Abstract
About 2.5 million people die of Plasmodium falciparum malaria every year. Fatalities are associated with systemic and organ-specific inflammation initiated by a parasite toxin. Recent studies show that glycosylphosphatidylinositol (GPI) functions as the dominant parasite toxin in the context of infection. GPIs also serve as membrane anchors for several of the most important surface antigens of parasite invasive stages. GPI anchoring is a complex posttranslational modification produced through the coordinated action of a multicomponent biosynthetic pathway. Here we present eight new genes of P. falciparum selected for encoding homologs of proteins essential for GPI synthesis: PIG-A, PIG-B, PIG-M, PIG-O, GPI1, GPI8, GAA-1, and DPM1. We describe the experimentally verified mRNA and predicted amino acid sequences and in situ localization of the gene products to the parasite endoplasmic reticulum. Moreover, we show preliminary evidence for the PIG-L and PIG-C genes. The biosynthetic pathway of the malaria parasite GPI offers potential targets for drug development and may be useful for studying parasite cell biology and the molecular basis for the pathophysiology of parasitic diseases.
Plasmodium falciparum malaria ranks along with human immunodeficiency virus disease and tuberculosis as one the most serious infectious diseases of humanity (81). It infects 5 to 10% of the global population and kills approximately 2.5 million people annually. Malarial fatalities are strongly associated with an exacerbated systemic or organ-specific inflammatory cascade, which is initiated by a parasite toxin. This toxin induces the expression of proinflammatory cytokines tumor necrosis factor alpha and interleukin-1 from macrophages and may directly activate the vascular endothelium. (56). Furthermore, both cytokines and malarial toxins can each directly induce the expression of other proinflammatory loci such as that for inducible nitric oxide synthase (66), thereby raising levels of nitric oxide, which may be a regulator of pathogenesis (8). The syndromes seen in severe P. falciparum malaria in African children and in nonimmune adults may consist of organ-specific as well as multiorgan and system disturbances, including fever, metabolic acidosis, hypoglycemia, shock, and jaundice as well as renal failure, pulmonary edema, and cerebral involvement, including seizures and coma. Despite their diversity, these various signs, symptoms, and syndromes are thought to represent in part manifestations of an underlying inflammatory cascade driven by a parasite toxin.
Recent studies suggest that the glycosylphosphatidylinositol (GPI) of parasite origin functions as the dominant malarial toxin in the context of infection (56, 57, 66, 67, 68). GPIs of Trypanosoma brucei have the same function (68). These findings have been confirmed by others (42) and extended to Trypanosoma cruzi (2). These data support the view (57) that GPIs of the parasitic protozoa are the dominant proinflammatory agents in this class of eukaryotic pathogen.
GPIs are ubiquitous among eukaryotes, having been described for Trypanosoma (1, 15, 23), Plasmodium (18, 19), Leishmania (43, 47, 55), and Toxoplasma (73), as well as yeast (13), fish, plants, and numerous mammalian sources (27, 53, 82). Structurally related to phosphatidylinositol (PI), a membrane phospholipid, they consist of a conserved core glycan (Manα1-2Manα1-6Manα1-4GlcNH2) linked to position 6 of the myo-inositol ring of PI. The non-N-acetylated glucosamine is a characteristic feature of GPIs. The synthesis consists of sequential additions to PI, and the mannose (Man) units are numbered accordingly. An ethanolamine phosphate (EtNP) is added at position 6 on Man3 and can be used for attachment to a polypeptide.
GPIs are built up in the endoplasmic reticulum (ER) by the sequential addition of sugar residues to PI by the action of glycosyltransferases (65). At some stage during this process, the maturing GPI is translocated across the membrane from the cytoplasmic to the luminal side of the ER by an undefined mechanism (75, 76). After the GPI glycolipid is completed, it may be exported to the cell surface, free or in covalent association with proteins. GPI can be coupled to a protein via an amide bond between the terminal EtNP of the GPI and the protein C terminus formed by the removal of the terminal amino acids of the protein chain, the GPI signal sequence (74). The tetrasaccharide core glycan may be further substituted with sugars, phosphates, and ethanolamine groups in a species- and tissue-specific manner. GPI fatty acid moieties can be either saturated or unsaturated diacylglycerols, alkylacylglycerols, monoalkylglycerols, or ceramides, with additional acyl modifications to the inositol ring, variously C14:0, C16:0, C18:0, C18:1, and C18:2. The overall picture is of a closely related family of glycolipids sharing certain core features but with a high level of variation in fatty acid composition and side chain modifications to the conserved core glycan. The significance of the structural differences among GPIs is not yet clear, although structure-activity studies on GPI toxicity demonstrate important functional differences among GPIs with different fatty acid compositions (2, 7, 67).
The structure of the GPI toxin of P. falciparum has been elucidated and shown to consist of EtNP-6(Manα1-2)Manα1-2Manα1-6Manα1-4GlcNα1-6(acyl-2)myoIns-1-P-(sn1,2 diacyl)-glycerol (18). The acyl components of GPIs are probably variable; in P. falciparum a preference for palmitoyl at the glycerol and myristoyl at the inositol was described (19). Some differences between GPIs of mammals and Plasmodium have been identified, as follows. (i) Plasmodium GPIs lack any modification of Man2 and Man3, while in mammals and yeast GPIs always carry an EtNP side chain at the position 2 on Man1 and sometimes also carry this side chain at position 6 on Man2. (ii) The Man4 is present in most Plasmodium anchors but on only a minority of mammalian GPIs. (iii) The lipid moiety is predominantly 1-alkyl, 2-acyl glycerol in mammalian erythrocytes but is invariably a diacylglycerol in intraerythrocytic Plasmodium. However, diacylglycerol-containing mammalian GPIs from kidney and spleen have been described.
Among the GPI-anchored proteins in Plasmodium, the most important are the circumsporozoite protein, which coats the sporozoites (49), and the merozoite surface proteins MSP-1 and MSP-2, two leading vaccine candidates that are thought to be of major importance for the infection of red blood cells (25, 26, 62). Only a four-mannose structure was detected for the GPI moieties of MSP-1 and MSP-2 which accumulate during schizogony (19), but a form lacking the fourth mannose might be used to anchor other, unidentified surface proteins (54).
Elucidating malarial GPI toxin biosynthesis is a research priority for several reasons. First, GPI biosynthetic pathways are suitable targets for drug development. It was shown that a block in GPI synthesis by disruption of the PIG-B gene makes blood stages of T. brucei nonviable (48), and specific inactivators have been proposed as possible targets for chemotherapy against sleeping sickness (59, 61). While GPI synthesis is also important in mammals, especially for embryogenesis (71), mammalian cells in culture have been shown to survive well in culture without it (46). An antimalarial drug could be searched for by screening inhibitors specific for a Plasmodium enzyme involved in GPI biosynthesis. Second, manipulation of GPI toxin biosynthesis has the potential to generate nonvirulent or hypervirulent forms of the parasite and lead to an understanding of GPI toxicity. Both lines of research can profit from the knowledge about the biosynthetic genes described here.
The pathway and the genes that participate in GPI synthesis in yeast and mammals have been identified with a range of approaches, which have recently been reviewed (14, 34, 35). In Plasmodium, GPIs are the sole or major carbohydrate modification in proteins (20), whereas N- and O-linked carbohydrates are virtually or totally absent (10, 11). The Plasmodium genome may be expected to contain genes for only a small number of mannosyltransferases, possibly only those needed for GPI synthesis. We therefore sought to identify the genes for GPI synthesis by database mining and then to confirm the mRNA sequence and expression in P. falciparum. Eight genes are described here.
MATERIALS AND METHODS
Database mining.
We aimed at identifying all of the genes involved in GPI biosynthesis in P. falciparum. Sequences of genes known to participate in this pathway in different species were used as probes to search systematically for orthologs in data provided by the consortium of the Malaria Genome Project. Similarity searches were run on the malaria-specific tblastn server provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Malaria). To avoid paralogs (homolog genes with a different function), sequences similar to the query were required to pass a “reverse blast test,” whereby the identified fragment must unequivocally return the query and its orthologs as the best matches in a similarity search against a comprehensive protein database. The genomic region containing the selected fragment was used for a precise prediction of the complete protein-coding sequence. Tools for ab initio gene prediction gave results that were mostly obviously incorrect and therefore could not be used. The prediction was done by hand, guided by the multiple alignment between the predicted protein sequence and its orthologs. The alignment always included well-characterized sequences from mammalian and yeast species. Occasionally, hypothetical orthologs from additional species (especially from Arabidopsis thaliana), drawn from databases or predicted by us, were added to improve the informative value of the multiple alignment and to get a better measure of evolutionary variability. The exons were predicted by interactively localizing matching protein subsequences and splicing sites and joining exons over probable introns so as to obtain an optimized multiple alignment. In the case of PIG-M the prediction was difficult, and we worked in parallel on the P. falciparum and Plasmodium yoelii genomic sequences to obtain it.
