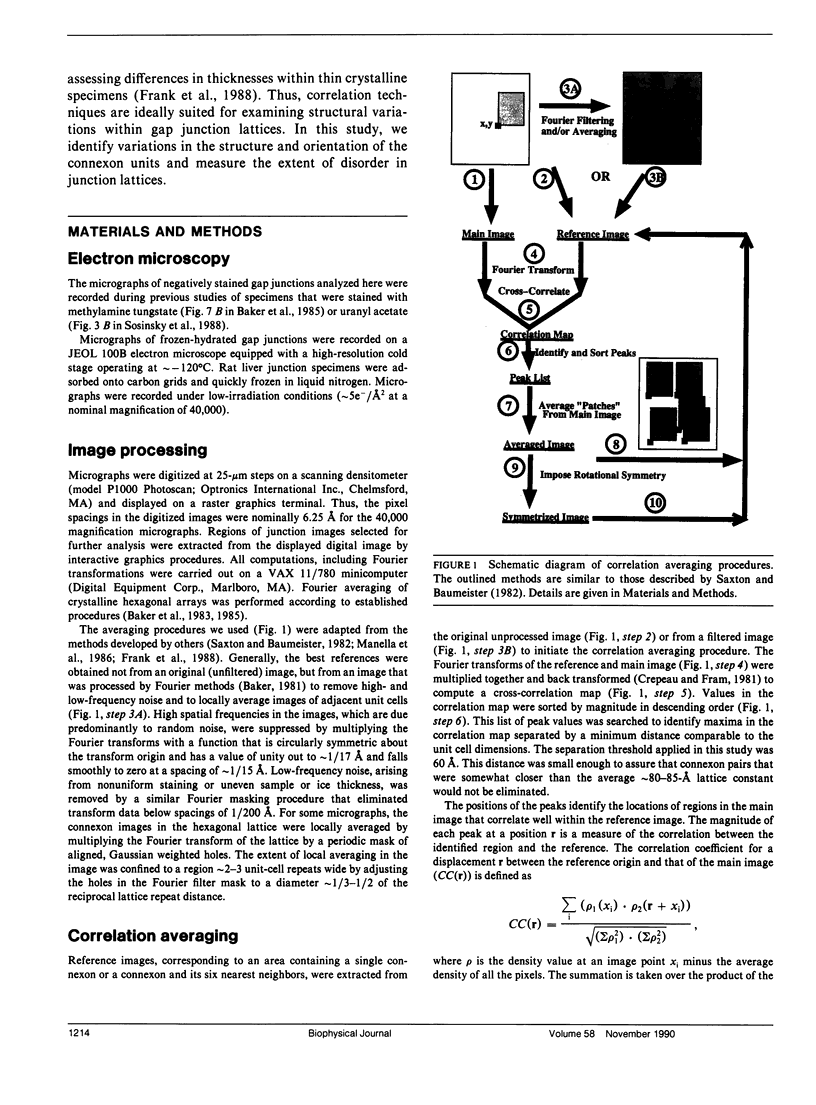
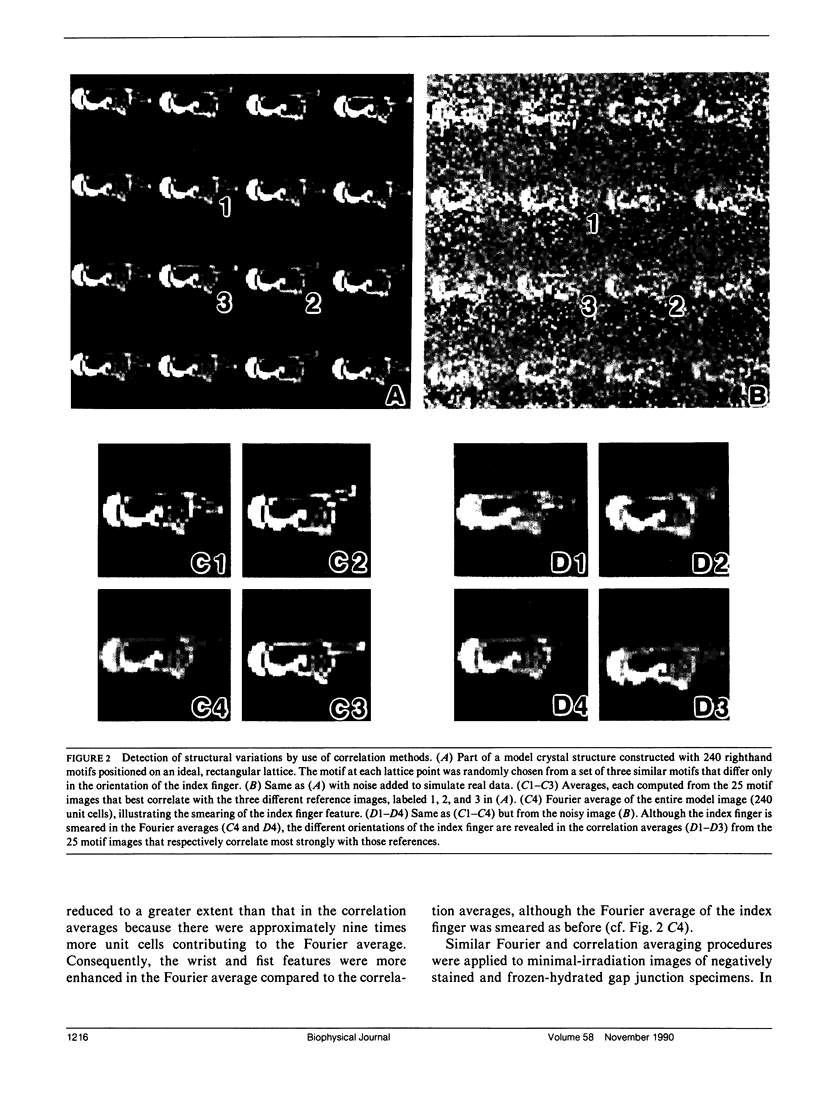
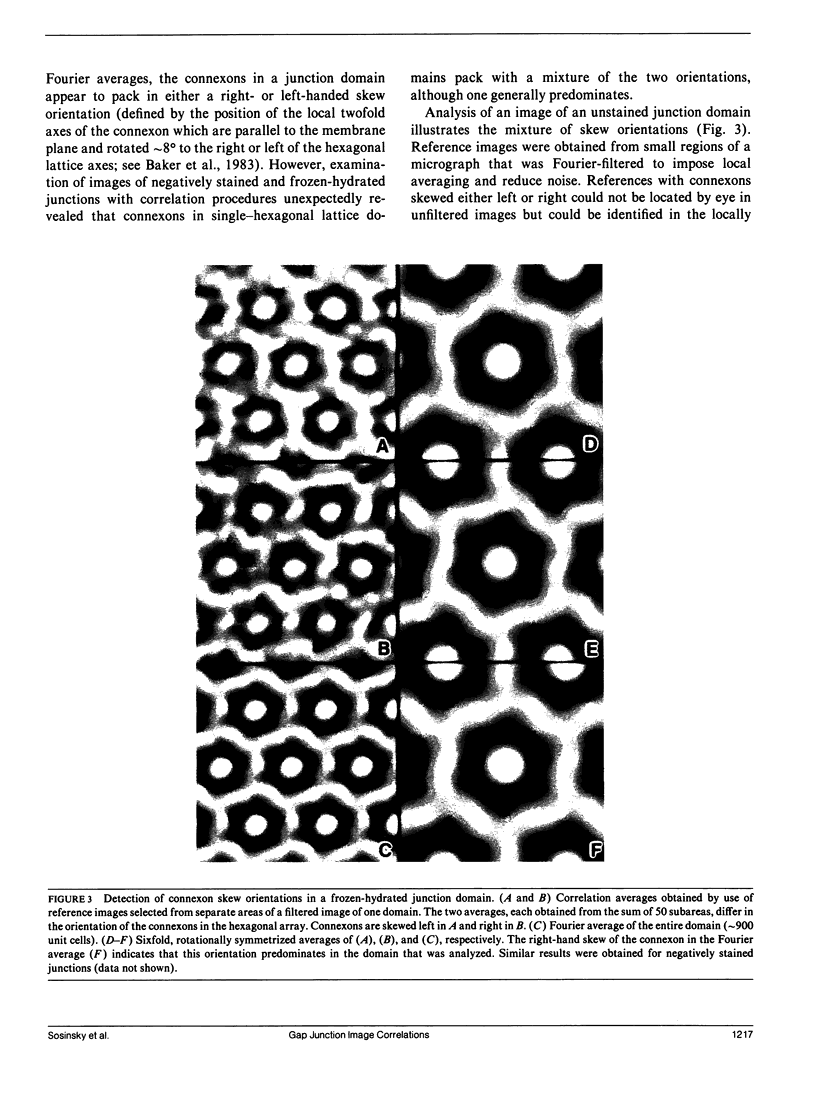
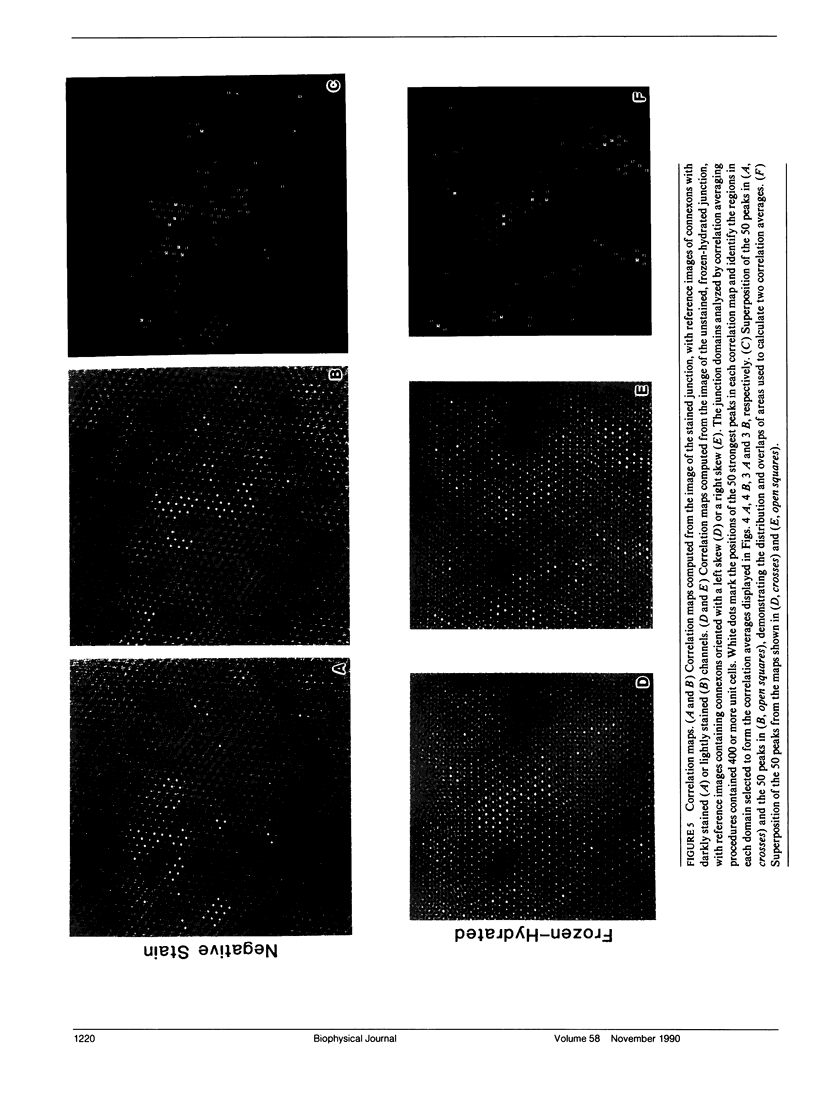
Abstract
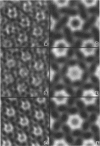
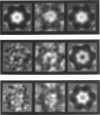
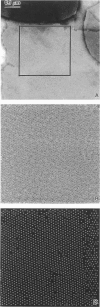
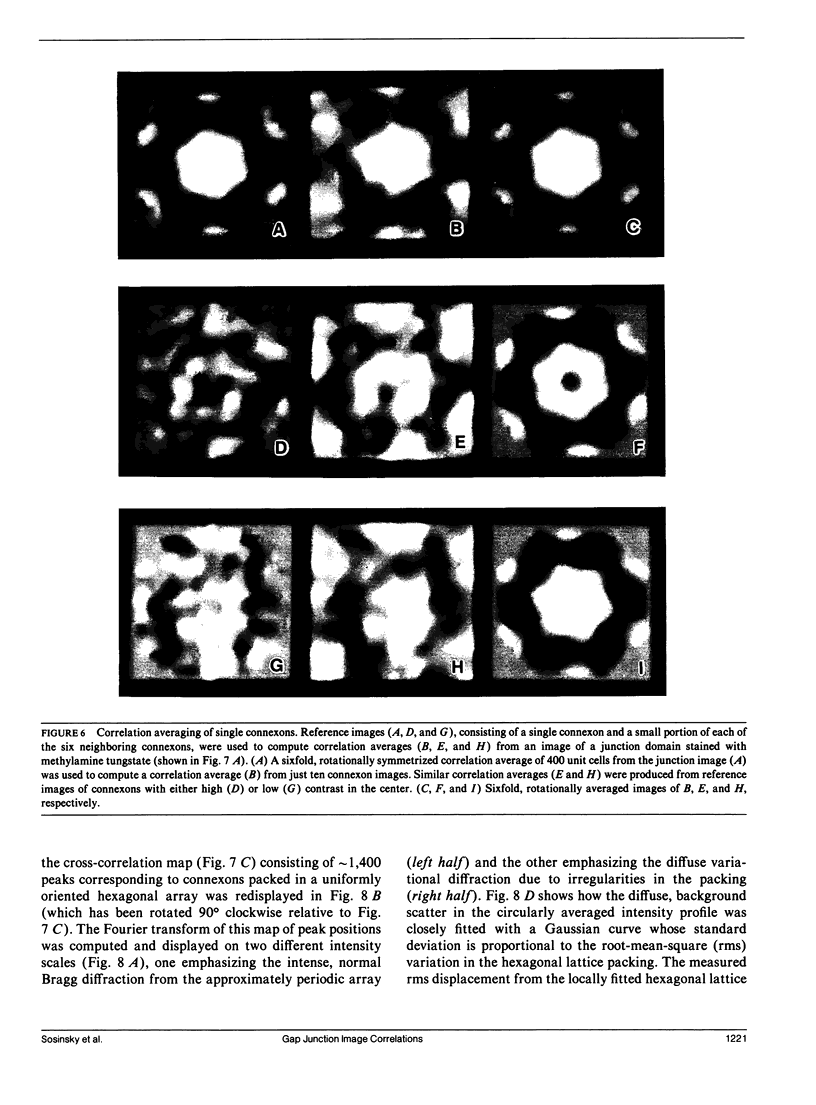
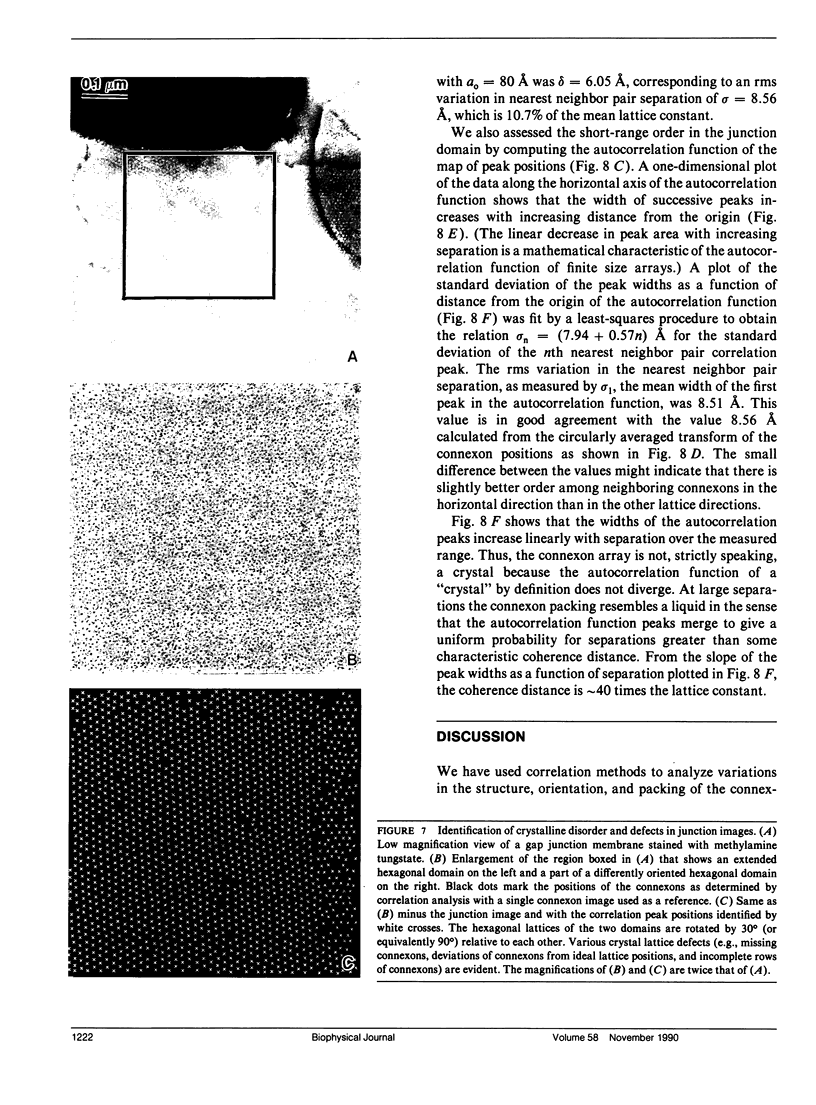
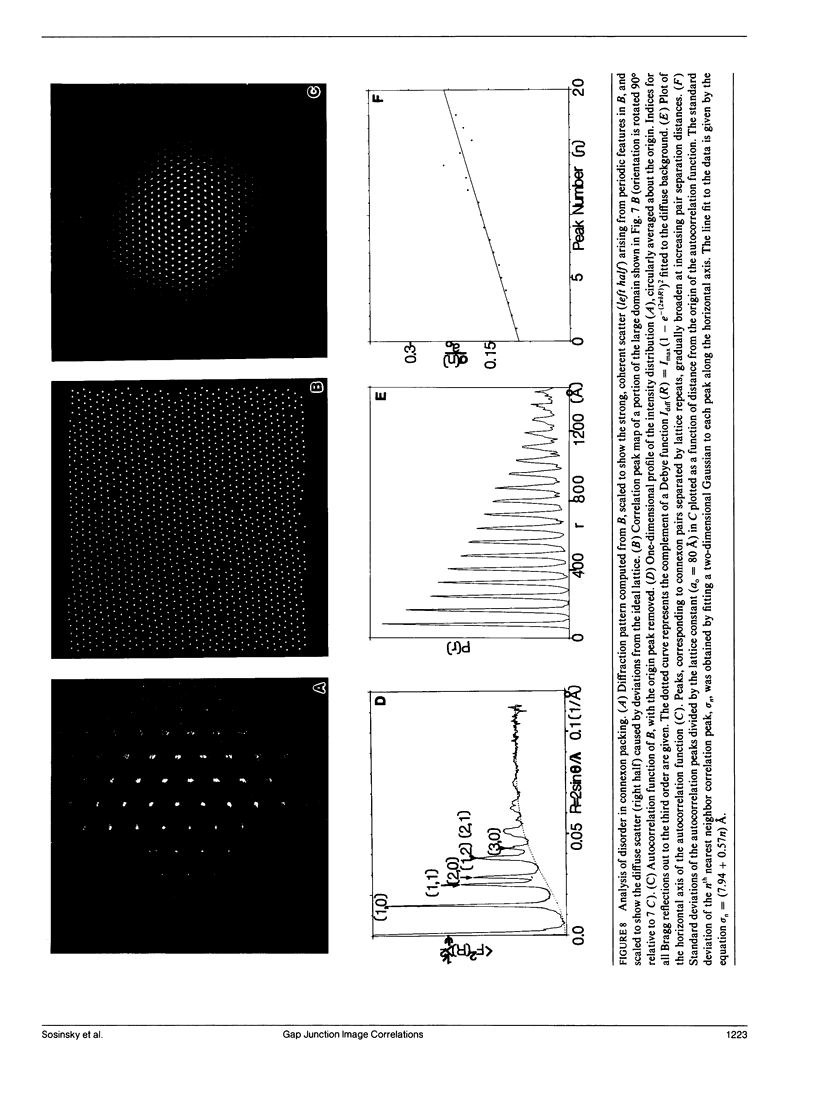
Fourier averages of connexon images computed from low-irradiation electron micrographs of isolated negatively stained gap junction domains exhibited differences in stain distribution and connexon orientation. To analyze these polymorphic structures, correlation averaging methods were applied to images from negatively stained and frozen-hydrated specimens. For the negatively stained specimens, separate averages over two subsets of connexons with differing degrees of stain accumulation in