Abstract
Analyses of low angle x-ray scattering from chromatin, isolated by identical procedures but from different species, indicate that fiber diameter and number of nucleosomes per unit length increase with the amount of nucleosome linker DNA. Experiments were conducted at physiological ionic strength to obtain parameters reflecting the structure most likely present in living cells. Guinier analyses were performed on scattering from solutions of soluble chromatin from Necturus maculosus erythrocytes (linker length 48 bp), chicken erythrocytes (linker length 64 bp), and Thyone briareus sperm (linker length 87 bp). The results were extrapolated to infinite dilution to eliminate interparticle contributions to the scattering. Cross-sectional radii of gyration were found to be 10.9 +/- 0.5, 12.1 +/- 0.4, and 15.9 +/- 0.5 nm for Necturus, chicken, and Thyone chromatin, respectively, which are consistent with fiber diameters of 30.8, 34.2, and 45.0 nm. Mass per unit lengths were found to be 6.9 +/- 0.5, 8.3 +/- 0.6, and 11.8 +/- 1.4 nucleosomes per 10 nm for Necturus, chicken, and Thyone chromatin, respectively. The geometrical consequences of the experimental mass per unit lengths and radii of gyration are consistent with a conserved interaction among nucleosomes. Cross-linking agents were found to have little effect on fiber external geometry, but significant effect on internal structure. The absolute values of fiber diameter and mass per unit length, and their dependencies upon linker length agree with the predictions of the double-helical crossed-linker model. A compilation of all published x-ray scattering data from the last decade indicates that the relationship between chromatin structure and linker length is consistent with data obtained by other investigators.
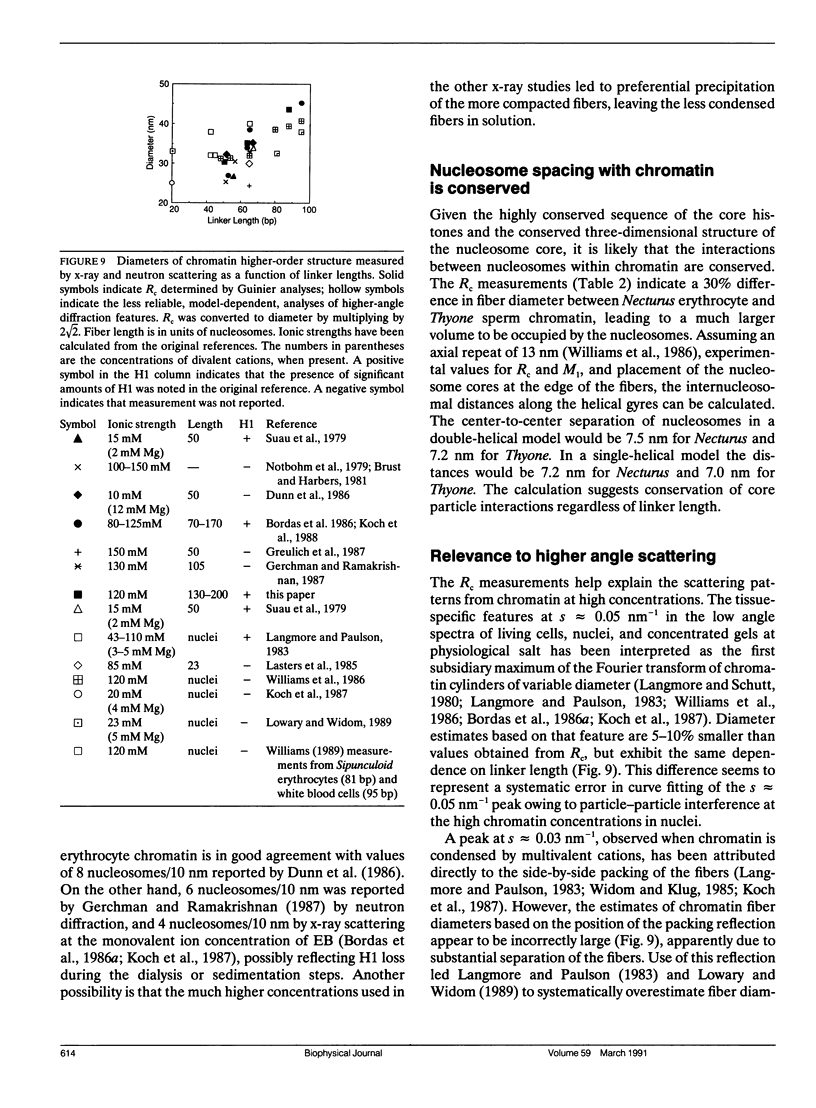
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alegre C., Subirana J. A. The diameter of chromatin fibres depends on linker length. Chromosoma. 1989 Jun;98(1):77–80. doi: 10.1007/BF00293338. [DOI] [PubMed] [Google Scholar]
- Athey B. D., Smith M. F., Rankert D. A., Williams S. P., Langmore J. P. The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J Cell Biol. 1990 Sep;111(3):795–806. doi: 10.1083/jcb.111.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Bernal J. D., Fankuchen I. X-RAY AND CRYSTALLOGRAPHIC STUDIES OF PLANT VIRUS PREPARATIONS : I. INTRODUCTION AND PREPARATION OF SPECIMENS II. MODES OF AGGREGATION OF THE VIRUS PARTICLES. J Gen Physiol. 1941 Sep 20;25(1):111–146. doi: 10.1085/jgp.25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. I. Synchrotron radiation X-ray scattering results. Eur Biophys J. 1986;13(3):157–173. doi: 10.1007/BF00542560. [DOI] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. II. Theoretical analysis of the X-ray scattering patterns and model calculations. Eur Biophys J. 1986;13(3):175–185. doi: 10.1007/BF00542561. [DOI] [PubMed] [Google Scholar]
- Brust R., Harbers E. Structural investigations on isolated chromatin of higher-order organisation. Eur J Biochem. 1981 Jul;117(3):609–615. doi: 10.1111/j.1432-1033.1981.tb06381.x. [DOI] [PubMed] [Google Scholar]
