE2 (Table 2), labeled with fluorescein (NEN life science product; NEN), and used as a probe to hybridize genomic DNA of ZA9 and IR715. Unmarked deletions of sipA, sptP, avrA, sspH1, and sopA (strains ZA10, ZA11, ZA13, ZA14, and ZA19, respectively) were constructed by allelic exchange. In brief, DNA regions upstream and downstream of the desired gene were amplified by PCR by using primers listed in Table 2. The primers were introduced into a restriction site (either XbaI or SacI) that was used to ligate the digested 3′ end of the upstream fragment to the digested 5′ end of the downstream fragment. The ligation product was reamplified by PCR and cloned into the vector pCR2.1 (Invitrogen). The insert was excised by restriction digestion with EcoRI and subcloned into the vector pRDH10 (26), a λpir-dependent suicide vector carrying sacB and a chloramphenicol resistance gene. The resulting plasmid was introduced into E. coli strain S17λ-pir and transferred into Salmonella serotype Typhimurium strain IR715 by conjugation. Exconjugants in which the suicide plasmid was integrated into the IR715 chromosome were selected on LB agar plates supplemented with chloramphenicol and nalidixic acid. A second recombination event, leading to the loss of plasmid sequences, was selected by growth in sucrose (5%) broth, followed by growth on sucrose agar plates at 30°C. Colonies that were sensitive to chloramphenicol were screened by PCR for deletion of the gene of interest. To confirm the unmarked deletions, Southern hybridization analyses were conducted. In brief, the primer pairs SipAF1 plus SipAB2, SptPF1 plus SptPB2, AvrAF1 plus AvrAB2, SspHF1 plus SspHB2, and SopAF1 plus SopAB2 were used to amplify sipA, sptP, avrA, sspH1, and sopA, respectively. The resulting PCR products were labeled with fluorescein (NEN) and used as probes to hybridize the digested genomic DNA of the corresponding mutant strains and a positive control (IR715). To generate ZA16 (sopBE2), sopE2::tetr was introduced from M119 into ZA15 (sopB) by phage P22-mediated transduction, and the mutation was confirmed by Southern hybridization. To create ZA17 (sopD), a DNA fragment containing nucleotides 101 to 621 of the sopD open reading frame was PCR amplified and cloned into plasmid pEP185.2, a suicide vector coding for chloramphenicol resistance (25). The resulting plasmid, pZA17, was introduced into E. coli strain S17λ-pir and transferred into strain IR715 by conjugation. Exconjugants containing pZA17 inserted into the chromosome of strain IR715 were selected by growth on LB agar plates supplemented with chloramphenicol and nalidixic acid. The sopD::pEP185.2 insertion was then introduced into ZA16 by P22-mediated transduction to generate ZA18 (sopBDE2). The sopD::pEP185.2 insertion in strains ZA17 and ZA18 was confirmed by Southern hybridization by using a probe containing nucleotides 101 to 621 of the sopD gene. The sopA and sipA genes were deleted sequentially from strain ZA18 to obtain strains ZA20 (sopABDE2) and ZA21 (sipAsopABDE2). The unmarked deletions and selection processes used for ZA20 and ZA21 were the same as those described above for single-knockout mutants. The mutations in the sipA, sopA, sopD, and sopE2 genes of ZA21 were confirmed by Southern hybridization as described above for each single mutant. In addition, the insertional mutation in sopB of ZA21 was also confirmed by Southern hybridization. The SopB probe was generated by PCR by using primers SopB1 and SopB2 (Table 2). To generate ZA22 (sipAsopB), sopB::mudJ was introduced from ZA15 (sopB) into ZA10 (sipA) by phage P22-mediated transduction and selection for kanamycin resistance. The mutation was confirmed by Southern hybridization. For complementation, the sipA gene was amplified by PCR by using primers z-SipA1 and z-SipA2. The PCR product was cloned into a low-copy-number vector, pWSK29 (51), to create plasmid pSipA. The orientation of the sipA gene in pSipA was determined by sequence analysis. To complement the defect of mutants with mutations in induction of secretory and inflammatory responses, plasmid pSipA was introduced into strains ZA21, ZA10, and ZA26 by electroporation and selection for resistance to ampicillin. The genetic organization of effector genes involved in fluid accumulation is shown in Fig. 1.
TABLE 2.
Primers used in this studya
| Primer | Sequence |
|---|---|
| AvrF1 | AGAAGGCGTTATCTACTTGC |
| AvrB1 | GCTCTAGAGGACTTAGCATACTTTTCCCTC |
| AvrF2 | GCTCTAGACGTCTGTGTGGTGAAGAAC |
| AvrB2 | TTAGCGGTGAGTCTGTCAG |
| SipAF1 | AGGCGGCTACTAAAATCC |
| SipAB1 | GCTCTAGATACCTGGCATTATGACGGG |
| SipAF2 | GCTCTAGAGGTCATTACTCATCATCC |
| SipAB2 | CAAGCGAGAGAAAAATACTACAC |
| SopAF1 | AAAGATGGCTGGAGAGCGAG |
| SopAB1 | GCGAGCTCTGGTTATTTTTGAGGTGAG |
| SopAF2 | GCGAGCTCGAACAGTTTACCGAGTGGTC |
| SopAB2 | TGAATGCGTCTGGCGAAAGC |
| SopB1 | CCCGTATTTGGTTCTGAATCTCC |
| SopB2 | AGCCTGAAACTGGTATCCGTGG |
| SopDF | GCGATATCTGGGGGGTTGGGATAAAGTC |
| SopDB | GCTCTAGATAAGCGAGTCCTGCCATTCG |
| SopE1 | GACTAACATAACACTATCCACC |
| SopE2 | TCAGGAGGCATTCTGAAGATAC |
| SptpF1 | GCGAAAAAGTATCAAGACATTG |
| SptpB1 | GCTCTAGATAAAGTCGGGCATCATTC |
| SptpF2 | GCTCTAGATTCATTGTCTGGGCGGAG |
| SptpB2 | ACGGTAAAATCTGAGAGAGG |
| SspHF1 | AGGCGTTGGGCGAATCTATC |
| SspHB1 | GCTCTAGATTCAGCGAGACACTGTTGC |
| SspHF2 | GCTCTAGACAACACTGGAACAGATTGC |
| SspHB2 | TACGCCCTGACTGAAGAAG |
| Z-SipA1 | GCGAGCTCACGACAACCTGGTAAAAG |
| Z-SipA2 | GCGAGCTCCTATCAACATCAACGGCA |
| LacZ1 | CTCTTCGCTATTACGCCA |
| LacZ2 | CATAAACCGACTACACAAA |
The restriction endonuclease sites incorporated into the primer sequences are underlined.
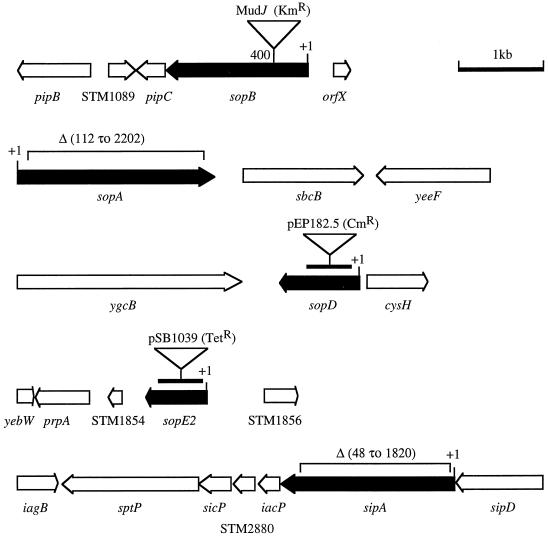
FIG. 1.