Multiple alignments were performed with Clustal (24); searches for partial matches to regular expressions derived for short motifs were performed with Mapwhere. Additional criteria were used to ascertain whether candidates could still be false-positive matches, for example, pseudogenes. For the correct genes, the overall sequence similarity should be in good agreement with the degree expected from the evolutionary distance between the species; blocks which are strongly conserved between other species should be present, and the sequence should potentially encode a complete protein. For a summary of the sequence analysis for the final protein sequence, see Table 1 and Fig. 3 to 9.
TABLE 1.
Lengths numbers of introns, and chromosomal locations of P. falciparum genes involved in GPI biosynthesis and sequence similarities to homologs in human and yeast
| Gene | Chromosomea | No. of introns | Product length (amino acids) | % Amino acid identity (similarity)b
|
||
|---|---|---|---|---|---|---|
| P. falciparum vs. H. sapiens | P. falciparum vs. S. cerevisiae | S. cerevisiae vs. H. sapiens | ||||
| PIG-A | 10 | 6 | 461 | 46 (66) | 39 (64) | 45 (67) |
| GPI1 | Blob | 8 | 669 | 20 (46) | 23 (45) | 24 (47) |
| PIG-M | 12 | 7 | 441 | 37 (62) | 39 (61) | 39 (64) |
| PIG-B | 13 | 0 | 786 | 25 (50) | 24 (51) | 28 (51) |
| PIG-O | 12 | 2 | 1,238 | 24 (47) | 24 (46) | 30 (50) |
| GAA1 | 13 | 3 | 700 | 19 (40) | 20 (44) | 27 (48) |
| GPI8 | 11 | 0 | 493 | 30 (54) | 31 (56) | 47 (67) |
| DPMI | 11 | 0 | 259 | 49 (70) | 31 (48) | 32 (50) |
Chromosome assignments are based on database information, using the system provided by the Malaria Genome Consortium. Chromosomes 6 to 8 are not yet resolved and are referred to as the Blob.
Identity and similarity were computed for the optimal global pairwise alignment driven by scoring matrix BLOSUM45 with gap penalties 4 and 12. Similarity is defined by a positive score. The percentage is calculated with respect to the length of the shorter sequence. Accession numbers are provided in the figure legends.
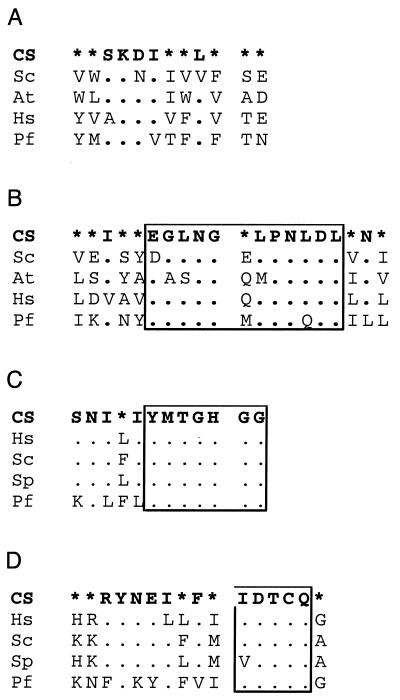
FIG. 3.
Best-conserved sequence blocks of PIG-A (A), GPI1 (B), and PIG-C (C). The top lines represent the consensus sequence (CS), which was defined when a residue occurred in at least 51% of the aligned sequences; otherwise, positions are indicated with asterisks. The species are H. sapiens (Hs), S. cerevisiae (Sc), S. pombe (Sp), A. thaliana (At), and P. falciparum (Pf). Dots in the alignments indicate the same amino acid as shown in the consensus; gaps are shown by dashes. The figure was prepared using ASAD (Keith Satterley, ftp://ftp.wehi.edu.au/pub/biology). (A) The best-conserved region in PIG-A. Of the 43 positions, 23 are invariant in the five species. Note that the sequence in P. falciparum adheres to the consensus to a similar degree as the other sequences. The protein from Arabidopsis has an insertion of three amino acids. Accession numbers are NP_002632 (Hs), NP_015150 (Sc), CAB09127 (Sp), and AAK62657 (At). (B) A conserved region of GPI1. These genes evolved faster, but a consensus can be defined for most positions in the conserved blocks. The sequence from P. falciparum again fits the consensus as expected. Some of the variable positions have retained the same character, for example, hydrophobicity. Accession numbers are AAC32661 (Hs), CAB10806 (Sp), AAB17870 (Sc), and NP_191276 (At). (C) A putative fragment of PIG-C in Plasmodium aligned with human and yeast PIG-C sequences. PIG-C is a short and not very strongly conserved protein. We have tentatively identified fragments of the possible homologs in P. falciparum and P. vivax (Pv), essentially based on the alignment shown. The two plasmodial sequences are almost identical over the 30 amino acids. Sources are the GSS sequence AZ573855 (Pv), the truncated cDNA AU087281 (Pf), and the protein submissions Q92535 (Hs) and AAB68262 (Sc).
FIG. 9.
Global alignment of the DPM1 protein sequences from A. thaliana (At), H. sapiens (Hs), and P. falciparum (Pf). The alignment shows a high degree of overall conservation. The only gaps are a short deletion in the Arabidopsis gene near the presumptive start codon and a one-amino-acid deletion at position 24 in the Plasmodium protein. The second aspartate in the boxed IDDGS motif and the first aspartate in the boxed MDAD motif are seen in a range of β-glycosyltransferases that transfer a single sugar unit and that have been suggested to participate in catalysis (53). Also boxed is a conserved GTRY motif, where the R is invariant in a broad family that includes sequences of unknown function annotated as DPM1-like proteins in databases. Accession numbers are AAH07073 (Hs) and NP_173481 (At). CS, consensus sequence. Symbols and figure preparation are as described in the legend to Fig. 3.
Determination of gene structure by sequencing.
Primers were designed to allow the generation by PCR of genomic and cDNA fragments, which could then be sequenced. For cDNA synthesis, RNA was extracted from parasite cultures consisting mainly of mid-stage trophozoites (plus 10 to 15% ring stages), using a Qiagen (Clifton Hill, Australia) RNeasy kit. After DNase treatment, first-strand cDNA was synthesized using an oligo(dT)15 primer and Superscript II reverse transcriptase (Life Technologies). Primers used in subsequent PCR experiments were 25 to 40 bp long and were used in pairs to amplify fragments of 300 to 1,200 bp. The same primers, along with some nested primers, were used to sequence the fragments, and the cDNA sequences were aligned with the genomic sequences to determine intron and exon positions. Some genes lacking introns and with clearly deducible amino acid sequences homologous to those of known genes were sequenced in less detail than more complicated genes. The sequence of DPM1 was not reconfirmed because no possibility for an intron was apparent, and cDNA sequences of GPI8 and GPI1 have been described by H. Shams-Eldin et al. in direct submissions to GenBank (accession numbers AJ401202 and AJ249657, respectively).
Antisera and immunohistochemistry.
To generate antisera, we determined oligopeptides with good predicted antigenicity on the basis of hydrophilicity (30, 52, 80) (at http://au.expasy.org/cgi-bin/protscale.pl) and a lack of cysteines. We chose the following sequences for peptide synthesis: anti-GAA1, YNNTNRIGKKIIRSST; anti-PIG-O, DKDKLKKNVNTLNEEN; anti-PIG-A, GKVKQENVKNILQTGH; anti-PIG-B, NEDNIKRNEKDENNGN; and anti-DPM1, HPKYIYNFIKKQREKN. Peptides were coupled to diphtheria toxoid, and 50 μg was emulsified in Freund's complete adjuvant and used to immunize mice, followed at 4-week intervals with two boosts of equal doses in Freund's incomplete adjuvant. Sera were collected into heparin and screened for reactivity to P. falciparum by an indirect fluorescent-antibody test. Thin films of parasites at mature stages were fixed in cold acetone and incubated with 1/80 dilutions of antibody. Localization to the parasite ER was determined by counterstaining with rabbit polyclonal antibodies to P. falciparum ERC1 (endoplasmic reticulum-located, calcium-binding protein [37]). The slides were washed extensively in phosphate-buffered saline and incubated with an appropriate mixture of fluorescein-conjugated anti-mouse and rhodamine-conjugated anti-rabbit antibodies (1:1,000). Preimmune sera served as negative controls. Slides were photographed under appropriate illumination for fluorescein isothiocyanate and rhodamine.