- Butler P. J. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. EMBO J. 1984 Nov;3(11):2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- CASPAR D. L. ASSEMBLY AND STABILITY OF THE TOBACCO MOSAIC VIRUS PARTICLE. Adv Protein Chem. 1963;18:37–121. doi: 10.1016/s0065-3233(08)60268-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Dimitrov S. I., Smirnov I. V., Makarov V. L. Optical anisotropy of chromatin. Flow linear dichroism and electric dichroism studies. J Biomol Struct Dyn. 1988 Apr;5(5):1135–1148. doi: 10.1080/07391102.1988.10506454. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J. J., Homo J. C., Lepault J., McDowall A. W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988 May;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerchman S. E., Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich K. O., Ausio J., Eisenberg H. Nucleosome core particle structure and structural changes in solution. J Mol Biol. 1985 Nov 5;186(1):167–173. doi: 10.1016/0022-2836(85)90266-9. [DOI] [PubMed] [Google Scholar]
- Greulich K. O., Wachtel E., Ausio J., Seger D., Eisenberg H. Transition of chromatin from the "10 nm" lower order structure, to the "30 nm" higher order structure as followed by small angle X-ray scattering. J Mol Biol. 1987 Feb 20;193(4):709–721. doi: 10.1016/0022-2836(87)90353-6. [DOI] [PubMed] [Google Scholar]
- KRATKY O. X-RAY SMALL ANGLE SCATTERING WITH SUBSTANCES OF BIOLOGICAL INTEREST IN DILUTED SOLUTIONS. Prog Biophys Mol Biol. 1963;13:105–173. doi: 10.1016/s0079-6107(63)80015-2. [DOI] [PubMed] [Google Scholar]
- Koch M. H., Sayers Z., Michon A. M., Marquet R., Houssier C., Willführ J. The superstructure of chromatin and its condensation mechanism. V. Effect of linker length, condensation by multivalent cations, solubility and electric dichroism properties. Eur Biophys J. 1988;16(3):177–185. doi: 10.1007/BF00261903. [DOI] [PubMed] [Google Scholar]
- Koch M. H., Vega M. C., Sayers Z., Michon A. M. The superstructure of chromatin and its condensation mechanism. III: Effect of monovalent and divalent cations X-ray solution scattering and hydrodynamic studies. Eur Biophys J. 1987;14(5):307–319. doi: 10.1007/BF00254896. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Paulson J. R. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983 Apr;96(4):1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmore J. P., Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980 Dec 11;288(5791):620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- Lasters I., Wyns L., Muyldermans S., Baldwin J. P., Poland G. A., Nave C. Scatter analysis of discrete-sized chromatin fragments favours a cylindrical organization. Eur J Biochem. 1985 Sep 2;151(2):283–289. doi: 10.1111/j.1432-1033.1985.tb09098.x. [DOI] [PubMed] [Google Scholar]
- Lowary P. T., Widom J. Higher-order structure of Saccharomyces cerevisiae chromatin. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8266–8270. doi: 10.1073/pnas.86.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARKHAM R., HITCHBORN J. H., HILLS G. J., FREY S. THE ANATOMY OF THE TOBACCO MOSAIC VIRUS. Virology. 1964 Mar;22:342–359. doi: 10.1016/0042-6822(64)90025-x. [DOI] [PubMed] [Google Scholar]