Mutations in effector genes (solid arrows) which are required for fluid accumulation in calves. The open arrows indicate the positions of genes in the surrounding DNA region. Numbers indicate the positions of insertions or deletions relative to the first nucleotide (+1) in the open reading frame. The bars indicate internal fragments of sopD and sopE2 that were cloned into suicide vectors to inactivate these genes.
Inverse PCR.
To determine the insertion site for transposon Tn5lacZYA in Salmonella serotype Typhimurium strain JLR137, transposon-flanking DNA was amplified by inverse PCR by using primers LacZ1 and LacZ2 (Table 2). Briefly, genomic DNA of JLR137 was digested with RsaI, and the fragments were ligated and used as templates for PCR amplification. The resulting PCR product was cloned into the vector pCR2.1 and sequenced.
Animal experiments.
Male Holstein calves that were 4 to 5 weeks old and weighed 45 to 55 kg were used. They were fed antibiotic-free milk replacer twice a day and given water ad libitum. Prior to experiments, the calves were screened for elevated total leukocyte count, rectal temperature, and fecal excretion of Salmonella serotypes. Salmonella serotypes in fecal swabs were detected by enrichment growth in tetrathionate broth (Difco) and then Rappaport-Vassiliadis R10 broth (Difco) and subsequent plating on brilliant green agar and XLT-4 agar (BBL) plates. Serum antibodies against Salmonella serotype Typhimurium were detected by an enzyme-linked immunosorbent assay by using wells coated with purified lipopolysaccharide from this serotype (Sigma).
For ligated ileal loop assays, calves were fasted for 48 h prior to surgery. Anesthesia was induced with Propofol (Propoflo; Abbot Laboratories, Chicago, Ill.) and maintained with Isofluorane (Isoflo; Abbot Laboratories) for the duration of the experiment (35). To prepare bacterial inocula, the desired bacterial strains were cultured in LB broth for 18 h at 37°C with shaking at 150 rpm. The resultant cultures were diluted 1:100 with sterile LB broth and incubated for additional 4 h. Bacteria in the logarithmic phase of growth were then harvested by centrifugation for 15 min at 1,500 × g and resuspended in fresh LB broth to obtain a final concentration of approximately 0.75 × 109 CFU/ml. After a laparotomy was performed and the ileum was exposed, ileal loops with lengths ranging from 6 to 9 cm were ligated with a 1-cm interposed loop between them. The loops were infected by intralumenal injection of 3 ml of a suspension of IR715 or a mutant strain in LB broth containing approximately 1 × 109 CFU of bacteria. The control loops received 3 ml of sterile LB broth. Following injection, the ileal loops were placed back into the abdominal cavity until the time for sample processing. At 8 h postinfection, the fluid accumulated in the loops was measured, and samples for bacteriologic culture and histopathological analysis were collected. Tissue samples from Peyer's patches were homogenized and serially diluted in phosphate-buffered saline and were plated on LB agar plates supplemented with appropriate antibiotics for enumeration of bacteria.
For oral infection, the optical densities at 600 nm of overnight cultures were determined in order to estimate the numbers of bacteria per milliliter. A volume containing 1 × 1010 CFU of bacteria was mixed with 50 ml of a suspension containing 5% magnesium trisilicate, 5% sodium bicarbonate, and 5% magnesium carbonate. Groups of four calves were infected with the following strains: IR715, ZA9, ZA17, ZA19, and ZA20. Rectal temperature and fecal score (for assessment of diarrhea) were recorded daily. Fecal shedding of Salmonella serotype Typhimurium was monitored by daily plating of fecal samples on brilliant green agar plates. Blood samples were taken prior to infection (zero time) and on days 1 and 2 postinfection. These time points were chosen because a previous study (48) demonstrated that calves inoculated orally with 1010 CFU of wild-type Salmonella serotype Typhimurium developed lethal signs of illness before 3 days postinfection. The concentrations of sodium in blood plasma samples were determined by the Texas Veterinary Medical Center, Texas A&M University. When the calves developed anorexia or were unable to stand, they were euthanized for humane reasons as described previously (23). At 10 days postinfection, the surviving calves were euthanized. At necropsy, tissue samples from livers, spleens, mesenteric lymph nodes, and Peyer's patches were collected, homogenized and serially diluted in phosphate-buffered saline, and plated on LB agar plates supplemented with appropriate antibiotics for enumeration of bacteria.
Histopathology.
The tissue samples were fixed in formalin, processed by using standard procedures for paraffin embedding, cut into 5-μm sections, and stained with hematoxylin and eosin. Inflammatory changes were scored by using a scale from 1 to 5 according to the following criteria: 1, intravascular infiltration of polymorphonuclear leukocytes (PMN); 2, margination and perivascular infiltration of PMN and/or mild diffuse infiltration of PMN at the tips of absorptive villi; 3, moderate diffuse infiltration of PMN in the mucosa and perivascular multifocal infiltration in the submucosa; 4, severe diffuse infiltration of PMN in the mucosa and mild to moderate infiltration in the submucosa; and 5, severe diffuse infiltration of PMN throughout the mucosa and submucosa associated with edema and necrosis of the mucosa.
Statistical analysis.
To compare the virulence characteristics of a particular mutant and wild-type strain IR715, data for fluid secretion (volume/length), bacterial colonization, and plasma concentration of electrolytes were analyzed by using Student's t test. To assess the effects of different mutations on secretory responses, fluid secretion data were expressed as percentages of the response elicited by wild-type strain IR715. Data then underwent arcsin transformation, and analysis of variance was performed. The averages were compared by using Duncan's multiple range test.
RESULTS
Evaluation of the role of SPI1 effector genes in eliciting secretory and inflammatory responses in bovine ligated ileal loops.
Previous studies have demonstrated that a mutation in sopA, sopB, or sopD of Salmonella serotype Dublin or in sopB of Salmonella serotype Typhimurium significantly reduces the ability to cause fluid accumulation in bovine ligated ileal loops (21, 35). To identify additional effector genes required for fluid accumulation in ligated ileal loops, Salmonella serotype Typhimurium strains having mutations in effector genes were constructed by using strain IR715, a spontaneous nalidixic acid-resistant derivative of bovine Salmonella serotype Typhimurium isolate ATCC 14028 (44). Derivatives of IR715 which have unmarked deletions of sopA (ZA20), sipA (ZA10), sptP (ZA11), avrA (ZA13), or sspH1 (ZA14) were generated by allelic exchange. A derivative of IR715 containing a transposon insertion in slrP (STN39) has been described previously and was also included in this study (49). A mutation in sopD was introduced into strain IR715 by integration of a suicide vector carrying an internal fragment of the sopD open reading frame (bp 101 to 621) into the chromosome by homologous recombination (ZA17). In addition, derivatives of ATCC 14028 and IR715 carrying a sopE2::tetr allele (M119 and ZA9, respectively) were tested in this study. The sopE1 gene was not included in our investigation, because it is not present in strain ATCC 14028.