RESULTS AND DISCUSSION
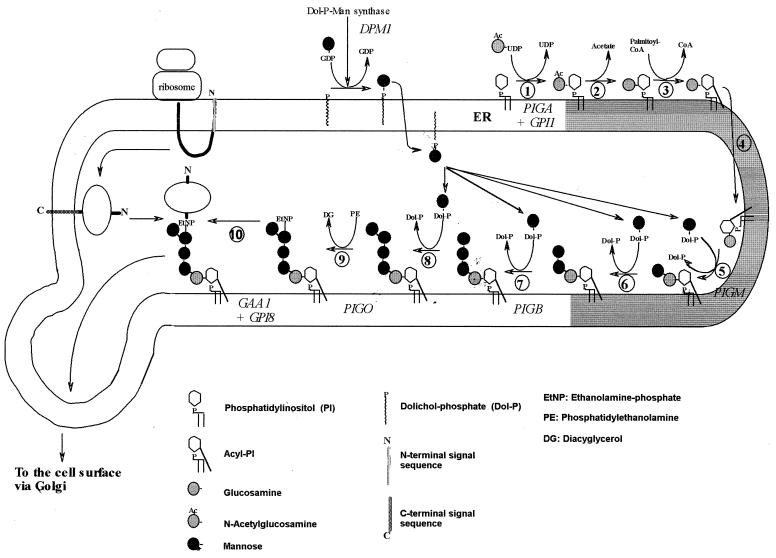
The aim of the project was to identify the full set of genes involved in GPI biosynthesis in Plasmodium. We summarize the present state of the evidence that we gathered for these genes. Kinoshita and Inoue (34) have subdivided mammalian GPI biosynthesis into 11 steps, and we have adapted their diagram for P. falciparum GPI biosynthesis (Fig. 1); based on the known structure of P. falciparum GPI, addition of EtNP to Man1 and Man2 is not expected, while an extra step for the addition of Man4 is included, resulting in 10 steps. Data suggest that steps 8 and 9 can occur in either order in P. falciparum. In this paper we present the sequences of proteins needed for steps 1 (PIG-A and GPI1), 5 (PIG-M), 7 (PIG-B), 9 (PIG-O), and 10 (GPI8 and GAA-1) and for the generation of the mannose donor dolichol-phosphate-mannose (Dol-P-Man) (DPM1). A candidate ortholog for step 2 (deacetylation of N-glucosamine by human PIG-L or yeast GPI12) was found very recently and has not yet been characterized. For steps 3 (inositol acylation), 4 (transport), and 6 (second mannosylation), no genes have been discovered in any species. No candidate gene for the addition of Man4 in step 8 (performed in yeast by Smp3) could be identified.
FIG. 1.
Proposed biosynthetic pathway of GPI in the ER of P. falciparum. The diagram is reproduced and modified with kind permission from Kinoshita and Inoue (34). In step 1, N-acetylglucosamine is added to PI, and the following steps involve deacetylation, lipidation of the inositol ring, and addition of mannoses. An EtNP on the third mannose enables the linkage of the completed GPI anchor to protein. There is some evidence that steps 8 and 9 can occur in the reverse order. Gene names are given in italics. (Reprinted from reference 34 with permission of the publisher.)
The protein and nucleotide sequences derived from our analysis are available at our website (http://www.wehi.edu.au/bioweb/Mauro/GPI). Table 1 gives information on the evolutionary conservation and lengths of the proteins, as well as the chromosome localization and structures of eight key genes. The proteins have evolved rather slowly, with a 40 to 70% similarity, which allows recognition by sequence, although nonconserved regions cannot be identified by similarity alone and required the exact determination of the position of the introns. PIG-A and DPM1 are strongly conserved proteins; GAA-1 is the most variable. Typically the similarities between the human and yeast sequences are, in accordance with the phylogenetic tree of the species, marginally higher than those with Plasmodium, except for DPM1 (Table 1). The PIG-O gene probably belongs to chromosome 12 but is also present in the BLOB file (chromosomes 6 to 8 as designated by the Malaria Genome Consortium).
The sequences of the experimentally determined intron-exon boundaries and their flanking sequences are shown in Table 2. The predictions based on the multiple alignment were almost perfectly correct. The localization of the start codon is based on the same alignment. The mapping of the exons proved that the genes are transcribed and spliced in a way compatible with coding for the predicted protein product. No verification was undertaken for GPI1 and GPI8, for which cDNA sequences are already available; for DPM1, where introns could be excluded with high confidence; and for PIG-L, whose genomic sequence was not available at the time. Two noncanonical splice sites were found (Table 2): a GC donor in the PIG-A gene and a very unusual CT-AC second intron in the PIG-O gene. This is possibly the first time a CT-AC splice site has been observed. In general, there is a high frequency of thymine in the last 40 bases of the introns (Table 2, splice acceptor), and a strong preference for adenine can be seen in the first 20 bases of the introns (Table 2, splice donor).
TABLE 2.
Sequences of intron-exon boundaries of the P. falciparum genes
| Gene | In- tron | Length (bp) | Sequencea
|
|
|---|---|---|---|---|
| Splice donor | Splice acceptor | |||
| GAA-1 | 1 | 172 | AAGTTATAGG gttagtaaag aaaaaaaaaa | atttgatata tctctttttt aatgtttcat acatttttag CGTTTTCTTA |
| 2 | 318 | ACTTTCTGAG gtacaaaaaa aaatatgaat | tttatttatt tatacatttc ttttgtgttt atccctttag AGAGAACATT | |
| 3 | 219 | ATTAAGTTCG gtaagtaacc ctaaaataaa | tatattttta tttttttttt ttatattcct ttttttttag TTATATAATT | |
| PIG-O | 1 | 160 | ATATACTAAA gtaatacaat ccacacatta | tacatttata tatatttata aatttttgtt ccttttgtag AGCCTTCATT |
| 2 | 225 | TGTAATACAT ctataaaaga aaaattttat | ttttttataa cgggacataa atttatttta aataacttac CACTAGTTTC | |
| PIG-A | 1 | 209 | ATAAAAAAGG gttatacata taaaaaaaaa | taatatttgt ttcaaatatt ttatttttat ttttttttag GTTTCAAGGT |
| 2 | 123 | TGGTCACCAG gtgcaagaac aaataaataa | tttatttatt tatttattat tattattatt atttttttag GCTACGTCAG | |
| 3 | 206 | CCACGAAAGG gtaaacatga gtttattaac | gttcatattt tttttttttt tttttttttt ttttatctag CTAACCAAAA | |
| 4 | 131 | GGAAAAGACG gtaaaatcac ataatacata | gtttttatgt ttattcttgt ttcattgttt tataatatag GAAAAGGTGT | |
| 5 | 86 | ACAGTTTTTA gtaatataaa aaataatata | tttatttata tatatatata tatattttcc attattgtag GCATAATTTA | |
| 6 | 147 | TCATGCCAAG gcaagattaa aaagaagaaa | atatataagc ctattcatgt ttcctttttt ttttttttag ACAAAATATT | |
| PIG-M | 1 | 150 | TCTATATCAT gtaagtattc aaatatatgt | ttttaaaaga atacattatt attttttttt tttatatcag ATATGGATAT |
| 2 | 124 | GAATTCAAAG gtaataataa taataaaaaa | tatatatata tatatatata tgttgttttt ttctttaaag ATTATTCCCT | |
| 3 | 78 | GTTTTTACAA gtatataata attaatatat | aatatatata tatatatata tatgtgtgtg ttttttttag ACTATTTCTT | |
| 4 | 179 | CACATCTCAG gtaaataaag gagaaattaa | ctacaaattt catataattt atatttaatt tattttttag TATTTCATTT | |
| 5 | 103 | CTTAAGCAAG gtaataaaaa aaaaagaaaa | atatatatat atatatatta tttttttttt tttaatgcag AGAAATATGT | |
| 6 | 169 | TGTGGCAAAA gtgggctcaa aaagatttct | tactacatat gtattatatt atattttatt ttatttttag TTGCATTGGC | |
| 7 | 85 | CTTTTTACAA gtaatgaatt tctttatttt | ttttataata tatattcttt tattctcaca tattttatag TTATTTTACT | |
| Consensus | ********AG gtaa*aaaaa aaa*aa*aaa | ttt**t***t t*t*t*tttt tttttttttt ttttttttag *********T | ||
The exon is in uppercase, and the intron is in lowercase. There are two noncanonical splice sites (boldface). A GC donor in the PIG-A gene and a very unusual CT-AC second intron in the PIG-O gene are shown. The last line shows the consensus across these 18 introns. The consensus was defined when residues occurred in at least 51% of the aligned sequences; otherwise, positions are indicated by asterisks.