- Makarov V., Dimitrov S., Smirnov V., Pashev I. A triple helix model for the structure of chromatin fiber. FEBS Lett. 1985 Feb 25;181(2):357–361. doi: 10.1016/0014-5793(85)80292-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980 Nov;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988 Jun;9(6):255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- Pearson E. C., Butler P. J., Thomas J. O. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2(8):1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley A. S., Pardon J. F., Richards B. M. The relation between the unit thread of chromosomes and isolated nucleohistone. J Mol Biol. 1974 Jan 5;85(4):533–549. doi: 10.1016/0022-2836(74)90314-3. [DOI] [PubMed] [Google Scholar]
- Richards B. M., Pardon J. F. The molecular structure of nucleohistone (DNH). Exp Cell Res. 1970 Sep;62(1):184–196. doi: 10.1016/0014-4827(79)90519-6. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Sen D., Crothers D. M. Condensation of chromatin: role of multivalent cations. Biochemistry. 1986 Apr 8;25(7):1495–1503. doi: 10.1021/bi00355a004. [DOI] [PubMed] [Google Scholar]
- Smith M. F., Athey B. D., Williams S. P., Langmore J. P. Radial density distribution of chromatin: evidence that chromatin fibers have solid centers. J Cell Biol. 1990 Feb;110(2):245–254. doi: 10.1083/jcb.110.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau P., Bradbury E. M., Baldwin J. P. Higher-order structures of chromatin in solution. Eur J Biochem. 1979 Jul;97(2):593–602. doi: 10.1111/j.1432-1033.1979.tb13148.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Butler P. J. Salt-induced folding of sea urchin sperm chromatin. Eur J Biochem. 1986 Jan 15;154(2):343–348. doi: 10.1111/j.1432-1033.1986.tb09403.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Rees C. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem. 1983 Jul 15;134(1):109–115. doi: 10.1111/j.1432-1033.1983.tb07538.x. [DOI] [PubMed] [Google Scholar]
- Wang S. W., Robins A. J., d'Andrea R., Wells J. R. Inverted duplication of histone genes in chicken and disposition of regulatory sequences. Nucleic Acids Res. 1985 Feb 25;13(4):1369–1387. doi: 10.1093/nar/13.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J., Finch J. T., Thomas J. O. Higher-order structure of long repeat chromatin. EMBO J. 1985 Dec 1;4(12):3189–3194. doi: 10.1002/j.1460-2075.1985.tb04064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J., Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985 Nov;43(1):207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- Widom J. Toward a unified model of chromatin folding. Annu Rev Biophys Biophys Chem. 1989;18:365–395. doi: 10.1146/annurev.bb.18.060189.002053. [DOI] [PubMed] [Google Scholar]
- Williams S. P., Athey B. D., Muglia L. J., Schappe R. S., Gough A. H., Langmore J. P. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986 Jan;49(1):233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L., Rattner J. B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984 Jul;99(1 Pt 1):42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf H., Franke W. W. Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunits containing different numbers of nucleosomes. J Cell Biol. 1984 Jul;99(1 Pt 1):272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]