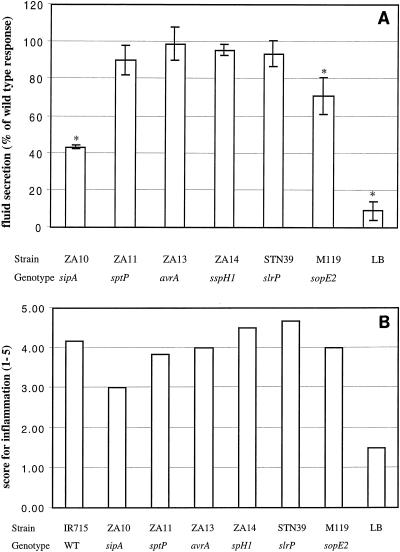
The virulence of these mutants with respect to secretory and inflammatory responses, as well as colonization of Peyer's patches, was assessed in bovine ligated ileal loops. An initial evaluation of these Salmonella serotype Typhimurium mutants demonstrated that mutations in sipA or sopE2, but not mutations in sptP, avrA, sspH1, or slrP, caused significant reductions in fluid secretion (P < 0.05) at 8 h postinfection (Fig. 2A). Inflammatory changes compared to uninfected control loops were scored by blinded examination of sections of the ileal mucosa. This analysis revealed that a mutation in sipA, but not a mutation in sopE2, sptP, avrA, sspH1, or slrP, caused a marked reduction in the severity of inflammation in the ileal mucosa (Fig. 2B). The secretory and inflammatory responses induced by ATCC 14028 were similar to those induced by its nalidixic acid-resistant derivative, IR715 (data not shown). No significant differences between loops infected with the wild type (IR715) and loops infected with the mutants described above were observed with regard to the numbers of bacteria recovered from Peyer's patches (data not shown). We next investigated the role of the effector proteins, which have previously been implicated in fluid accumulation caused by Salmonella serotype Dublin, in triggering fluid accumulation during Salmonella serotype Typhimurium infection. Compared to the Salmonella serotype Typhimurium wild type (IR715), strains having mutations in sopA, sopB, or sopD caused significantly less fluid accumulation in bovine ligated ileal loops (P < 0.05) (Fig. 3A). Strains IR715 (wild type), ZA19 (sopA), and ZA9 (sopD) induced moderate to severe diffuse PMN infiltration in the ileal mucosa and submucosa. The severity of inflammation was only slightly reduced in loops infected with ZA15 (sopB) (Fig. 3B). Collectively, these data suggested that five SPI1 secreted effector proteins (SopA, SopB, SopD, SopE2, and SipA) are required for fluid accumulation in ligated ileal loops. The genes encoding these effector proteins and mutations used in this study to inactivate the genes are shown in Fig. 1. Our data further suggest that four effector proteins (SptP, AvrA, SspH1, and SlrP) play no apparent role in eliciting fluid accumulation in bovine ligated ileal loops.
FIG. 2.
Ability of Salmonella serotype Typhimurium strains carrying mutations in SPI1 effector genes to induce secretory and inflammatory changes in bovine ligated ileal loops at 8 h postinfection. (A) Data for fluid accumulation in loops shown as percentages of the fluid secretion elicited by the wild type (ATCC 14028 for M119 and IR715 for ZA10, ZA11, ZA13, ZA14, and STN39). The 100% line indicates the amount of fluid accumulation elicited by the Salmonella serotype Typhimurium wild type. An asterisks indicates that the fluid accumulation was significantly lower than the wild-type response (P < 0.05). (B) Inflammatory responses scored on a scale from 1 to 5 according the criteria described in Materials and Methods. WT, wild type.
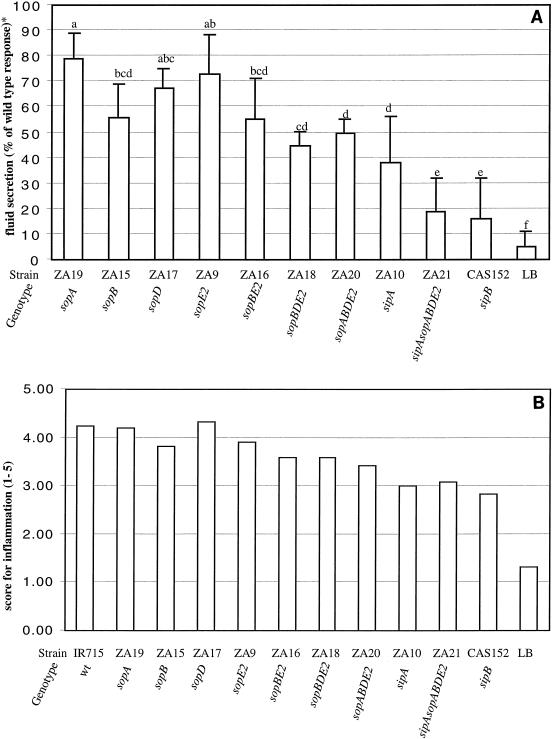
FIG. 3.
Ability of Salmonella serotype Typhimurium mutants carrying multiple mutations in SPI1 effector genes to induce secretory and inflammatory changes in bovine ligated ileal loops at 8 h postinfection. (A) Data for fluid accumulation in loops shown as percentages of the fluid secretion elicited by the wild type (IR715). The 100% line indicates the amount of fluid accumulation elicited by the Salmonella serotype Typhimurium wild type. All mutants shown elicited significantly less fluid accumulation than the wild type (IR715) (P < 0.05). The results are means from experiments performed with three animals, in which each strain was tested in two loops/animal. The bars indicate means ± standard deviations. The same letter above two bars indicates that the amounts of fluid accumulation elicited by the mutants are not significantly different. Different letters above two bars indicate that the amounts of fluid accumulation elicited by the mutants are significantly different (P < 0.05). (B) Inflammatory changes in infected loops assessed by microscopic examination of hematoxylin- and eosin-stained thin sections of infected tissue. wt, wild type.
SPI1 effector proteins act in concert to elicit fluid accumulation and inflammation.
SipB is a translocase required for delivery of SPI1 effector proteins into the host cell cytoplasm. A Salmonella serotype Typhimurium sipB mutant (CAS152) (48) caused less fluid accumulation in bovine ligated ileal loops than strains carrying a mutation in sipA, sopA, sopB, sopD, or sopE2 (Fig. 3A). We reasoned that the drastic reduction in fluid accumulation caused by a mutation in sipB could be accounted for by the fact that translocation of SPI1 effector proteins involved in enteropathogenicity, including SipA, SopA, SopB, SopD, and SopE2, is prevented. To test this hypothesis, we introduced mutations in sipA, sopA, sopB, sopD, and sopE2 one by one into the Salmonella serotype Typhimurium wild type (IR715) to generate a series of mutants, one of which carried mutations in all five effector genes (sipAsopABDE2 mutant). The virulence of these multiple-knockout mutants in bovine ligated ileal loops was compared to that of the isogenic wild-type strain (IR715) and that of strains carrying mutations in sipB (CAS152) and in individual effector genes (Fig. 3A).
The sopBE2 double mutant (ZA16) caused fluid accumulation at a level similar to that caused by the sopB mutant (ZA15). The sopBDE2 triple mutant (ZA18) induced less fluid secretion than strains having mutations in either sopB (ZA15), sopD (ZA17), or sopE2 (ZA9), but the difference was not statistically significant. The sopABDE2 quadruple mutant (ZA20) caused significantly less fluid secretion than strains carrying mutations in either sopA (ZA19), sopD (ZA17), or sopE2 (ZA9) (P < 0.05). However, the amounts of fluid secretion elicited by the sopABDE2 quadruple mutant (ZA20) and the sopB mutant (ZA15) were not statistically different. The sopABDE2 quadruple mutant (ZA20) elicited significantly more fluid secretion than the sipB mutant (CAS152). In contrast, the sipAsopABDE2 quintuple mutant (ZA21) induced fluid secretion at a level similar to that caused by the sipB mutant (CAS152). Furthermore, the sipAsopABDE2 quintuple mutant (ZA21) caused significantly less fluid secretion than the sopABDE2 quadruple mutant (ZA20), the sopBDE2 triple mutant (ZA18), the sopBE2 double mutant (ZA16), or strains carrying single mutations in sipA (ZA10), sopA (ZA19), sopB (ZA15), sopD (ZA17), or sopE2 (ZA9). These data demonstrated that inactivation of sipB and simultaneous inactivation of sipA, sopA, sopB, sopD, and sopE2 caused similar reductions in the ability of Salmonella serotype Typhimurium to cause fluid accumulation in bovine ligated ileal loops. Hence, these data supported the idea that the main function of SipB in eliciting fluid accumulation is the translocation of SipA, SopA, SopB, SopD, and SopE2 into host cells.