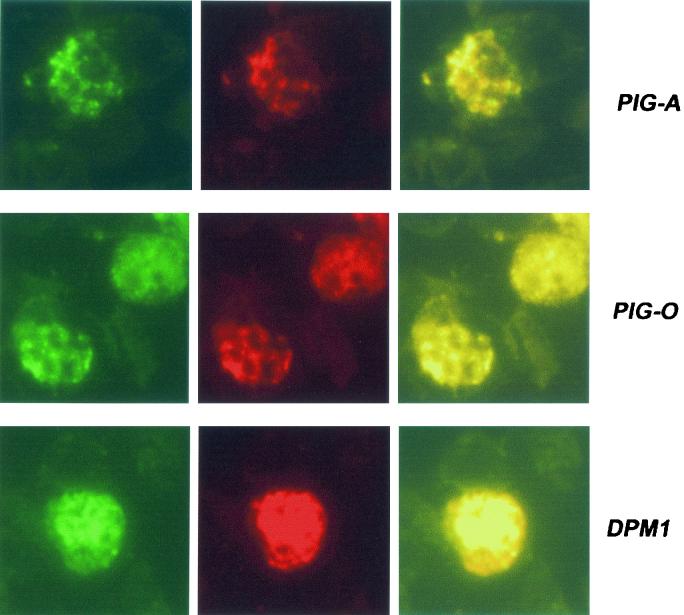
To determine whether the genes were expressed in blood stages of the parasite, antibodies were raised against synthetic peptides for the proteins PIG-A, PIG-B, PIG-O, GAA-1, and DPM-1. Immunofluorescence assays detected each of the proteins in late stages of erythrocyte infection (Fig. 2), suggesting that we have successfully identified the correct protein-encoding genes. PIG-A, PIG-O, and DPM1 colocalized with an ER-located, calcium-binding protein, ERC1 (Fig. 2). Similar results were obtained with antibodies raised against GAA1. PIG-B also colocalized with ERC1, but in a more restricted fluorescence pattern (data not shown).
FIG. 2.
Detection of proteins in blood stage parasites was performed using indirect fluorescent-antibody tests. Thin films of parasites at mature stages were fixed in cold acetone and incubated sequentially with 1/80 dilutions of sera from peptide-immunized mice plus rabbit anti-ERC1 (1:200) to determine localization to the parasite ER. These slides were washed and incubated with an appropriate mixture of fluorescein-conjugated anti-mouse and rhodamine-conjugated anti-rabbit (1:1.000) antibodies. Preimmune sera served as negative controls (data not shown). Slides were photographed under appropriate illumination for fluorescein isothiocyanate (first column, showing PIG-A, PIG-O, or DPM1 localization) and rhodamine (second column, showing ERC1 localization), and photographs were overlaid (third column).
Genes identified for each step of the GPI biosynthetic pathway. (i) Step 1: transfer of GlcNAc from UDP-GlcNAc to PI to form GPI.
In mammalian cells, at least six proteins have been linked to the transfer of GlcNAc from UDP-GlcNAc to PI to form GPI (29, 32, 45, 78). The catalytic center is probably provided by PIG-A, whose yeast ortholog (GPI3) has been shown to bind the substrate UDP-GlcNAc (36). Its sequence reveals motifs shared with a large family of glycosyltransferases, which transfer activated sugars from different nucleotide carriers to a variety of substrates, including bacterial lipopolysaccharides. These motifs are also encoded in the P. falciparum gene that we have identified. The best conserved block, possibly encompassing the active site, is shown in Fig. 3A.
The GPI1 gene of P. falciparum was identified by its ability to complement a GPI1-defective yeast strain (cDNA clone with GenBank accession number AJ249657) (58). A comparison between the cDNA and the genomic sequence reveals eight introns and the apparent existence of a microexon 11 nucleotides in length (not shown).
With the exception of what might be a fragment of PIG-C (Fig. 3C), no homolog for the other mammalian genes implicated in this step (PIG-H, PIG-P, and DPM2) could be found. The roles of these genes are not yet understood. The genes themselves are not vertebrate specific, as we found convincing sequence homologs for all of them in nonvertebrate and nonanimal species (not shown), such as Schizosaccharomyces pombe. DPM2 was first identified as a gene required for assisting the transfer of mannose units from dolichol phosphate by the catalytic DPM1 (40), suggesting that it actually plays an indirect accessory role in both reactions. As a homolog gene has not been identified in the completed sequence of the genome of Saccharomyces cerevisiae, DPM2 appears to be dispensable for GPI synthesis in some species, so it is plausible that Plasmodium may not require the PIG-H, PIG-P, and DPM2 genes, although a final judgment is not yet possible.
(ii) Step 2: deacetylation to GlcN-PI.
The second reaction also takes place on the cytoplasmic side of the ER, and the catalyzing enzyme, N-acetylglucosaminylphosphatidylinositol de-N-acetylase, is encoded by the PIG-L gene in humans (79). When we started our work we found only a short expressed sequence tag sequence of Plasmodium berghei that potentially encodes 60 amino acids with significant similarity to yeast and human PIG-L, particularly in a dodecamer motif, AHPDDEXMFFXP (Fig. 4). Recently, genomic sequences that contain this presumptive PIG-L gene have been made available in databases for P. yoelii, P. falciparum, and Plasmodium knowlesi. The best local alignment extends the one shown in Fig. 4 by about 50 amino acids (across an intron) and contains a second motif, YGVSGHPNHIS, which is invariant in the three Plasmodium species (not shown).
FIG. 4.
The identification of PIG-L in plasmodia is based on an expressed sequence tag sequence (BF295271) from P. berghei (Pb) and a genomic fragment from P. yoelii (Py). Their sequences are aligned to the PIG-L proteins of H. sapiens (Hs) and S. cerevisiae (Sc), with accession numbers BAA74775 (Hs) and BAA74776 (Sc). The numbers indicate the length of a poorly aligned region in the middle which is not shown. CS, consensus sequence. Symbols and figure preparation are as described in the legend to Fig. 3.
(iii) Steps 5, 7, and 8: addition of mannoses.
The three mannose units in the GPI core are linked by α-1,4, α-1,6. and α-1,2 bonds. The three enzymes are not expected to be closely related. The fourth mannose is added in α-1,2 linkage like the third one. While the human Man3 transferase (PIG-B) has been known for some time (69), the Man1 and Man4 transferases, called PIG-M and Smp3 (yeast), respectively, have just recently been discovered (21, 41). However, no transferase for Man2 has been described so far. In P. falciparum we found genes encoding putative Man1 and Man3 transferases (PIG-B and PIG-M) (Fig. 5), which align very well to their counterparts, but not an ortholog of Smp3 (the most similar gene is PIG-B). Very close relatives of Smp3 can easily be found for various species (S. pombe, Drosophila melanogaster, and Homo sapiens). Assuming that a similar degree of conservation extends to Plasmodium, the failure to identify an Smp3 homolog could be due to its absence in the database to date. The P. falciparum genome sequence is thought to be almost complete, but substantial fragments that are difficult to clone could still be missing, at least for chromosomes 6 to 8 (the “Blob”). Alternatively, a different gene might perform this function in Plasmodium.
FIG. 5.
