Abstract
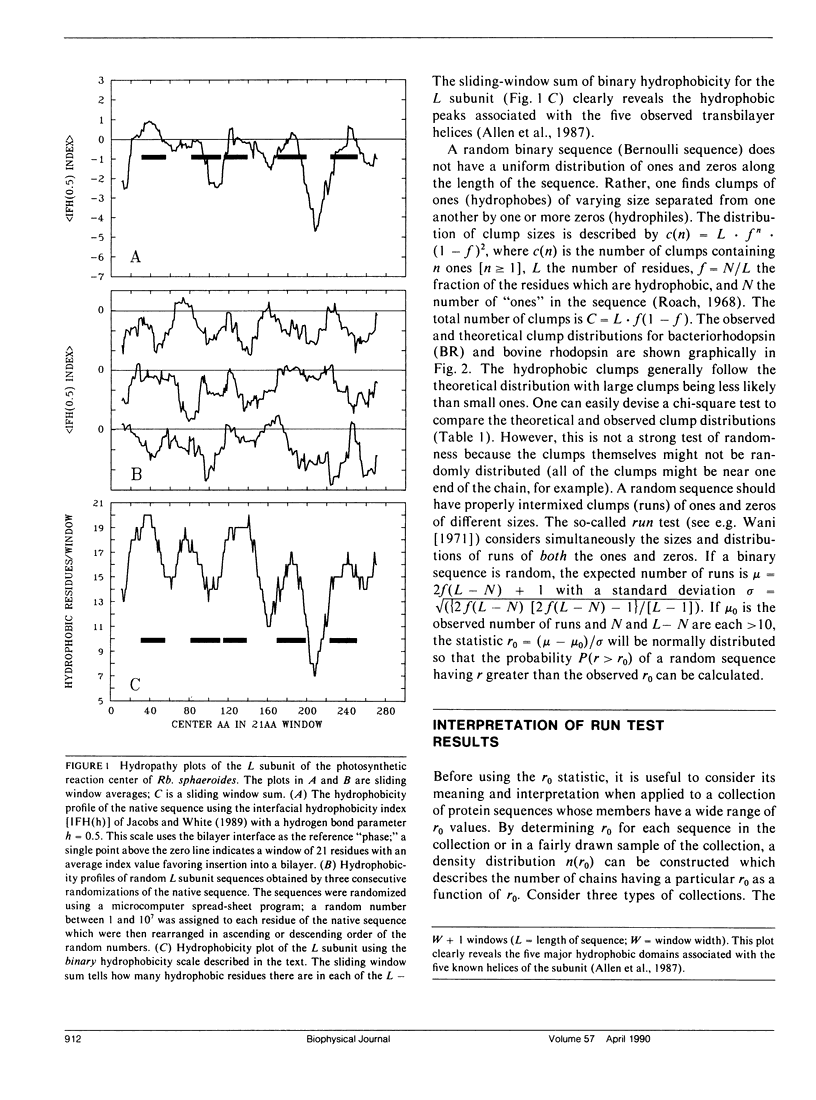
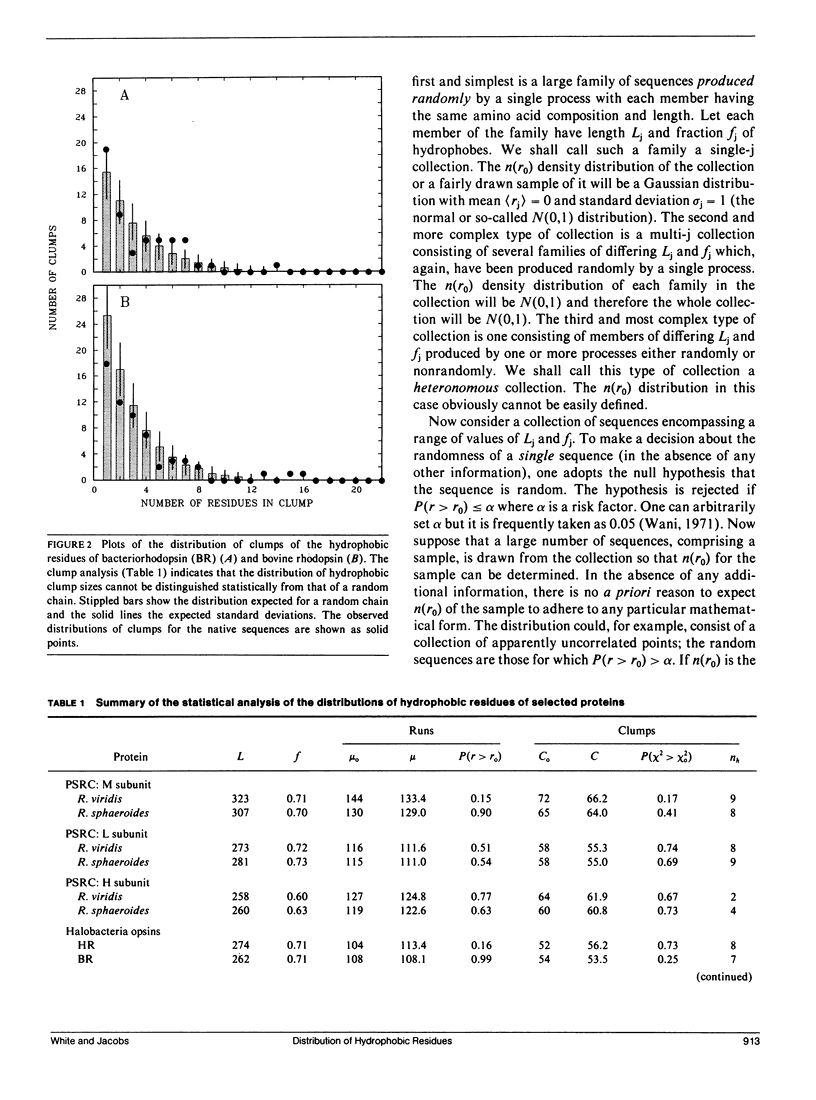
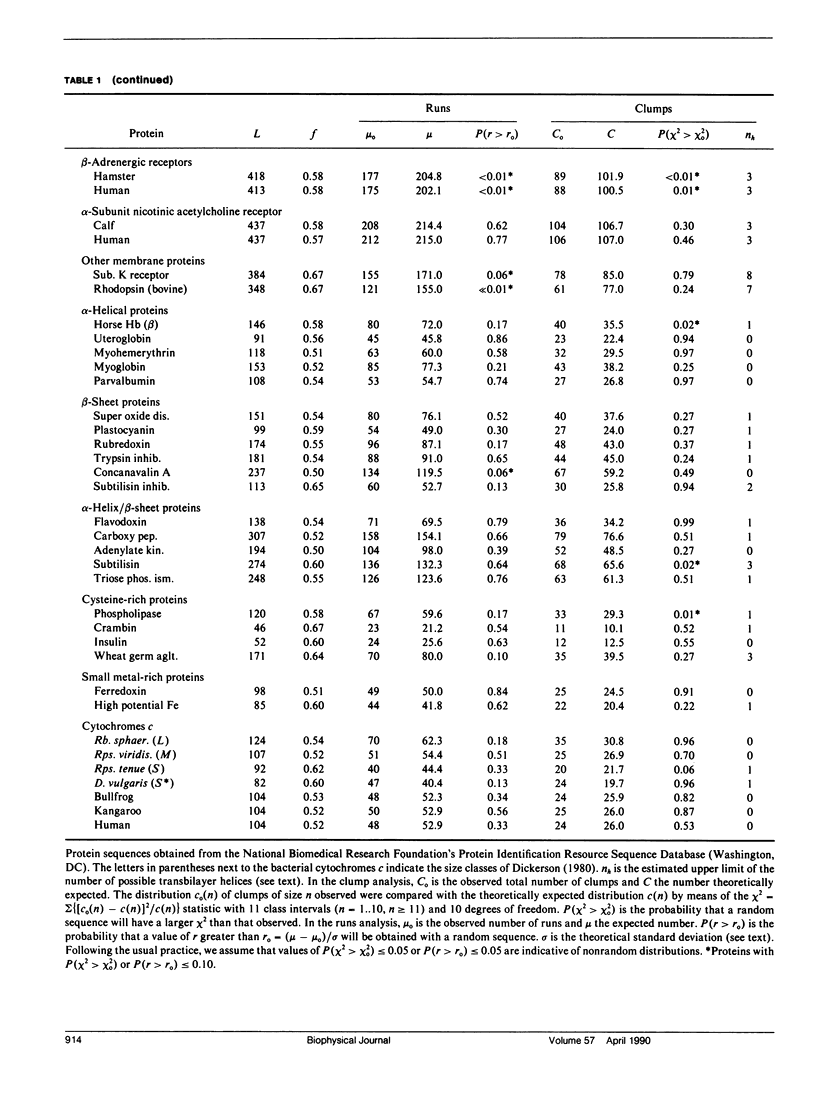
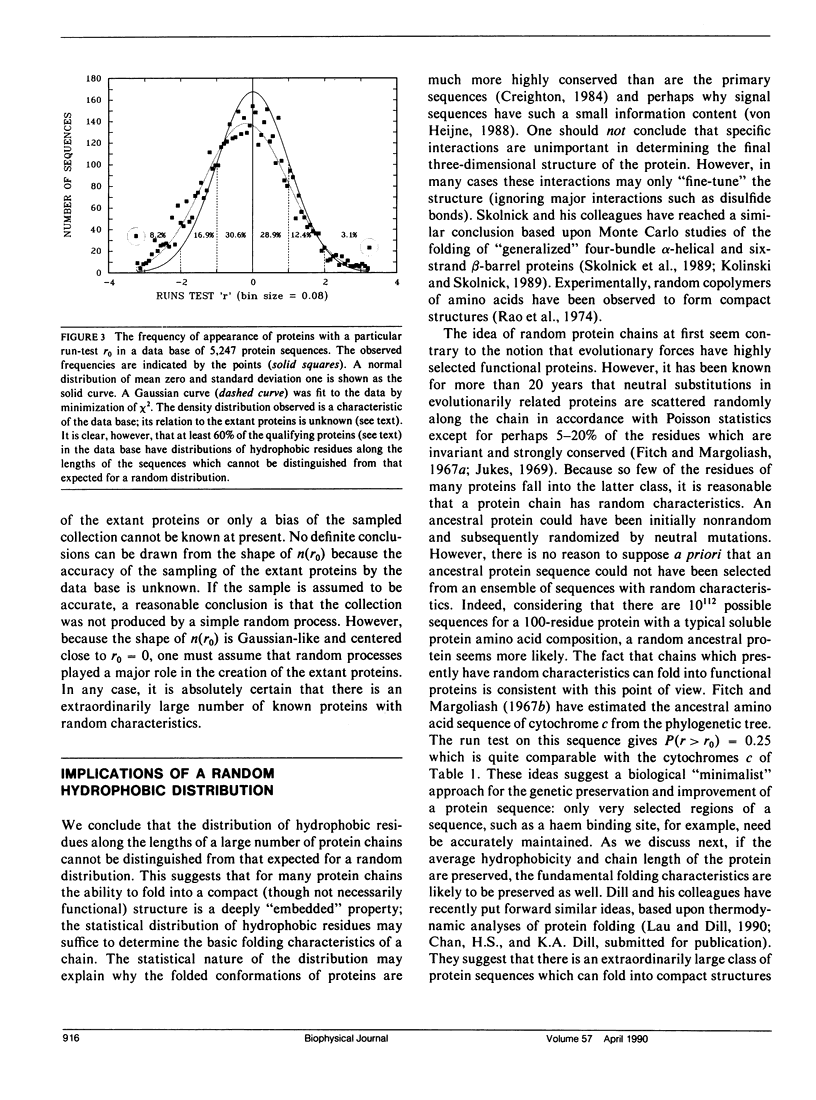
We consider in this paper the statistical distribution of hydrophobic residues along the length of protein chains. For this purpose we used a binary hydrophobicity scale which assigns hydrophobic residues a value of one and non-hydrophobes a value of zero. The resulting binary sequences are tested for randomness using the standard run test. For the majority of the 5,247 proteins examined, the distribution of hydrophobic residues along a sequence cannot be distinguished from that expected for a random distribution. This suggests that (a) functional proteins may have originated from random sequences, (b) the folding of proteins into compact structures may be much more permissive with less sequence specificity than previously thought, and (c) the clusters of hydrophobic residues along chains which are revealed by hydrophobicity plots are a natural consequence of a random distribution and can be conveniently described by binomial statistics.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6162–6166. doi: 10.1073/pnas.84.17.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Oesterhelt D. The halo-opsin gene. II. Sequence, primary structure of halorhodopsin and comparison with bacteriorhodopsin. EMBO J. 1987 Jan;6(1):265–273. doi: 10.1002/j.1460-2075.1987.tb04749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Ptitsyn O. B. Why do globular proteins fit the limited set of folding patterns? Prog Biophys Mol Biol. 1987;50(3):171–190. doi: 10.1016/0079-6107(87)90013-7. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. A method for estimating the number of invariant amino acid coding positions in a gene using cytochrome c as a model case. Biochem Genet. 1967 Jun;1(1):65–71. doi: 10.1007/BF00487738. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Jacobs R. E., White S. H. The nature of the hydrophobic binding of small peptides at the bilayer interface: implications for the insertion of transbilayer helices. Biochemistry. 1989 Apr 18;28(8):3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Evolutionary pattern of specificity regions in light chains of immunoglobulins. Biochem Genet. 1969 Apr;3(2):109–117. doi: 10.1007/BF00520347. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kerlavage A. R., Fraser C. M., Chung F. Z., Venter J. C. Molecular structure and evolution of adrenergic and cholinergic receptors. Proteins. 1986 Dec;1(4):287–301. doi: 10.1002/prot.340010403. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lau K. F., Dill K. A. Theory for protein mutability and biogenesis. Proc Natl Acad Sci U S A. 1990 Jan;87(2):638–642. doi: 10.1073/pnas.87.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978 Oct 3;17(20):4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- Lodish H. F. Multi-spanning membrane proteins: how accurate are the models? Trends Biochem Sci. 1988 Sep;13(9):332–334. doi: 10.1016/0968-0004(88)90101-6. [DOI] [PubMed] [Google Scholar]