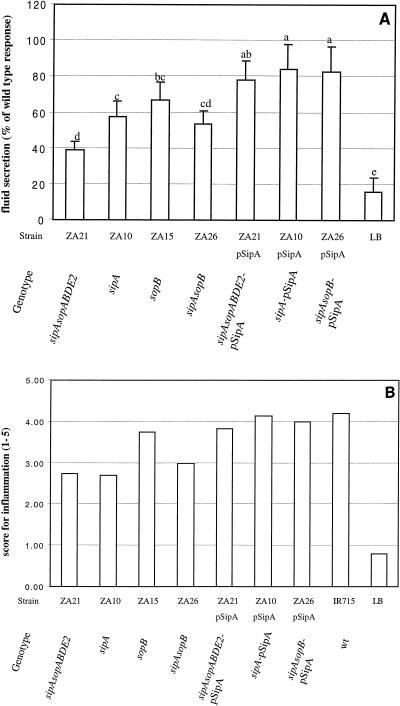
Although mutations in sipA (ZA10), sopA (ZA19), sopB (ZA15), sopD (ZA17), or sopE2 (ZA9) caused a significant reduction in fluid accumulation compared to the fluid accumulation observed with the wild type (IR715), it was not clear to what degree mutations in individual effector genes contributed to the phenotype observed for the sipAsopABDE2 quintuple mutant (ZA21) (Fig. 2A and 3A). Inactivation of sipA (ZA10) or sopB (ZA15) resulted in a more pronounced reduction in fluid accumulation than mutations in sopA (ZA19), sopD (ZA17), or sopE2 (ZA9). Furthermore, the amount of fluid accumulated in loops infected with the sopB (ZA15) mutant was not significantly different from the amount accumulated in loops infected with the sopBE2 double mutant (ZA16), the sopBDE2 triple mutant (ZA18), or the sopABDE2 quadruple mutant (ZA20). Only the sipAsopABDE2 quintuple mutant (ZA21) caused significantly less fluid secretion than the sopB mutant (ZA15) or the sipA mutant (ZA10). It was, therefore, tempting to speculate that simultaneous inactivation of sipA and sopB may reduce fluid accumulation to the level elicited by the sipAsopABDE2 quintuple mutant (ZA21). To test this idea, we constructed a sipAsopB double mutant (ZA26) and assessed its virulence in bovine ligated ileal loops (Fig. 4A). As in the previous experiment, the amount of fluid accumulated in loops infected with the sopB mutant (ZA15) or the sipA mutant (ZA10) was significantly larger than the amount accumulated in loops infected with the sipAsopABDE2 quintuple mutant (ZA21) (P < 0.05). The sipAsopB double mutant (ZA26) caused less fluid accumulation than the sopB mutant (ZA15) or the sipA mutant (ZA10) and more fluid accumulation than the sipAsopABDE2 quintuple mutant (ZA21), but the differences were not statistically significant (Fig. 4A). Thus, the sipAsopB double mutant (ZA26) had a phenotype intermediate between that of the sipAsopABDE2 quintuple mutant (ZA21) and that of the single mutants (ZA15 and ZA10).
FIG. 4.
In vivo complementation in bovine ligated ileal loops at 8 h postinfection of strains carrying a sipA deletion with the cloned sipA gene. (A) Data for fluid accumulation in loops shown as percentages of the fluid secretion elicited by the wild type (IR715). The results are means from three experiments in which each strain was tested in at least two loops/animal. The bars indicate means ± standard deviations. The same letter above two bars indicates that the amounts of fluid accumulation elicited by the mutants are not significantly different. Different letters above two bars indicate that the amounts of fluid accumulation elicited by the mutants are significantly different (P < 0.05). (B) Inflammatory changes in infected loops assessed by microscopic examination of hematoxylin- and eosin-stained thin sections of infected tissue. wt, wild type.
SipA has not previously been implicated in eliciting fluid accumulation during infection of bovine ligated ileal loops with Salmonella serotype Typhimurium or Salmonella serotype Dublin. The sipA mutant (ZA10) used in this study carries an unmarked deletion (Fig. 1). Although unlikely, the possibility exists that this mutation has a polar effect on expression of the downstream iacP gene (Fig. 1). To establish that the in vivo phenotype of the sipA mutant (ZA10) was not caused by a polar effect, complementation experiments were performed by using pSipA, a plasmid carrying an intact sipA gene. Infection of bovine ileal loops with the sipAsopABDE2 quintuple mutant (ZA21), the sipA mutant (ZA10), or the sipAsopB double mutant (ZA26) resulted in significantly less fluid secretion than infection with the corresponding complemented strains, ZA21(pSipA), ZA10(pSipA), and ZA26(pSipA), respectively (Fig. 4A). Collectively, these data demonstrated that sipA plays an important role in eliciting host secretory and inflammatory responses during Salmonella serotype Typhimurium infection of calves.
To evaluate the local inflammatory response to infections with Salmonella serotype Typhimurium mutants, hematoxylin- and eosin-stained sections of Peyer's patches were examined by light microscopy, and the inflammatory changes were scored on a scale from 1 to 5 according to the criteria described in Materials and Methods (Fig. 2B). The inflammatory lesions caused by strains ZA16 (sopBE2), ZA18 (sopBDE2), and ZA20 (sopABDE2) were less severe than those caused by the Salmonella serotype Typhimurium wild type (IR715). Consistent with the results obtained during the initial characterization of mutants (Fig. 2B), deletion of sipA caused a considerable reduction in the local inflammatory response (Fig. 3B). Introduction of plasmid pSipA into strains ZA10, ZA21, and ZA26 resulted in elevated inflammatory responses in bovine ligated ileal loops (Fig. 4B). In sections of Peyer's patches infected with the sipAsopABDE2 quintuple mutant (ZA21) or the sipB mutant (CSA152), a reduced inflammatory response with mild perivascular to coalescent infiltration of PMN in the mucosa and submucosa was observed. Focal intravascular and/or perivascular infiltration of PMN in the mucosa was also observed in some sections from ileal loops inoculated with sterile LB broth, which may have been a result of the surgical procedure. To investigate whether the reduced ability of Salmonella serotype Typhimurium mutants to cause fluid accumulation or inflammation in loops correlates with their ability to colonize bovine Peyer's patches, the number of bacteria recovered from infected tissue was determined. We found that mutations in sipA (ZA10), sopA (ZA19), sopB (ZA15), sopD (ZA17), and sopE2 (ZA9) or simultaneous inactivation of sipAsopABDE2 (ZA21) did not significantly reduce the bacterial numbers recovered from bovine Peyer's patches at 8 h postinfection compared to the numbers observed with the wild type (IR715). In contrast, the number of sipB mutant (CSA152) cells recovered from Peyer's patches was approximately 100-fold less than the number of wild-type (IR715) cells (data not shown). These data suggested that SipB is required for colonization of the bovine mucosa, either directly through its interaction with host proteins (caspase 1) or through translocation of AvrA, SlrP, SspH1, SptP, or other as-yet-unidentified SPI1 secreted effector proteins into cells of the bovine host.
Role of SopB, SopE2, SopA, SopD, and SipA in causing diarrhea and lethal morbidity after oral infection of calves.