Two well-conserved sequence blocks characteristic of the α-1,2-mannosyltransferase PIG-B. The top line represents the consensus sequence (CS). The species are A. thaliana (At), H. sapiens (Hs), S. cerevisiae (Sc), S. pombe (Sp), T. brucei (Tb), Zymomonas mobilis (Z), and P. falciparum (Pf). The Zymomonas protein is a bacterial sequence homolog of unproven function. It serves to tentatively identify the best-conserved positions, which are more likely to be essential for function. (A) The best-conserved sequence block in PIG-B follows the first transmembrane domain and is one of the few hydrophilic segments. Of the 40 positions, only 8 are invariant (12 without Zymomonas), but the consensus can be defined for most positions (27). The bottom line shows that a homologous block of sequence is also present in the Smp3 glycosyltransferase of yeast, with most residues similar and 11 identical to the consensus sequence. (B) The second well-conserved block of PIG-B and Smp3 is also present in other α-1,2-mannosyltransferases, for example, in yeast ALG9 (accession number NP_014180). Accession numbers for PIG-B are CAC01884 (At), BAA07709 (Hs), CAA96854 (Sc), CAB53078 (Sp), BAA94863 (Tb), and AAD53921 (Z); that for yeast Smp3 is NP_014792. Symbols and figure preparation are as described in the legend to Fig. 3.
The mannosyltransferases form a superfamily. Interestingly, the general organization of the PIG-M protein is similar to that of PIG-B and Smp3, which is presumably a sign of a common origin, although conservation at the amino acid level has been lost completely over evolutionary time. The PIG-B, Smp3, and PIG-M proteins all have multiple potential transmembrane domains interspersed with short hydrophilic loops, most of which are very short. The first transmembrane domain is followed by the longest hydrophilic region (about 30 amino acids). As an exception, the P. falciparum PIG-B has two additional long hydrophilic insertions. This tendency to have additional hydrophilic stretches has been observed in other proteins of P. falciparum, and we have verified that these are not intronic sequences. The first transmembrane domain contains a characteristic arginine in the middle, which is conserved in 17 of 18 PIG-B/PIG-M/Smp-3 homologs that we have aligned (not shown). In each of the three families, other hydrophobic stretches also show one conserved charged or hydrophilic residue. These residues might be engaged in strong inter- or intramolecular contacts in the membrane. The presence of one or several hydrophilic residues within a transmembrane domain is a common feature of ER-resident proteins that contributes to their localization, and the efficacy is stronger for amino acids D and R and when the hydrophilic residues are positioned in the middle of a transmembrane domain (6, 38, 39).
There are structural similarities between the α-1,2-mannosyltransferases. Database analysis suggests that the Smp3 gene is very conserved, with five or six well-defined motifs (not shown). Two of them correspond to the only two blocks of strong similarity in the PIG-B genes (Fig. 5). Among many others, three strongly related proteins, with National Center for Biotechnology Information identifiers 1302525 (YNR030w, S. cerevisiae), 3738170 (S. pombe), and 12804615 (H. sapiens), whose function seems to be unknown, also have a similar motif and a general similarity to the PIG-B sequences and therefore probably form another family of related but yet-uncharacterized glycosyltransferases.
In the PIG-M α-1,4-mannosyltransferases, a few sequence blocks are very well conserved and likely encode a critical function. The PIG-M proteins are slightly shorter than PIG-B and Smp3. Again, the first transmembrane domain, with the conserved R, is followed by one of the best conserved motifs (Fig. 6), embedded in a relatively hydrophilic short loop. In contrast to the case for PIG-B, there is also some degree of conservation in the terminal parts of the PIG-M proteins. Most notably, all of six presumptive PIG-M sequences (those in Fig. 6 and one from Caenorhabditis elegans), have a lysine as the third-to-last amino acid (not shown). Lysines near the carboxy terminus and particularly at position −3 have been implicated in the ER retention of proteins, especially a dilysine motif in type Ia ER membrane proteins (33). The lysines are preferentially located at the third- and fourth-to-last positions (KKXX-COOH motif), but variations in position and some replacements with arginine are not uncommon (60, 72). A second positively charged K or R residue is indeed present at position −4 or −5 in many (but not all) PIG-M proteins, reinforcing the hypothesis of a function as a retention signal. In contrast to the similarities at the protein level, the gene structures of P. falciparum PIG-B and PIG-M differ considerably in that the first gene has no introns while the second has seven.
FIG. 6.
Conserved sequence blocks of the α-1,4-mannosyltransferase PIG-M. A. The first well-conserved block of PIG-M is one of the few hydrophilic regions and closely follows the first transmembrane domains (like in PIG-B). The sequence aligns very poorly with that of the α-1,2-mannosyltransferases. It is strongly conserved among the orthologs, with 15 invariant positions in 36 amino acids (20 when the Trypanosoma sequence, which deviates most from the consensus, is omitted). The D[I,V]D motif (boxed) was identified as essential for activity. (B and C) Two more blocks are strongly conserved, suggesting a tight requirement for a function that remains to be identified. The species and accession numbers are as follows: A. thaliana (At), CAC34506; H. sapiens (Hs), BAB18567; S. cerevisiae (Sc), CAB89880; T. brucei (Tb), BAB20994; and P. falciparum (Pf). For Saccharomyces the nucleotide entry Z49907 was used, as the corresponding protein entries such as CAB89880 are probably based on the wrong start codon. CS, consensus sequence. Symbols and figure preparation are as described in the legend to Fig. 3.
(iv) Step 8: addition of EtNP.
Preliminary evidence produced in different laboratories indicates that the three enzymes that add the EtNP groups are PIG-N/MCD4 (EtNP addition to Man1), Gpi7 (Man2), and PIG-O/GPI13 (Man3) (4, 16, 17, 28, 70, 83). In P. falciparum only the terminal EtNP that serves as bridge to the protein is present. In addition, mammals also require PIG-F (31), which has an unknown biochemical function. In a phylogenetic tree with sequences from H. sapiens, S. cerevisiae, S. pombe, C. elegans, and D. melanogaster, the phosphoesterases cluster according to orthologous genes rather than taxonomic group (not shown). This suggests that development of the ability to add EtNP to all three mannoses preceded the evolutionary splitting of yeasts and animals, since convergent evolution seems implausible. In P. falciparum we identified, as expected, exactly one gene of the family. It clusters with the PIG-O subfamily. This reinforces the view that PIG-O is the Man3 phosphoesterase. It also suggests that the three genes existed before the splitting of Opisthokonta, Apicomplexa, and Plantae in early eukaryote evolution (3) and that two genes were lost in the evolutionary history of Plasmodium. A loss could be explained either as an adaptation to parasitic life and rapid growth or as a more specific selection of some biochemical property with functional importance.
The three phosphoesterases are all large proteins of approximately 100 to 120 kDa and 800 to 1100 amino acids, and they share a common organization, with a short hydrophobic segment (probably a signal sequence) near the N-terminal end, a large hydrophilic N-terminal half, and a hydrophobic second half that includes many potential transmembrane domains. Short conserved sequence motifs have been identified in the hydrophilic half (4, 16). The order of the three motifs is conserved, but in P. falciparum the highest similarity to the third one is shifted by about 100 amino acids due to a hydrophilic insertion. Motifs 1 and 2 (Fig. 7) are part of pattern 01663 in the Pfam motif library (63) derived from type I phosphodiesterases and nucleotide pyrophosphatases that catalyze the cleavage of phosphodiester bonds in NAD, deoxynucleotides, and nucleotide sugars. One might expect that motif 1 is involved in binding the EtNP and that motif 2 is involved in catalysis. Motif 3 is more specific for the phosphodiesterases of GPI synthesis (Fig. 7C). A partially similar block can be found in some other enzymes, i.e., phosphohexose mutase, phosphoglucomutase, and nucleoside diphosphate kinase (not shown). The meaning of this observation is unclear, but it might suggest a role in binding the GPI substrate. The second of many conserved hydrophobic stretches in the PIG-O gene products has very strongly conserved D and R residues with consensus O6-D-(GA)-L-R-O-D-O3, where O represents a hydrophobic amino acid. As noted above for the mannosyltransferases, this might be a transmembrane domain with charged residues engaged in protein interactions and/or involved in ER retention.
FIG. 7.
Sequence motifs in the GPI phosphoesterases. The species used are H. sapiens (Hs), S. cerevisiae (Sc), S. pombe (Sp), and P. falciparum (Pf). The sequences of yeast Gpi7 (CAA89353) and MCD4 (NP_012756) were added to compare regions that are common to all GPI phosphoesterases. MCD4 aligns poorly with the first block and is omitted there. (A) A region of strong homology between the phosphoesterases of the PIG-O family. The box highlights a PTX[ST]X8TGX2P motif present in many alkaline phosphatases. The plasmodial sequence deviates more from the consensus, but most of its substitutions are conservative. The number 5 represents an insertion of five residues in Gpi7. (B) The second block includes the boxed decamer motif HXLGXDHXGH that is also present in many peroxidases. (C) The third block is specific for the GPI phosphoesterases (see text) and may contact the GPI substrate. Accession numbers are AAC07985 (Hs), BAA21454 (Sp), and NP_013069 (Sc). CS, consensus sequence. Symbols and figure preparation are as described in the legend to Fig. 3.
