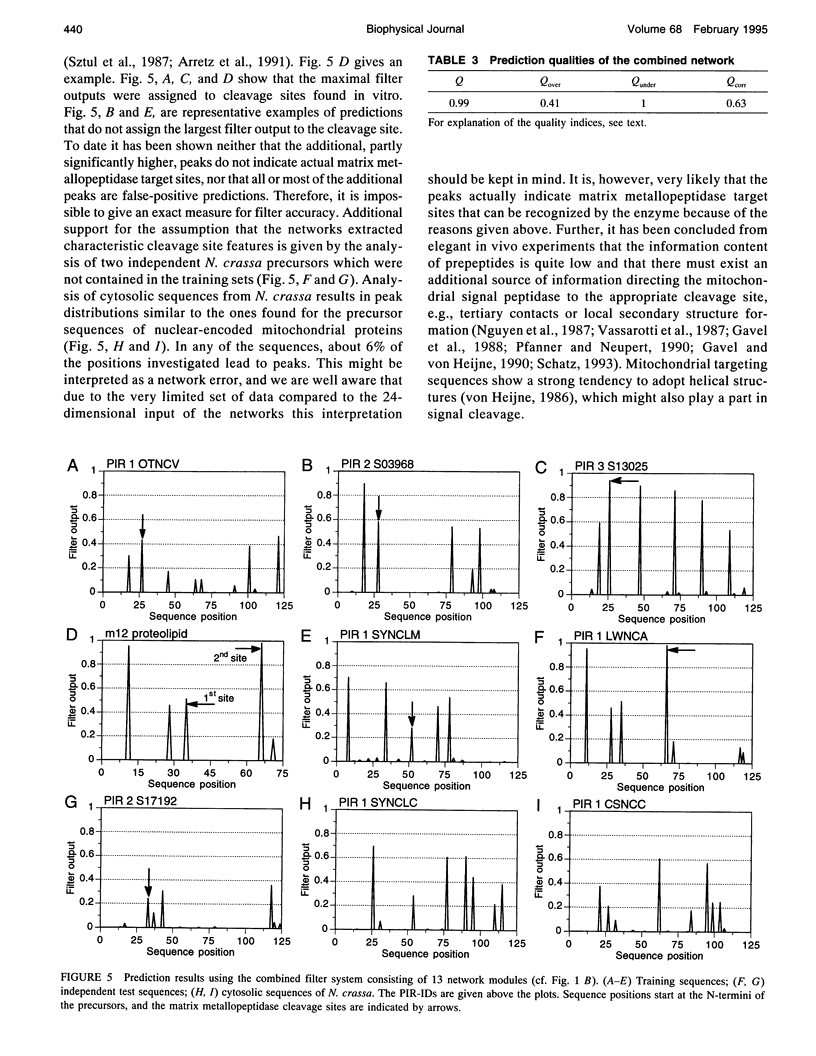
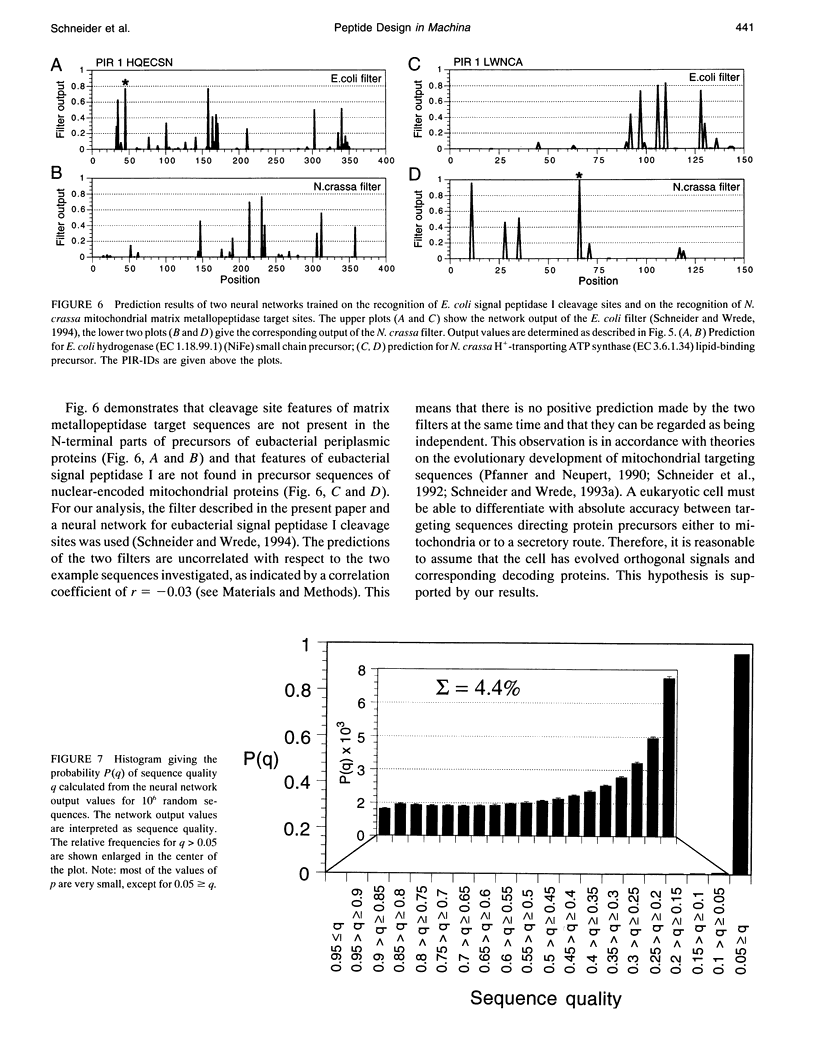
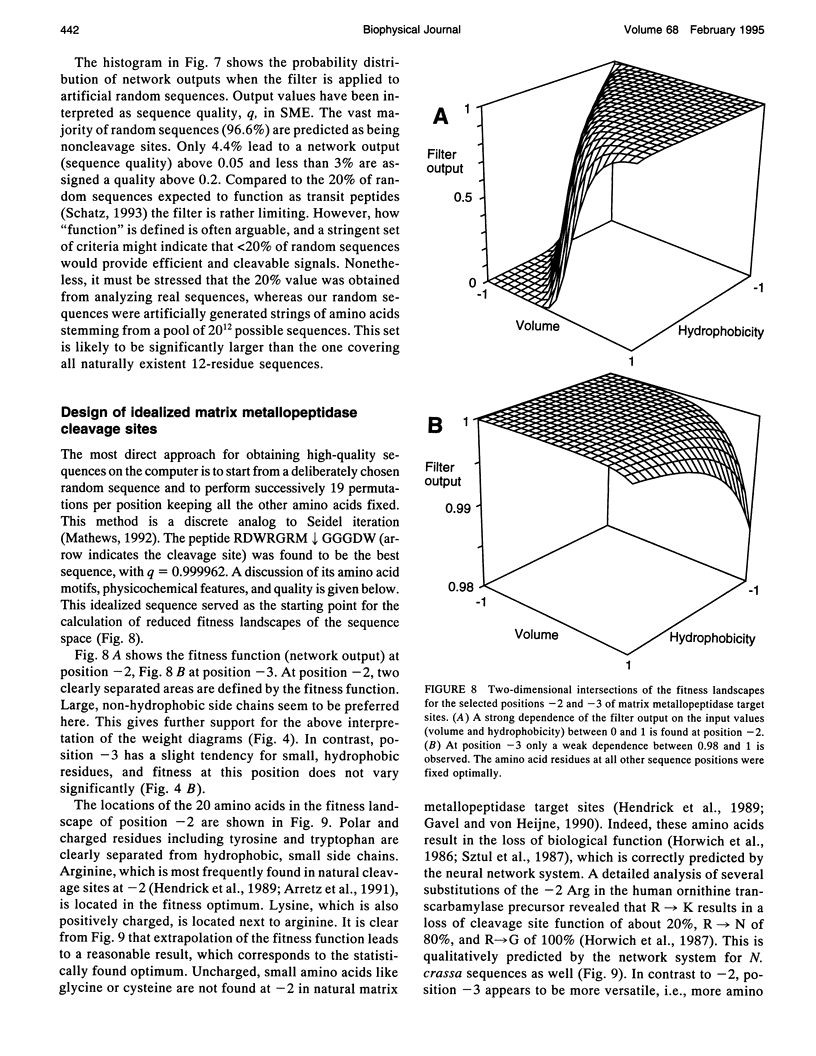
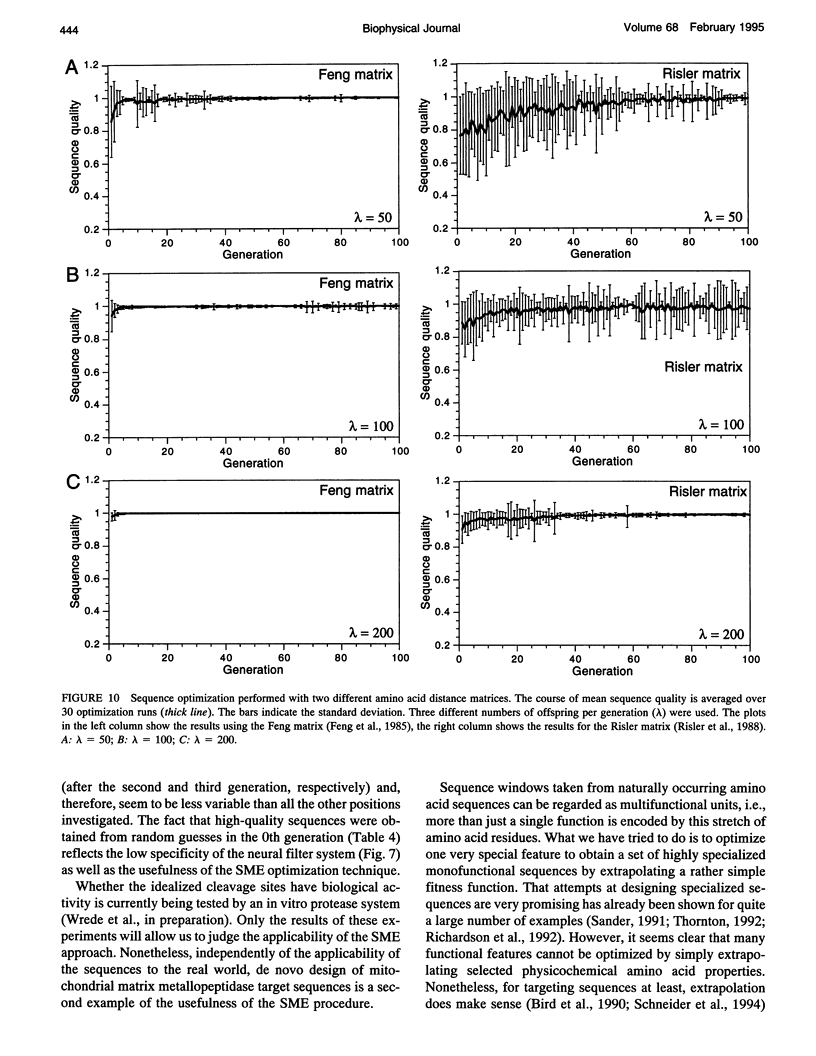
Abstract
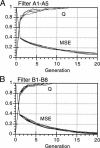
Artificial neural networks were used for extraction of characteristic physiochemical features from mitochondrial matrix metalloprotease target sequences. The amino acid properties hydrophobicity and volume were used for sequence encoding. A window of 12 residues was employed, encompassing positions -7 to +5 of precursors with cleavage sites. Two sets of noncleavage site examples were selected for network training which was performed by an evolution strategy. The weight vectors of the optimized networks were visualized and interpreted by Hinton diagrams. A neural filter system consisting of 13 perceptron-type networks accurately classified the data. It served as the fitness function in a simulated molecular evolution procedure for sequence-oriented de novo design of idealized cleavage sites. A detailed description of the strategy is given. Several putative high-quality cleavage sites were obtained revealing the critical nature of the residues in the positions -2 and -5. Charged residues seem to have a major influence on cleavage site function.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arretz M., Schneider H., Wienhues U., Neupert W. Processing of mitochondrial precursor proteins. Biomed Biochim Acta. 1991;50(4-6):403–412. [PubMed] [Google Scholar]
- Barker W. C., George D. G., Mewes H. W., Tsugita A. The PIR-International Protein Sequence Database. Nucleic Acids Res. 1992 May 11;20 (Suppl):2023–2026. doi: 10.1093/nar/20.suppl.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird P., Gething M. J., Sambrook J. The functional efficiency of a mammalian signal peptide is directly related to its hydrophobicity. J Biol Chem. 1990 May 25;265(15):8420–8425. [PubMed] [Google Scholar]