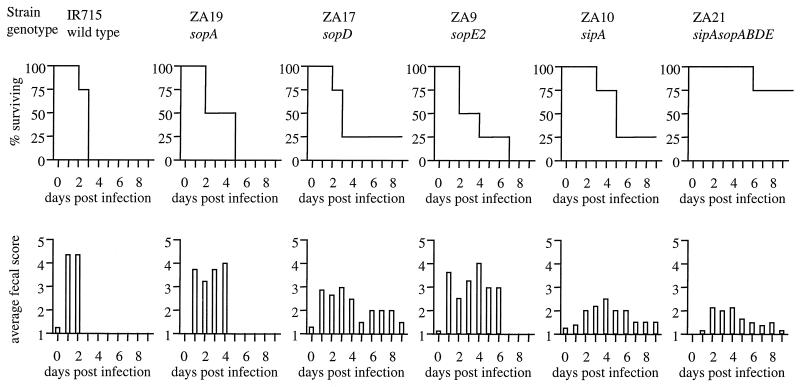
We have shown previously that a Salmonella serotype Typhimurium sopB mutant causes diarrhea and intestinal lesions at wild-type levels when calves are infected with a dose of 1010 CFU/animal (47), whereas a Salmonella serotype Typhimurium sipB mutant (CSA152) causes no diarrhea or only mild diarrhea and no lethal morbidity at this dose (48). One possible explanation for these observations is that inactivation of only one effector gene (i.e., sopB) does not result in marked attenuation during oral infection, because SopB acts in concert with SopE2, SopA, SopD, and SipA to cause diarrhea in calves. This hypothesis further implies that a sipB mutant is markedly attenuated, because it is unable to translocate SopB, SopE2, SopA, SopD, and SipA. To determine whether SopB, SopE2, SopA, SopD, and SipA act in concert during oral infection of calves, groups of four calves were inoculated orally with strain IR715 (wild type), ZA10 (sipA), ZA19 (sopA), ZA17 (sopD), ZA9 (sopE2), or ZA21 (sipAsopABDE2) by using a dose of 1 × 1010 CFU/animal. Infection of calves with the wild type (IR715), the sopE2 mutant (ZA9), or the sopA mutant (ZA19) induced aqueous diarrhea with feces containing various combinations of blood, fibrin, and mucus at day 1 postinfection (Fig. 5). All calves infected with the Salmonella serotype Typhimurium wild type (IR715) were euthanized between 24 and 72 h postinfection, because they had signs of terminal illness, such as severe anorexia and an inability to stand. Similarly, infection with the sopE2 mutant (ZA9) or the sopA mutant (ZA19) caused lethal morbidity in all calves; however, the time to death was delayed compared to the time of death of animals infected with the wild type (IR715). The severity of diarrhea was reduced in calves infected with the sopD mutant (ZA17), while calves infected with the sipA mutant (ZA10) or the sipAsopABDE2 quintuple mutant (ZA21) had only mild transient diarrhea with a delayed onset (starting on day 2 or 3 postinfection). While the sipA mutant (ZA10) and the sopD mutant (ZA17) caused lethal morbidity in three of four calves, the sipAsopABDE2 quintuple mutant (ZA21) caused lethal morbidity only in one of four calves. The data obtained with ZA10, the sipA mutant used throughout this study, were not identical to the data obtained previously with a different Salmonella serotype Typhimurium sipA mutant, strain JLR137 (48). Strain JLR137 causes reduced mortality but is able to cause diarrhea in calves. Construction of JLR137, which is described in a previous report (48), was performed by transducing the Tn5lacZY marker from EE633, a strain in which the transposon is inserted close to or within the sipA gene, as indicated by Southern hybridization (19). To further investigate why the phenotype reported for JLR137 is different from that found for the sipA mutant (ZA10) used throughout this study, we cloned DNA flanking the Tn5lacZY insertion in JLR137. Sequence analysis of the transposon-flanking DNA revealed that the transposon in JLR137 was inserted into the glpD gene, suggesting that a transposition event had occurred during transduction of Tn5lacZY from EE633. Furthermore, Southern hybridization with a probe specific for the sipA gene confirmed that Tn5lacZY was not inserted into or close to the sipA gene in strain JLR137. These results indicated that JLR137 carries a mutation in glpD, but not a mutation in sipA. In contrast, strain ZA10 carries a sipA deletion, as indicated by Southern hybridization and complementation analysis (Fig. 4A).
FIG. 5.
Ability of SPI1 mutants to cause diarrhea and lethal morbidity in calves infected orally with a dose of 1010 CFU per animal. Lethal morbidity caused by the wild type or by Salmonella serotype Typhimurium mutants over time is shown in the upper graphs. The lower graphs show the severity of diarrhea over time, which was scored on a scale from 1 to 5, as follows: 1, normal feces; 2, soft feces with loss of distinct conformation; 3, diarrhea, loose feces with reduced solid matter; 4, diarrhea, aqueous feces with markedly reduced or little solid matter, or fibrin; 5, diarrhea, aqueous feces with no solid matter, fibrin and blood.
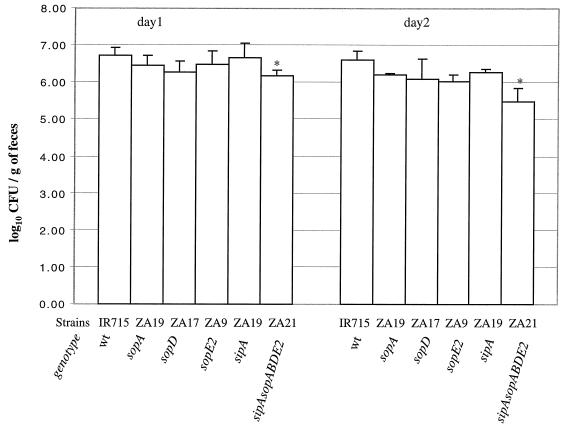
To assess the severity of the clinical signs of disease, the plasma concentration of sodium was analyzed at zero time and on days 1 and 2 postinfection for each animal, and the rectal temperature and fecal shedding of Salmonella serotype Typhimurium were recorded daily. On day 2 postinfection, the sodium plasma concentrations of all infected calves except one animal infected with the sipAsopABDE2 quintuple mutant (ZA21) were below the normal range, 135 to 153 meq/liter (Fig. 6). The reduction in the plasma sodium levels at 2 days postinfection compared to the values recorded prior to infection was significantly greater in calves infected with the wild type (IR715) than in animals infected with the sipAsopABDE2 quintuple mutant (ZA21) (P < 0.05). The rectal temperature of all infected calves was elevated (39.3 to 40°C) on days 1 and 2 postinfection, and there were not significant differences among treatment groups (data not shown). In the calves that survived infection, the temperature gradually returned to the normal range, which was determined by taking the temperature of uninfected animals (the normal range was between 37.9 and 38.3°C). All calves shed high numbers of bacteria (≥106 CFU/g of feces) on day 1 postinfection. The numbers of bacteria recovered from feces of calves infected with the wild type (IR715) were significantly higher (P < 0.05) than the numbers of bacteria recovered from animals infected the sipAsopABDE2 quintuple mutant (ZA21) (Fig. 7). The difference became more pronounced on day 2 postinfection, when calves infected with the wild type (IR715) shed approximately 10-fold more bacteria than animals infected with the sipAsopABDE2 quintuple mutant (ZA21) (P < 0.05). These data suggested that the number of bacteria shed in the feces may increase with the severity of diarrhea; however, this was not investigated further.
FIG. 6.