the axial channel were obtained. Two populations of connexons with opposite skew orientations were distinguishable within a single junctional domain of a frozen-hydrated specimen. Correlation maps calculated using the left- and right-skewed references showed that the selected connexons tend to locally cluster. Using correlation methods to analyze packing disorder in a typical connexon lattice, we estimated the root-mean-square variation in the nearest neighbor pair separation to be approximately 11% of the lattice constant. Displacements of the connexons relative to each other increased with increasing pair separation in the lattice, rather like a liquid, although long-range orientation order was conserved as in a crystal. These results support the hypothesis that the hexagonal ordering of the connexons results from short-range repulsive forces.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abney J. R., Braun J., Owicki J. C. Lateral interactions among membrane proteins. Implications for the organization of gap junctions. Biophys J. 1987 Sep;52(3):441–454. doi: 10.1016/S0006-3495(87)83233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. S., Caspar D. L., Hollingshead C. J., Goodenough D. A. Gap junction structures. IV. Asymmetric features revealed by low-irradiation microscopy. J Cell Biol. 1983 Jan;96(1):204–216. doi: 10.1083/jcb.96.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. S., Sosinsky G. E., Caspar D. L., Gall C., Goodenough D. A. Gap junction structures. VII. Analysis of connexon images obtained with cationic and anionic negative stains. J Mol Biol. 1985 Jul 5;184(1):81–98. doi: 10.1016/0022-2836(85)90045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V., Goodenough D. A. Gap junctions, electrotonic coupling, and intercellular communication. Neurosci Res Program Bull. 1978 Sep;16(3):1–486. [PubMed] [Google Scholar]