(v) Step 10: covalent linking to the protein (transamidation).
The transamidation reaction requires at least two proteins, GAA-1 and GPI8 (5, 22, 84), which form a complex in mammalian cells (50). For both proteins we have identified the P. falciparum ortholog.
GPI8 is most probably the catalytic subunit, as it associates with substrate proteins (64, 77). It has homologies to proteinases (5, 12), and it is related to the caspase family of cysteine proteases. Mutational analysis has identified a cysteine and a histidine as being likely components of the active site and essential for the transamidation reaction (44, 50). Another common feature is the potential for a transmembrane domain near the C terminus. A cDNA for GPI8 which complements a GPI8 mutant yeast strain was obtained (H. Shams-Eldin et al., unpublished data). It has the expected amino acid sequence features (Fig. 8). A comparison with genomic DNA reveals that the gene has no introns.
FIG. 8.
(A and B) Alignment of the two best-conserved blocks characteristic of GAA-1. The protein evolved considerable divergence at the amino acid level, and only seven positions are invariant in the two aligned blocks (taken together) with the species A. thaliana (At), H. sapiens (Hs), S. cerevisiae (Sc), and P. falciparum (Pf). Accession numbers are CAA55944 (Sc), NP_197414 (At), and AAH03171 (Hs). (C and D). The histidine (C) and cysteine (D) in the boxed motifs of GPI8 are essential for the transamidation reaction (see text) in other species and are conserved in P. falciparum. Accession numbers are AAB81597 (Hs), P49018 (Sc), T40853 (S. pombe [Sp]), and AJ401202 (Pf). CS, consensus sequence. Symbols and figure preparation are as described in the legend to Fig. 3.
P. falciparum GAA1 is a gene with three introns that encodes a protein of 700 amino acids. GAA1 proteins are probably required for correct localization of GPI8 and are very hydrophobic. While this property is conserved, their sequence has diverged considerably, indicating a rather unspecific biochemical role. In a multiple alignment, only one or two short conserved blocks could be identified (Fig 8).
Two additional components of the transamidase complex were recently identified in humans and yeast: PIG-S/GPI17 and PIG-T/GPI16 (50). Homologous sequences can easily be found for animal and yeast species. The sequence of PIG-T appears to be under strong selection pressure, as it is highly conserved. A coding region with similarity to human PIG-T can be found in plasmodia. It is strongly conserved between P. falciparum, P. knowlesi, and P. yoelii, but the level of similarity to the other sequences is too low to identify it as a likely PIG-T ortholog (data not shown).
(vi) Auxiliary step: synthesis of the mannose donor Dol-P-Man.
DPM1 is the Dol-P-Man synthase that catalyzes the production of Dol-P-Man (51). The protein has been well characterized for many species. There is a P. falciparum sequence with strong similarity (Fig. 9). A very good alignment with almost no gaps is obtained with a conceptual translation of the genomic sequence, strongly suggesting that the gene has no introns. An uncertainty remains as to the transcriptional start site, as there are two AUG codons in an appropriate position, the first of which has been used for Fig. 9. As noted before (9), a phylogenetic tree curiously splits the proteins in two clusters (not shown), with those with a presumptive C-terminal transmembrane domain (Saccharomyces, Ustilago, Leishmania, and Trypanosoma) on one side and those without it (Homo, Caenorhabditis, Schizosaccharomyces, Arabidopsis, and Plasmodium) on the other side, while each species seems to have only one gene. This pattern is not easy to interpret, as Plasmodium is evolutionarily closer to Kinetoplastida and Schizosaccharomyces would be expected to cluster with other yeasts and fungi. It suggests convergent evolution in different evolutionary lines.
Conclusions.
To identify systematically the genes involved in the GPI pathway in Plasmodium, we used critical evaluation of amino acid sequence similarity data, exon prediction, confirmation by PCR, and protein localization. With the genome sequences at hand, it is thus possible rapidly to identify many candidates for further research. Except for Smp3, it seems that the genes for which no ortholog was identified are those likely to have a role more in stabilization or regulation than in catalysis. Either their sequences have diverged to the point of escaping similarity searches or the proteins are not required or are still absent in the Plasmodium databases. The sequence conservation between the genes in other species generally suggests that the Plasmodium sequence should be recognizable, assuming a relative evolutionary rate similar to those in the other genes. It seems plausible that the machinery for GPI synthesis has been reduced to minimal requirements in plasmodia: EtNP is not added to Man1 or Man2, and inositol is not deacylated. The only exception is the addition of Man4, which would not seem to be essential but may have a role in toxin activity. On the whole, fast-growing parasites such as Plasmodium might be under pressure to lose nonessential genes, in contrast to higher eukaryotes, where fine-tuned regulation is much more important. Targeted gene disruption and other genetic manipulations will now be possible and will help elucidate the function of parasite GPI in malaria, and some of the proteins that we have identified may be dissimilar enough to the human counterparts to be potential drug targets.
Acknowledgments
Genomic sequence data for P. falciparum were obtained from the Malaria Genome Project. Preliminary sequence data for P. falciparum chromosomes 10 and 11 were obtained from The Institute for Genomic Research (www.tigr.org), which was supported by an award from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md. Sequence data for P. falciparum chromosome 12 were obtained from the Stanford Genome Technology Center website (http://www-sequence.stanford.edu/group/malaria). Sequencing of P. falciparum chromosome 12 was accomplished as part of the Malaria Genome Project with support by the Burroughs Wellcome Fund. Sequence data for P. falciparum chromosomes 6 to 8 and 13 were obtained from The Sanger Institute website (http://www.sanger.ac.uk/Projects/P_falciparum/) with support by The Wellcome Trust.
This work was supported by a program grant from the Human Frontiers of Science Program, Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, Stiftung P. E. Kempkes, the UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases, NIH grant AI-45548, and a program grant from the NH&MRC. M.D. was supported in part by Schweizerischer Nationalfondsprojekt 20-50686.97. H.S.-E. thanks the Wilhelm Schaumann Foundation for a doctoral fellowship. L.S. is an International Research Scholar of the Howard Hughes Medical Institute.