- Cornette J. L., Cease K. B., Margalit H., Spouge J. L., Berzofsky J. A., DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987 Jun 5;195(3):659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Dalbey R. E., Von Heijne G. Signal peptidases in prokaryotes and eukaryotes--a new protease family. Trends Biochem Sci. 1992 Nov;17(11):474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Johnson M. S., Doolittle R. F. Aligning amino acid sequences: comparison of commonly used methods. J Mol Evol. 1984;21(2):112–125. doi: 10.1007/BF02100085. [DOI] [PubMed] [Google Scholar]
- Fontana W, Stadler PF, Bornberg-Bauer EG, Griesmacher T, Hofacker IL, Tacker M, Tarazona P, Weinberger ED, Schuster P. RNA folding and combinatory landscapes. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1993 Mar;47(3):2083–2099. doi: 10.1103/physreve.47.2083. [DOI] [PubMed] [Google Scholar]
- Gavel Y., Nilsson L., von Heijne G. Mitochondrial targeting sequences. Why 'non-amphiphilic' peptides may still be amphiphilic. FEBS Lett. 1988 Aug 1;235(1-2):173–177. doi: 10.1016/0014-5793(88)81257-2. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990 Oct;4(1):33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. Mutation data matrix and its uses. Methods Enzymol. 1990;183:333–351. doi: 10.1016/0076-6879(90)83022-2. [DOI] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974 Sep 6;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Harpaz Y., Gerstein M., Chothia C. Volume changes on protein folding. Structure. 1994 Jul 15;2(7):641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990 Feb 23;247(4945):930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988 Jun 3;53(5):795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hodges P. E., Rosenberg L. E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A. L., Kalousek F., Fenton W. A., Furtak K., Pollock R. A., Rosenberg L. E. The ornithine transcarbamylase leader peptide directs mitochondrial import through both its midportion structure and net positive charge. J Cell Biol. 1987 Aug;105(2):669–677. doi: 10.1083/jcb.105.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A. L., Kalousek F., Fenton W. A., Pollock R. A., Rosenberg L. E. Targeting of pre-ornithine transcarbamylase to mitochondria: definition of critical regions and residues in the leader peptide. Cell. 1986 Feb 14;44(3):451–459. doi: 10.1016/0092-8674(86)90466-6. [DOI] [PubMed] [Google Scholar]