Change in the plasma sodium concentration during oral infection of calves with Salmonella serotype Typhimurium strains. The graph shows the difference in the plasma sodium concentration between a sample collected on day 2 postinfection and a preinfection sample. Solid circles indicate that the plasma sodium concentration on day 2 postinfection was below the normal range. The open circle indicates that the plasma sodium concentration on day 2 postinfection was within the normal range. WT, wild type.
FIG. 7.
Recovery of Salmonella serotype Typhimurium strains from the feces of calves after oral infection with a dose of 1010 CFU per animal. The bars indicate the mean ± standard deviation for four calves. An asterisks indicates that the difference between the wild type (IR715) and an individual mutant was significant. wt, wild type.
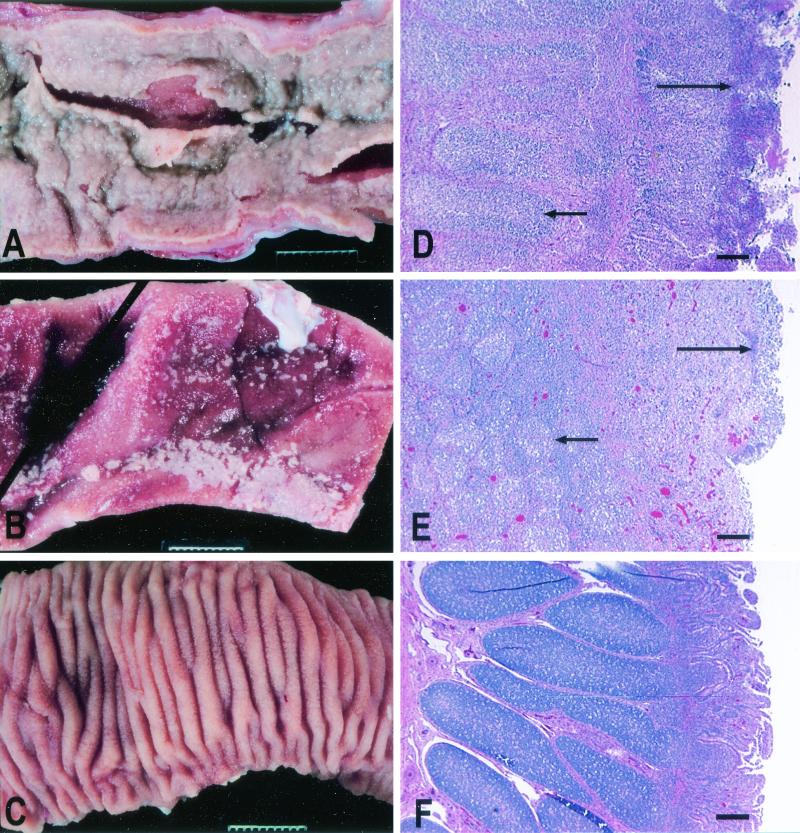
At necropsy, a gross pathological examination was performed, and tissue samples from mesenteric lymph nodes and Peyer's patches were collected for histopathologic evaluation. Calves which had signs of lethal morbidity after infection with strains IR715, ZA19 (sopA), ZA9 (sopE2), and ZA17 (sopD) and one of the calves infected with strain ZA10 (sipA) developed severe acute fibrinopurulent necrotizing enteritis with purulent exudate and segmental or continuous pseudomembrane deposition in the terminal ileum (Fig. 8). Two calves that developed lethal morbidity after infection with the sipA mutant (ZA10) had reduced severity of intestinal lesions (marked subacute fibrinopurulent necrotizing enteritis confined to the Peyer's patches). No gross pathological lesions were detected in calves infected with ZA21 (sipAsopABDE2) and calves that survived infection with ZA10 (sipA) or ZA17 (sopD).
FIG. 8.
Representative examples of the gross pathology and histopathology of Peyer's patches and the terminal ileum of calves inoculated orally with 1010 CFU of different Salmonella serotype Typhimurium strains. (A) Severe acute fibrinopurulent necrotizing enteritis with segmental or continuous pseudomembrane formation in a calf infected with wild-type strain IR715 (similar pathological changes were observed in calves infected with the sopA mutant, the sopD mutant, or the sopE2 mutant). Bar = 1 cm. (B) Marked subacute fibrinopurulent necrotizing enteritis often confined to the Peyer's patches of the terminal ileum of a calf infected with strain ZA10 (sipA). Bar = 1 cm. (C) Normal Peyer's patches and ileum of a calf infected with strain ZA21 (sipAsopABDE). Bar = 1 cm. (D to F) Hematoxylin- and eosin-stained sections of Peyer's patches of calves infected with IR715, ZA10 (sipA), and ZA21 (sipAsopABDE), respectively. The short arrows indicate areas of lymphoid depletion; the long arrows indicate various degrees of fibrinopurulent necrotizing ileitis at the mucosal surface. Bars = 200 μm.
Microscopic examination also revealed severe necrotizing enteritis in calves infected with IR715, ZA9 (sopE2), and ZA19 (sopA), as well as the three calves that developed lethal signs of disease after infection with ZA17 (sopD). The histopathologic lesions in the terminal ileum of these calves included extensive necrosis of the ileal mucosa, neutrophilic infiltration in mucosa and submucosa, accumulation of fibrin and necrotic debris at the lumenal surface, and marked depletion of lymphoid cells from the lymphoid follicles of Peyer's patches. Strain ZA10 (sipA) caused intestinal lesions of reduced severity, such as necrosis of the upper mucosa surrounded by a dense neutrophilic infiltrate and moderate lymphoid depletion in lymphoid follicles. In addition, lymphoid depletion was observed in the germinal centers of mesenteric lymph nodes of calves infected with IR715 (wild type) or the single-knockout mutants. The intestinal lesions were either absent or negligible in the terminal ileum of the four calves infected with ZA21 (sipAsopABDE2), despite the fact that one calf developed lethal morbidity. Similarly, intestinal lesions were either absent or negligible in the terminal ileum of calves that survived infection with strain ZA10 (sipA) or ZA17 (sopD).
DISCUSSION
In this study we determined which effector proteins secreted by the invasion-associated type III secretion system are required by Salmonella serotype Typhimurium to elicit fluid accumulation in the bovine ligated ileal loop model. In addition to the SPI1 translocation complex formed by SipB, SipC, and SipD, the bovine Salmonella serotype Typhimurium isolate ATCC 14028 encodes nine effector proteins, including SlrP, SspH1, SptP, AvrA, SipA, SopA, SopB, SopD, and SopE2. Here we show that Salmonella serotype Typhimurium strains carrying mutations in sipA, sopA, sopB, sopD, or sopE2 exhibited reduced enteropathogenicity but elicited more fluid accumulation than a Salmonella serotype Typhimurium sipB mutant (Fig. 3A). In contrast, mutations in slrP, avrA, sspH1, or sptP did not reduce the ability of Salmonella serotype Typhimurium to elicit fluid secretion in bovine ligated loops (Fig. 2A). Furthermore, a Salmonella serotype Typhimurium sipAsopABDE2 mutant elicited fluid accumulation at levels similar to those elicited by a Salmonella serotype Typhimurium sipB mutant (Fig. 3A). These data suggested that SopA, SopB, SopD, SopE2, and SipA are the major Salmonella serotype Typhimurium virulence factors involved in eliciting fluid accumulation in bovine ligated ileal loops. Furthermore, these data support the idea that the main function of SipB in eliciting fluid accumulation is the translocation of SipA, SopA, SopB, SopD, and SopE2 into host cells.