- Caspar D. L., Goodenough D. A., Makowski L., Phillips W. C. Gap junction structures. I. Correlated electron microscopy and x-ray diffraction. J Cell Biol. 1977 Aug;74(2):605–628. doi: 10.1083/jcb.74.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepeau R. H., Fram E. K. Reconstruction of imperfectly ordered zinc-induced tubulin sheets using cross-correlation and real space averaging. Ultramicroscopy. 1981;6(1):7–17. doi: 10.1016/s0304-3991(81)80173-8. [DOI] [PubMed] [Google Scholar]
- Dorset D. L., Massalski A. K., Rosenbusch J. P. In-plane phase transition of an integral membrane protein: nucleation of the OmpF matrix porin rectangular polymorph. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6143–6147. doi: 10.1073/pnas.86.16.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Chiu W., Degn L. The characterization of structural variations within a crystal field. Ultramicroscopy. 1988;26(4):345–360. doi: 10.1016/0304-3991(88)90234-3. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Heuser J. The inside and outside of gap-junction membranes visualized by deep etching. Cell. 1982 Sep;30(2):395–406. doi: 10.1016/0092-8674(82)90237-9. [DOI] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Baker T. S., Goodenough D. A. Gap junction structures. VI. Variation and conservation in connexon conformation and packing. Biophys J. 1984 Jan;45(1):208–218. doi: 10.1016/S0006-3495(84)84149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C. A., Frank J. Negative staining characteristics of arrays of mitochondrial pore protein: use of correspondence analysis to classify different staining patterns. Ultramicroscopy. 1984;13(1-2):93–102. doi: 10.1016/0304-3991(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Ribeiro A., Frank J. Structure of the Channels in the Outer Mitochondrial Membrane: Electron Microscopic Studies of the Periodic Arrays Induced by Phospholipase a(2) Treatment of the Neurospora membrane. Biophys J. 1986 Jan;49(1):307–317. doi: 10.1016/s0006-3495(86)83643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CA, Van Winkle DH Experimental observation of two-stage melting in a classical two-dimensional screened Coulomb system. Phys Rev Lett. 1987 Mar 23;58(12):1200–1203. doi: 10.1103/PhysRevLett.58.1200. [DOI] [PubMed] [Google Scholar]
- Saxton W. O., Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982 Aug;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Sosinsky G. E., Jésior J. C., Caspar D. L., Goodenough D. A. Gap junction structures. VIII. Membrane cross-sections. Biophys J. 1988 May;53(5):709–722. doi: 10.1016/S0006-3495(88)83152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]