Editor: R. N. Moore
REFERENCES
- 1.Acosta-Serrano, A., S. Schenkman, N. Yoshida, A. Mehlert, J. M. Richardson, and M. A. J. Ferguson. 1995. The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J. Biol. Chem. 270:27244-27253. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, I. C., M. M. Camargo, D. O. Procopio, L. S. Silva, A. Mehlert, L. R. Travassos, R. T. Gazzinelli, and M. A. Ferguson. 2000. Highly purified glycosylphosphatidylinositols from Trypanosoma cruzi are potent proinflammatory agents. EMBO J. 19:1476-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 4.Benachour, A., G. Sipos, I. Flury, F. Reggiori, E. Canivenc-Gansel, C. Vionnet, A. Conzelmann, and M. Benghezal. 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274:15251-15261. [DOI] [PubMed] [Google Scholar]
- 5.Benghezal, M., A. Benachour, S. Rusconi, M. Aebi, and A. Conzelmann. 1996. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15:6575-6583. [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S., P. Cosson, N. Shah, and R. D. Klausner. 1991. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 10:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camargo, M. M., A. C. Andrade, I. C. Almeida, L. R. Travassos, and R. T. Gazzinelli. 1997. Glycoconjugates isolated from Trypanosoma cruzi but not from Leishmania species membranes trigger nitric oxide synthesis as well as microbicidal activity in IFN-γ-primed macrophages. J. Immunol. 159:6131-6139. [PubMed] [Google Scholar]
- 8.Clark, I. A., and K. A. Rockett. 1996. Nitric oxide and parasitic disease. Adv. Parasitol. 37:1-56. [DOI] [PubMed] [Google Scholar]
- 9.Colussi, P. A., C. H. Taron, J. C. Mack, and P. Orlean. 1997. Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94:7873-7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieckmann-Schuppert, A., E. Bause, and R. T. Schwarz. 1993. Studies on O-glycans of Plasmodium falciparum-infected human erythrocytes: evidence for O-GlcNAc and O-GlcNAc-transferase in malaria parasites. Eur. J. Biochem. 216:779-788. [DOI] [PubMed] [Google Scholar]
- 11.Dieckmann-Schuppert, A., S. Bender, M. Odenthal-Schnittler, E. Bause, and R. T. Schwarz. 1992. Apparent lack of N-glycosylation in the asexual intraerythrocytic stage of Plasmodium falciparum. Eur. J. Biochem. 205:815-825. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhaber, B., P. Bork, and F. Eisenhaber. 2001. Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng. 14:17-25. [DOI] [PubMed] [Google Scholar]
- 13.Fankhauser, C., S. W. Homans, J. E. Thomas-Oates, M. J. McConville, C. Desponds, A. Conzelmann, and M. A. J. Ferguson. 1993. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J. Biol. Chem. 268:26365-26374. [PubMed] [Google Scholar]
- 14.Ferguson, M. A. 1999. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112:2799-2809. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, M. A., S. W. Homans, R. A. Dwek, and T. W. Rademacher. 1988. Glycosylphosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239:723-728. [DOI] [PubMed] [Google Scholar]
- 16.Flury, I., A. Benachour, and A. Conzelmann. 2000. YLL031c belongs to a novel family of membrane proteins involved in the transfer of ethanolaminephosphate onto the core structure of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 275:24458-24465. [DOI] [PubMed] [Google Scholar]
- 17.Gaynor, E. C., G. Mondesert, S. J. Grimme, S. I. Reed, P. Orlean, and S. D. Emr. 1999. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 10:627-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerold, P., A. Dieckmann-Schuppert, and R. T. Schwarz. 1994. Glycosylphosphatidylinositols synthesized by asexual erythrocytic stages of the malarial parasite, Plasmodium falciparum. Candidates for plasmodial glycosylphosphatidylinositol membrane anchor precursors and pathogenicity factors. J. Biol. Chem. 269:2597-2606. [PubMed] [Google Scholar]
- 19.Gerold, P., L. Schofield, M. Blackman, A. A. Holder, and R. T. Schwarz. 1996. Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol. Biochem. Parasitol. 75:131-143. [DOI] [PubMed] [Google Scholar]
- 20.Gowda, D. C., P. Gupta, and E. A. Davidson. 1997. Glycosylphosphatidylinositol anchors represent the major carbohydrate modification in proteins of intraerythrocytic stage Plasmodium falciparum. J. Biol. Chem. 272:6428-6439. [DOI] [PubMed] [Google Scholar]
- 21.Grimme, S. J., B. A. Westfall, J. M. Wiedman, C. H. Taron, and P. Orlean. 2001. The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 276:27731-27739. [DOI] [PubMed] [Google Scholar]
- 22.Hamburger, D., M. Egerton, and H. Riezman. 1995. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 129:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heise, N., M. L. Cardoso de Almeida, and M. A. J. Ferguson. 1995. Characterization of the lipid moiety of the glycosylphosphatidylinositol anchor of Trypanosoma cruzi 1G7-antigen. Mol. Biochem. Parasitol. 70:71-84. [DOI] [PubMed] [Google Scholar]
- 24.Higgins, D. G. 1994. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 25:307-318. [DOI] [PubMed] [Google Scholar]
- 25.Holder, A. A., and R. R. Freeman. 1981. Immunization against blood-stage rodent malaria using purified parasite antigens. Nature 294:361-364. [DOI] [PubMed] [Google Scholar]
- 26.Holder, A. A., M. J. Lockyer, K. G. Odink, J. S. Sandhu, M. V. Riveros, L. S. Davey, M. L. V. Tizard, R. T. Schwarz, and R. R. Freeman. 1985. Primary structure of the precursor to the three major surface antigens of Plasmodium falciparum merozoites. Nature 317:270-273. [DOI] [PubMed] [Google Scholar]
- 27.Homans, S. E., M. A. J. Ferguson, R. A. Dwek, T. W. Rademacher, R. Anand, and A. F. Williams. 1988. Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333:269-272. [DOI] [PubMed] [Google Scholar]
- 28.Hong, Y., Y. Maeda, R. Watanabe, N. Inoue, K. Ohishi, and T. Kinoshita. 2000. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275:20911-20919. [DOI] [PubMed] [Google Scholar]
- 29.Hong, Y., K. Ohishi, R. Watanabe, Y. Endo, Y. Maeda, and T. Kinoshita. 1999. GPI1 stabilizes an enzyme essential in the first step of glycosylphosphatidylinositol biosynthesis. J. Biol. Chem. 274:18582-18588. [DOI] [PubMed] [Google Scholar]
- 30.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue, N., T. Kinoshita, T. Orii, and J. Takeda. 1993. Cloning of a human gene, PIG-F, a component of glycosylphosphatidylinositol anchor biosynthesis, by a novel expression cloning strategy. J. Biol. Chem. 268:6882-6885. [PubMed] [Google Scholar]
- 32.Inoue, N., R. Watanabe, J. Takeda, and T. Kinoshita. 1996. PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem. Biophys. Res. Commun. 226:193-199. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita, T., and N. Inoue. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632-638. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita, T., K. Ohishi, and J. Takeda. 1997. GPI-anchor synthesis in mammalian cells: genes, their products, and a deficiency. J. Biochem. (Tokyo) 122:251-257. [DOI] [PubMed] [Google Scholar]
- 36.Kostova, Z., D. M. Rancour, A. K. Menon, and P. Orlean. 2000. Photoaffinity labelling with P3-(4-azidoanilido)uridine 5′-triphosphate identifies Gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem. J. 350:815-822. [PMC free article] [PubMed] [Google Scholar]
- 37.La Greca, N., A. R. Hibbs, C. Riffkin, M. Foley, and L. Tilley. 1997. Identification of an endoplasmic reticulum-resident calcium-binding protein with multiple EF-hand motifs in asexual stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 89:283-293. [DOI] [PubMed] [Google Scholar]
- 38.Lankford, S. P., P. Cosson, J. S. Bonifacino, and R. D. Klausner. 1993. Transmembrane domain length affects charge-mediated retention and degradation of proteins within the endoplasmic reticulum. J. Biol. Chem. 268:4814-4820. [PubMed] [Google Scholar]
- 39.Letourneur, F., and P. Cosson. 1998. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J. Biol. Chem. 273:33273-33278. [DOI] [PubMed] [Google Scholar]
- 40.Maeda, Y., S. Tomita, R. Watanabe, K. Ohishi, and T. Kinoshita. 1998. DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1, and binding of dolichol phosphate. EMBO J. 17:4920-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeda, Y., R. Watanabe, C. L. Harris, Y. Hong, K. Ohishi, K. Kinoshita, and T. Kinoshita. 2001. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20:250-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magez, S., B. Stijlemans, M. Radwanska, E. Pays, M. A. Ferguson, and P. DeBaetselier. 1998. The glycosyl-inositol-phosphate and dimyristoylglycerol moieties of the glycosylphosphatidylinositol anchor of the trypanosome variant-specific surface glycoprotein are distinct macrophage-activating factors. J. Immunol. 160:1949-1956. [PubMed] [Google Scholar]
- 43.McConville, M. J., S. W. Homans, J. E. Thomas-Oates, A. Dell, and A. Bacic. 1990. Structures of the glycoinositolphospholipids from Leishmania major: a family of novel galactofuranose-containing glycolipids. J. Biol. Chem. 265:7385-7394. [PubMed] [Google Scholar]
- 44.Meyer, U., M. Benghezal, I. Imhof, and A. Conzelmann. 2000. Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39:3461-3471. [DOI] [PubMed] [Google Scholar]