- Horwich A. L., Kalousek F., Rosenberg L. E. Arginine in the leader peptide is required for both import and proteolytic cleavage of a mitochondrial precursor. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4930–4933. doi: 10.1073/pnas.82.15.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- King R. D., Sternberg M. J. Machine learning approach for the prediction of protein secondary structure. J Mol Biol. 1990 Nov 20;216(2):441–457. doi: 10.1016/S0022-2836(05)80333-X. [DOI] [PubMed] [Google Scholar]
- Laforet G. A., Kendall D. A. Functional limits of conformation, hydrophobicity, and steric constraints in prokaryotic signal peptide cleavage regions. Wild type transport by a simple polymeric signal sequence. J Biol Chem. 1991 Jan 15;266(2):1326–1334. [PubMed] [Google Scholar]
- Lohmann R., Schneider G., Behrens D., Wrede P. A neural network model for the prediction of membrane-spanning amino acid sequences. Protein Sci. 1994 Sep;3(9):1597–1601. doi: 10.1002/pro.5560030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975 Oct 20;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Miura S., Amaya Y., Mori M. A metalloprotease involved in the processing of mitochondrial precursor proteins. Biochem Biophys Res Commun. 1986 Feb 13;134(3):1151–1159. doi: 10.1016/0006-291x(86)90371-2. [DOI] [PubMed] [Google Scholar]
- Miyata T., Miyazawa S., Yasunaga T. Two types of amino acid substitutions in protein evolution. J Mol Evol. 1979 Mar 15;12(3):219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- Nguyen M., Argan C., Sheffield W. P., Bell A. W., Shields D., Shore G. C. A signal sequence domain essential for processing, but not import, of mitochondrial pre-ornithine carbamyl transferase. J Cell Biol. 1987 May;104(5):1193–1198. doi: 10.1083/jcb.104.