- McCrea P. D., Engelman D. M., Popot J. L. Topography of integral membrane proteins: hydrophobicity analysis vs. immunolocalization. Trends Biochem Sci. 1988 Aug;13(8):289–290. doi: 10.1016/0968-0004(88)90120-x. [DOI] [PubMed] [Google Scholar]
- Nolan C., Margoliash E. Comparative aspects of primary structures of proteins. Annu Rev Biochem. 1968;37:727–790. doi: 10.1146/annurev.bi.37.070168.003455. [DOI] [PubMed] [Google Scholar]
- Orcutt B. C., George D. G., Dayhoff M. O. Protein and Nucleic Acid Sequence Database Systems. Annu Rev Biophys Bioeng. 1983;12:419–441. doi: 10.1146/annurev.bb.12.060183.002223. [DOI] [PubMed] [Google Scholar]
- Pittsyn O. B. Invariant features of globin primary structure and coding of their secondary structure. J Mol Biol. 1974 Sep 15;88(2):287–300. doi: 10.1016/0022-2836(74)90482-3. [DOI] [PubMed] [Google Scholar]
- Ptitsyn O. B., Finkelstein A. V. Similarities of protein topologies: evolutionary divergence, functional convergence or principles of folding? Q Rev Biophys. 1980 Aug;13(3):339–386. doi: 10.1017/s0033583500001724. [DOI] [PubMed] [Google Scholar]
- Rao S. P., Carlstrom D. E., Miller W. G. Collapsed structure polymers. A scattergun approach to amino acid copolymers. Biochemistry. 1974 Feb 26;13(5):943–952. doi: 10.1021/bi00702a019. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rose G. D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978 Apr 13;272(5654):586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Roy S. Hydrophobic basis of packing in globular proteins. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4643–4647. doi: 10.1073/pnas.77.8.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski A., Skolnick J. Monte Carlo simulation of equilibrium globular protein folding: alpha-helical bundles with long loops. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2668–2672. doi: 10.1073/pnas.86.8.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick J., Kolinski A., Yaris R. Dynamic Monte Carlo study of the folding of a six-stranded Greek key globular protein. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1229–1233. doi: 10.1073/pnas.86.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R. Helical length distribution from protein crystallographic data. Indian J Biochem Biophys. 1976 Jun;13(2):192–193. [PubMed] [Google Scholar]
- Sternberg M. J., Thornton J. M. On the conformation of proteins: an analysis of beta-pleated sheets. J Mol Biol. 1977 Feb 25;110(2):285–296. doi: 10.1016/s0022-2836(77)80073-9. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz P., Płochocka D., Rabczenko A. The formation of protein secondary structure. Its connection with amino acid sequence. Biophys Chem. 1988 Aug;31(1-2):139–142. doi: 10.1016/0301-4622(88)80018-8. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]