The mechanism by which SopA, SopB, SopD, SopE2, and SipA contribute to fluid accumulation may be related to the ability of these proteins to stimulate an inflammatory response. For instance, SipA elicits the production of chemoattractants that promote transepithelial PMN migration in a tissue culture model (28). A nonpolar deletion of sipA drastically reduced the ability of Salmonella serotype Typhimurium to elicit fluid accumulation and an inflammatory response in bovine ligated ileal loops (Fig. 2 and 3), and this defect could be partially complemented in vivo by introducing the cloned gene on a plasmid (Fig. 4A). A role for sopA and sopD in eliciting inflammation is suggested by the finding that mutations in these genes reduce the ability of Salmonella serotype Dublin to cause PMN influx into ligated ileal loops (21, 54). SopB and SopE2 interfere with intracellular signaling pathways (30, 42, 57) by acting as inositol phosphate phosphatase (31) and a guanine nucleotide exchange factor for Cdc42 (43), respectively. Nuclear responses triggered by SopB and SopE2 may result in the expression of proinflammatory cytokines that attract PMN (6). Thus, all effector proteins implicated in eliciting fluid accumulation in bovine ligated ileal loops (i.e., SopA, SopB, SopD, SopE2, and SipA) have also been implicated in eliciting the production of proinflammatory mediators in cell culture models in vitro. The PMN infiltrate elicited by Salmonella serotype Typhimurium may cause fluid secretion by an exudative mechanism since it results in increased vascular permeability and injury to the permeability barrier formed by the intestinal epithelium. Epithelial damage is first observed at 3 h postinfection and coincides with the appearance of fluid accumulation in bovine ligated ileal loops (35).
SPI1 was originally discovered because mutations in this region reduce the ability of Salmonella serotype Typhimurium to invade epithelial cells in vitro (12). The translocation of several effector proteins into host cells has been associated with this invasion phenotype. The main mechanism for SPI1-mediated invasion of tissue culture cells is the induction of actin cytoskeleton rearrangements triggered by SopB, SopE1, and SopE2 (30, 57). It is necessary to inactivate the genes encoding these three effector proteins to reduce the invasiveness of Salmonella serotype Typhimurium SL1344 to the level of a mutant with a defective type III secretion system (i.e., an invA or invG mutant) (30, 57). In addition, SipA may play a minor role in bacterial uptake in vitro since entry is delayed at early time points in a Salmonella serotype Typhimurium sipA mutant (58). Since Salmonella serotype Typhimurium strain ATCC 14028, a strain used in this study, does not carry the sopE1 gene, we anticipated that inactivation of sopB, sopE2, and sipA would reduce its invasiveness to the level of a sipB mutant. However, at 8 h after infection of bovine ligated ileal loops, the numbers of cells of both the wild type and the sipAsopABDE2 mutant recovered from Peyer's patch tissue were approximately 100-fold higher than the numbers of cells of a Salmonella serotype Typhimurium sipB mutant recovered (data not shown). Since this study was aimed at investigating fluid accumulation, a phenotype that develops several hours after inoculation, our experimental design was not optimal for studying bacterial invasion, which starts within minutes after infection (9). Hence, the fact that higher numbers of cells of the sipAsopABDE2 mutant than of a sipB mutant were recovered from Peyer's patches may be unrelated to the ability of the former strain to invade epithelial cells but rather may reflect its ability to survive or multiply within bovine tissue.
This study revealed several differences between Salmonella serotype Dublin and Salmonella serotype Typhimurium with regard to phenotypes caused by inactivation of genes encoding effector proteins. For instance, the fluid secretion elicited by a Salmonella serotype Typhimurium sopB mutant was not significantly different from that elicited by a Salmonella serotype Typhimurium sopB sopD sopE2 mutant (Fig. 2A). In contrast, a Salmonella serotype Dublin sopB sopD mutant elicits less fluid accumulation in bovine ligated ileal loops than a Salmonella serotype Dublin sopB mutant or a Salmonella serotype Dublin sopD mutant (21). Furthermore, mutations in sopA and sipB cause similar reductions in the ability of Salmonella serotype Dublin to elicit fluid accumulation in bovine ligated ileal loops (54). In Salmonella serotype Typhimurium, on the other hand, a mutation in sopA caused a comparatively small reduction in fluid secretion, and a Salmonella serotype Typhimurium sipB mutant caused significantly less fluid accumulation than a sopA mutant (Fig. 3A). It may not be surprising that not all data obtained with Salmonella serotype Dublin can be extrapolated to Salmonella serotype Typhimurium since these organisms cause divergent disease manifestations in cattle. Although Salmonella serotype Typhimurium and Salmonella serotype Dublin infections in young calves commonly result in diarrhea, Salmonella serotype Dublin infections are more invasive, and meningoencephalitis, polyarthritis, osteomyelitis, or pneumonia may eventually occur in the absence of diarrhea (32). Unlike Salmonella serotype Typhimurium, Salmonella serotype Dublin may cause abortion in pregnant cows and heifers with no other clinical signs of infection (15, 32). Furthermore, Salmonella serotype Dublin and Salmonella serotype Typhimurium are associated with distinct human disease syndromes (bacteremia and enterocolitis, respectively) (8, 27, 41); that is, diarrhea is the prominent sign of disease caused by Salmonella serotype Typhimurium, and only 1% of human isolates are from blood (46). In contrast, only about one-third of Salmonella serotype Dublin human patients develop diarrhea, while bacteria are cultured from blood in 75 to 91% of the cases (8, 45).
The ligated ileal loop model is a valuable tool for characterizing virulence factors involved in enteropathogenesis of Salmonella serotypes (50). However, the loop model restricts investigation of Salmonella serotype Typhimurium host interactions to the ileum, while severe pathological changes during an oral infection are observed in both the ileum and the colon (47). Furthermore, the loop model is suitable only for studying host-pathogen interactions at early times (<12 h) postinfection. We therefore characterized Salmonella serotype Typhimurium mutants further by performing oral infection experiments with calves. Compared to the wild type, the sipA mutant (ZA10) caused greatly reduced diarrhea. These results appeared to contradict our previous finding that the sipA mutation in strain JLR137 reduced mortality but not the severity of diarrhea during oral infection of calves (48). Analysis of the strain used in the previous study (JLR137) revealed that it carried a mutation in glpD but not a mutation in sipA. In contrast, the sipA mutant used throughout this study (ZA10) carries a deletion in sipA (Fig. 1), as demonstrated by Southern hybridization and complementation analysis (Fig. 4A). The data show that sipA encodes an effector protein required for diarrhea. A qualitative assessment of the discharge from infected animals suggested that a mutation in sipA caused a more pronounced reduction in the severity of diarrhea than mutations in sopA, sopD, and sopE2 (Fig. 5). However, the difference was not statistically significant when electrolyte loss was assessed by determining the decrease in blood sodium concentration associated with Salmonella serotype Typhimurium infection (Fig. 6). The sipAsopABDE2 mutant caused less mortality (Fig. 5) and was shed in lower numbers with the feces (Fig. 7) than the wild type or strains carrying mutations in sipA, sopA, sopD, or sopE2. Furthermore, we have shown previously that a mutation in sopB does not reduce mortality or the severity of diarrhea during oral infection of calves (47). These findings further support the concept that the secreted effector proteins act in concert during the pathogenesis of Salmonella serotype Typhimurium-induced enterocolitis in calves.
Acknowledgments
The work in the laboratory of L. Garry Adams was supported by Texas Agricultural Experiment Station project 8409 and by Public Health Service grant AI44170. Work in the laboratory of Andreas J. Bäumler was supported by the Texas Advanced Research (Technology) Program under grant 000089-0051-1999 and by Public Health Service grants AI40124 and AI44170. Renée M. Tsolis was supported by USDA/NRICGP grant 9702568. Renato L. Santos was supported by CAPES, Brasília, Brazil, and by Universidade Federal de Minas Gerais, Belo Horizonte, Brazil.