- 45.Miyata, T., J. Takeda, Y. Iida, N. Yamada, N. Inoue, M. Takahashi, K. Maeda, T. Kitani, and T. Kinoshita. 1993. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science 259:1318-1320. [DOI] [PubMed] [Google Scholar]
- 46.Mohney, R. P., J. J. Knez, L. Ravi, D. Sevlever, T. L. Rosenberry, S. Hirose, and M. E. Medof. 1994. Glycoinositol phospholipid anchor-defective K562 mutants with biochemical lesions distinct from those in Thy-1− murine lymphoma mutants. J. Biol. Chem. 269:6536-6542. [PubMed] [Google Scholar]
- 47.Murray, P. J., T. W. Spithill, and E. Handman. 1989. The PSA-2 glycoprotein complex of Leishmania major is a glycosylphosphatidylinositol-linked promastigote surface antigen. J. Immunol. 143:4221-4226. [PubMed] [Google Scholar]
- 48.Nagamune, K., T. Nozaki, Y. Maeda, K. Ohishi, T. Fukuma, T. Hara, R. T. Schwarz, C. Sutterlin, R. Brun, H. Riezman, and T. Kinoshita. 2000. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 97:10336-10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nussenzweig, V., and R. S. Nussenzweig. 1985. Circumsporozoite proteins of malaria parasites. Cell 42:401-403. [DOI] [PubMed] [Google Scholar]
- 50.Ohishi, K., Y. Kurimoto, N. Inoue, Y. Endo, J. Takeda, and T. Kinoshita. 1996. Cloning and characterization of the murine GPI anchor synthesis gene Pigf, a homologue of the human PIG-F gene. Genomics 34:340-346. [DOI] [PubMed] [Google Scholar]
- 51.Orlean, P., C. Albright, and P. W. Robbins. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263:17499-17507. [PubMed] [Google Scholar]
- 52.Parker, J. M., D. Guo, and R. S. Hodges. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425-5432. [DOI] [PubMed] [Google Scholar]
- 53.Roberts, W. L., S. Santikarn, V. N. Reinhold, and T. L. Rosenberry. 1988. Structural characterization of the glycoinositol phospholipid membrane anchor of human erythrocyte acetylcholinesterase by fast atom bombardment mass spectrometry. J. Biol. Chem. 263:18776-18784. [PubMed] [Google Scholar]
- 53a.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt, A., R. T. Schwarz, and P. Gerold. 1998. Plasmodium falciparum: asexual erythrocytic stages synthesize two structurally distinct free and protein-bound glycosylphosphatidylinositols in a maturation-dependent manner. Exp. Parasitol. 88:95-102. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, P., M. A. J. Ferguson, M. J. McConville, A. Mehlert, S. W. Homans, and C. Bordier. 1990. Structure of the glycosyl-phosphatidylinositol membrane anchor of the Leishmania major promastigote surface protease. J. Biol. Chem. 265:16955-16964. [PubMed] [Google Scholar]
- 56.Schofield, L., and F. Hackett. 1993. Signal transduction in host cells by a glycosylphosphatidylinositol toxin of malaria parasites. J. Exp. Med. 177:145-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schofield, L., and S. D. Tachado. 1996. Regulation of host cell function by glycosylphosphatidylinositols of the parasitic protozoa. Immunol. Cell. Biol. 74:555-563. [DOI] [PubMed] [Google Scholar]
- 58.Shams-Eldin, H., N. Azzouz, M. H. Kedees, P. Orlean, T. Kinoshita, and R. T. Schwarz. 2002. The GPI1 homologue from Plasmodium falciparum complements a Saccharomyces cerevisiae GPI anchoring mutant. Mol. Biochem. Parasitol. 120:73-81. [DOI] [PubMed] [Google Scholar]
- 59.Sharma, D. K., T. K. Smith, C. T. Weller, A. Crossman, J. S. Brimacombe, and M. A. J. Ferguson. 1999. Differences between the trypanosome and human GlcNA-PI de-N-acetylases of glycosylphosphatidylinositol membrane anchor biosynthesis. Glycobiology 9:415-422. [DOI] [PubMed] [Google Scholar]
- 60.Shin, J., R. L. J. Dunbrack, S. Lee, and J. L. Strominger. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc. Natl. Acad. Sci. USA 88:1918-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, T. K., D. K. Sharma, A. Crossman, A. Dix, J. S. Brimacombe, and M. A. J. Ferguson. 1997. Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J. 16:6667-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smythe, J. A., R. L. Coppel, G. V. Brown, R. Ramasamy, D. J. Kemp, and R. F. Anders. 1988. Identification of two integral membrane proteins of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 85:5195-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 64.Spurway, T. D., J. A. Dalley, S. High, and N. J. Bulleid. 2001. Early events in glycosylphosphatidylinositol anchor addition. Substrate proteins associate with the transamidase subunit gpi8p. J. Biol. Chem. 276:15975-15982. [DOI] [PubMed] [Google Scholar]
- 65.Stevens, V. L. 1995. Biosynthesis of glycosylphosphatidylinositol membrane anchors. Biochem. J. 310:361-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tachado, S. D., P. Gerold, M. J. McConville, T. Baldwin, D. Quilici, R. T. Schwarz, and L. Schofield. 1996. Glycosylphosphatidylinositol toxin of plasmodium induces nitric oxide synthase expression in macrophages and vascular endothelial cells by a protein tyrosine kinase-dependent and protein kinase C-dependent signaling pathway. J. Immunol. 156:1897-1907. [PubMed] [Google Scholar]
- 67.Tachado, S. D., P. Gerold, R. Schwarz, S. Novakovic, M. McConville, and L. Schofield. 1997. Signal transduction in macrophages by glycosylphosphatidylinositols of Plasmodium, Trypanosoma and Leishmania: activation of protein tyrosine kinases and protein kinase C by inositolglycan and diacylglycerol moieties. Proc. Natl. Acad. Sci. USA 94:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tachado, S. D., and L. Schofield. 1994. Glycosylphosphatidylinositol toxin of Trypanosoma brucei regulates IL-1α and TNF-α expression in macrophages by protein tyrosine kinase mediated signal transduction. Biochem. Biophys. Res. Commun. 205:984-991. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi, M., N. Inoue, K. Ohishi, Y. Maeda, N. Nakamura, Y. Endo, T. Fujita, J. Takeda, and T. Kinoshita. 1996. PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 15:4254-4261. [PMC free article] [PubMed] [Google Scholar]
- 70.Taron, C. H., J. M. Wiedman, S. J. Grimme, and P. Orlean. 2000. Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell 11:1611-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tarutani, M., S. Itami, M. Okabe, M. Ikawa, T. Tezuka, K. Yoshikawa, T. Kinoshita, and J. Takeda. 1997. Tissue-specific knockout of the mouse Pig-A gene reveals important roles for GPI-anchored proteins in skin development. Proc. Natl. Acad. Sci. USA 94:7400-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 73.Tomavo, S., R. T. Schwarz, and J. F. Dubremetz. 1989. Evidence for glycosyl-phosphatidylinositol anchoring of Toxoplasma gondii major surface antigens. Mol. Cell. Biol. 9:4576-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Udenfriend, S., and K. Kodukula. 1995. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64:563-591. [DOI] [PubMed] [Google Scholar]
- 75.Vidugiriene, J., and A. K. Menon. 1993. Early lipid intermediates in glycosylphosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol. 121:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vidugiriene, J., and A. K. Menon. 1994. The GPI anchor of cell-surface proteins is synthesized on the cytoplasmic face of the endoplasmic reticulum. J. Cell Biol. 127:333-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vidugiriene, J., S. Vainauskas, A. E. Johnson, and A. K. Menon. 2001. Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment: photocrosslinking studies in a cell-free system. Eur. J. Biochem. 268:2290-2300. [DOI] [PubMed] [Google Scholar]
- 78.Watanabe, R., N. Inoue, B. Westfall, C. H. Taron, P. Orlean, J. Takeda, and T. Kinoshita. 1998. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 17:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe, R., K. Ohishi, Y. Maeda, N. Nakamura, and T. Kinoshita. 1999. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 339:185-192. [PMC free article] [PubMed] [Google Scholar]
- 80.Welling, G. W., W. J. Weijer, R. van der Zee, and S. Welling-Wester. 1985. Prediction of sequential antigenic regions in proteins. FEBS Lett. 188:215-218. [DOI] [PubMed] [Google Scholar]
- 81.World Health Organization. 1992. World malaria situation 1990. World Health Stat. Q. 45:257-266. [PubMed] [Google Scholar]
- 82.Xia, M.-Q., G. Hale, M. R. Lifely, M. A. J. Ferguson, D. Campbell, L. Packman, and H. Waldmann. 1993. Structure of the CAMPATH-1 antigen, a glycosylphosphatidylinositol-anchored glycoprotein which is an exceptionally good target for complement lysis. Biochem. J. 293:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yada, T., R. Sugiura, A. Kita, Y. Itoh, Y. Lu, Y. Hong, T. Kinoshita, H. Shuntoh, and T. Kuno. 2001. Its8, a fission yeast homolog of Mcd4 and Pig-n, is involved in GPI anchor synthesis and shares an essential function with calcineurin in cytokinesis. J. Biol. Chem. 276:13579-13586. [DOI] [PubMed] [Google Scholar]
- 84.Yu, J., S. Nagarajan, J. J. Knez, S. Udenfriend, R. Chen, and M. E. Medof. 1997. The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl. Acad. Sci. USA 94:12580-12585. [DOI] [PMC free article] [PubMed] [Google Scholar]