We thank Roberta Pugh and Christine Curry for technical assistance, Alan Patranella for care of the calves, and Ivan Sampaio for statistical analysis.
Editor: J. T. Barbieri
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Altmeyer, R. M., J. K. McNern, J. C. Bossio, I. Rosenshine, B. B. Finlay, and J. E. Galán. 1993. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol. Microbiol. 7:89-98. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 4.Bakshi, C. S., V. P. Singh, M. W. Wood, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behlau, I., and S. J. Miller. 1993. A PhoP repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L. M., S. Hobbie, and J. E. Galán. 1996. Requirement of cdc42 for salmonella-induced cytoskeletal and nuclear responses. Science 274:2115-2118. [DOI] [PubMed] [Google Scholar]
- 7.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 8.Fang, F. C., and J. Fierer. 1991. Human infection with Salmonella dublin. Medicine (Baltimore) 70:198-207. [DOI] [PubMed] [Google Scholar]
- 9.Frost, A. J., A. P. Bland, and T. S. Wallis. 1997. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet. Pathol. 34:369-386. [DOI] [PubMed] [Google Scholar]
- 10.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 11.Galan, J. E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2:46-50. [DOI] [PubMed] [Google Scholar]
- 12.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galyov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Watson, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25:903-912. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, E. A. 1961. Salmonellosis in calves. Vet. Rec. 73:1284-1296. [PubMed] [Google Scholar]
- 15.Hall, G. A., P. W. Jones, K. R. Parsons, N. Chanter, and M. M. Aitken. 1979. Studies of the virulence of Salmonella dublin in experimental infections of cattle and rats. Br. Vet. J. 135:243-248. [DOI] [PubMed] [Google Scholar]
- 16.Hardt, W. D., and J. E. Galan. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94:9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hueck, C. J., M. J. Hantman, V. Bajaj, C. Johnston, C. A. Lee, and S. I. Miller. 1995. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol. Microbiol. 18:479-490. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, L. E., E. A. Gibson, H. E. Roberts, E. T. Davies, G. Davies, and W. J. Sojka. 1971. Bovine salmonellosis in England and Wales. Br. Vet. J. 127:225-238. [DOI] [PubMed] [Google Scholar]
- 21.Jones, M. A., M. W. Wood, P. B. Mullan, P. R. Watson, T. S. Wallis, and E. E. Galyov. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun. 66:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaniga, K., D. Trollinger, and J. E. Galán. 1995. Identification of two targets of the type III secretion system encoded in inv and spa loci of Salmonella typhimurium that share homology to IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galán. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaniga, K., J. Uralil, J. B. Bliska, and J. E. Galán. 1996. A secreted tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol. Microbiol. 21:633-641. [DOI] [PubMed] [Google Scholar]
- 25.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R− M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley, R. A., R. Reissbrodt, W. Rabsch, J. M. Ketley, R. M. Tsolis, P. Everest, G. Dougan, A. J. Bäumler, M. Roberts, and P. H. Williams. 1999. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol. 65:1610-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, M. D., B. Amatya, and Y. Kimula. 1999. Histopathology of typhoid enteritis: morphologic and immunophenotypic findings. Mod. Pathol. 12:949-955. [PubMed] [Google Scholar]
- 28.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. USA 97:12283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Bäumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 30.Mirold, S., K. Ehrbar, A. Weissmuller, R. Prager, H. Tschape, H. Russmann, and W. D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 183:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norris, F. A., M. P. Wilson, T. S. Wallis, E. E. Galyov, and P. W. Majerus. 1998. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA 95:14057-14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rings, D. M. 1985. Salmonellosis in calves. Vet. Clin. N. Am. Food Anim. Pract. 1:529-539. [DOI] [PubMed] [Google Scholar]
- 33.Rothenbacher, H. 1965. Mortality and morbidity in calves with salmonellosis. J. Am. Vet. Med. Assoc. 147:1211-1214. [PubMed] [Google Scholar]
- 34.Santos, R. L., R. M. Tsolis, A. J. Bäumler, R. Smith 3rd, and L. G. Adams. 2001. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 69:2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos, R. L., R. M. Tsolis, S. Zhang, T. A. Ficht, A. J. Bäumler, and L. G. Adams. 2001. Salmonella-induced cell death is not required for enteritis in calves. Infect. Immun. 69:4610-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Bäumler. 2001. Animal models of Salmonella infections: enteritis vs. typhoid fever. Microb. Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 37.Schesser, K., J. M. Dukuzumuremyi, C. Cilio, S. Borg, T. S. Wallis, S. Pettersson, and E. E. Galyov. 2000. The salmonella YopJ-homologue AvrA does not possess YopJ-like activity. Microb. Pathog. 28:59-70. [DOI] [PubMed] [Google Scholar]
- 38.Smith, B. P., L. DaRoden, M. C. Thurmond, G. W. Dilling, H. Konrad, J. A. Pelton, and J. P. Picanso. 1994. Prevalence of salmonellae in cattle and in the environment of California dairies. J. Am. Vet. Med. Assoc. 205:467-471. [PubMed] [Google Scholar]
- 39.Smith, B. P., F. Habasha, M. Reina-Guerra, and A. J. Hardy. 1979. Bovine salmonellosis: experimental production and characterization of the disease in calves, using oral challenge with Salmonella typhimurium. Am. J. Vet. Res. 40:1510-1513. [PubMed] [Google Scholar]
- 40.Sojka, W. J., C. Wray, J. Shreeve, and A. J. Benson. 1977. Incidence of salmonella infection in animals in England and Wales, 1968-1974. J. Hyg. 78:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprinz, H., E. J. Gangarosa, M. Williams, R. B. Hornick, and T. E. Woodward. 1966. Histopathology of the upper small intestines in typhoid fever. Biopsy study of experimental disease in man. Am. J. Dig. Dis. 11:615-624. [DOI] [PubMed] [Google Scholar]
- 42.Steele-Mortimer, O., L. A. Knodler, S. L. Marcus, M. P. Scheid, B. Goh, C. G. Pfeifer, V. Duronio, and B. B. Finlay. 2000. Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector sigD. J. Biol. Chem. 275:37718-37724. [DOI] [PubMed] [Google Scholar]
- 43.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 44.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, D. N., J. M. Bied, J. S. Munro, and R. A. Feldman. 1982. Salmonella dublin infections in the United States, 1979-1980. J. Infect. Dis. 146:322-327. [DOI] [PubMed] [Google Scholar]
- 46.Threlfall, E. J., M. L. Hall, and B. Rowe. 1992. Salmonella bacteraemia in England and Wales, 1981-1990. J. Clin. Pathol. 45:34-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Bäumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsolis, R. M., L. G. Adams, M. J. Hantman, C. A. Scherer, T. Kimborough, R. A. Kingsley, T. A. Ficht, S. I. Miller, and A. J. Bäumler. 2000. SspA is required for lethal Salmonella enterica serovar Typhimurium infections in calves but is not essential for diarrhea. Infect. Immun. 68:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 52.Watson, P. R., E. E. Galyov, S. M. Paulin, P. W. Jones, and T. S. Wallis. 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect. Immun. 66:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, P. R., A. V. Gautier, S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 2000. Salmonella enterica serovars Typhimurium and Dublin can lyse macrophages by a mechanism distinct from apoptosis. Infect. Immun. 68:3744-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, M. W., M. A. Jones, P. R. Watson, A. M. Siber, B. A. McCormick, S. Hedges, R. Rosqvist, T. S. Wallis, and E. E. Galyov. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell Microbiol. 2:293-303. [DOI] [PubMed] [Google Scholar]
- 55.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 56.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 57.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092-2095. [DOI] [PubMed] [Google Scholar]