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Fox T. D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993 Dec 24;262(5142):1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- Qian N., Sejnowski T. J. Predicting the secondary structure of globular proteins using neural network models. J Mol Biol. 1988 Aug 20;202(4):865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C., Tweedy N. B., Gernert K. M., Quinn T. P., Hecht M. H., Erickson B. W., Yan Y., McClain R. D., Donlan M. E. Looking at proteins: representations, folding, packing, and design. Biophysical Society National Lecture, 1992. Biophys J. 1992 Nov;63(5):1185–1209. [PMC free article] [PubMed] [Google Scholar]
- Risler J. L., Delorme M. O., Delacroix H., Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988 Dec 20;204(4):1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- Schatz G. The protein import machinery of mitochondria. Protein Sci. 1993 Feb;2(2):141–146. doi: 10.1002/pro.5560020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Behrens M., Scherer P., Pratje E., Michaelis G., Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991 Feb;10(2):247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Röhlk S., Wrede P. Analysis of cleavage-site patterns in protein precursor sequences with a perceptron-type neural network. Biochem Biophys Res Commun. 1993 Jul 30;194(2):951–959. doi: 10.1006/bbrc.1993.1913. [DOI] [PubMed] [Google Scholar]
- Schneider G., Schuchhardt J., Wrede P. Artificial neural networks and simulated molecular evolution are potential tools for sequence-oriented protein design. Comput Appl Biosci. 1994 Dec;10(6):635–645. doi: 10.1093/bioinformatics/10.6.635. [DOI] [PubMed] [Google Scholar]
- Schneider G., Wrede P. Development of artificial neural filters for pattern recognition in protein sequences. J Mol Evol. 1993 Jun;36(6):586–595. doi: 10.1007/BF00556363. [DOI] [PubMed] [Google Scholar]
- Schneider G., Wrede P. The rational design of amino acid sequences by artificial neural networks and simulated molecular evolution: de novo design of an idealized leader peptidase cleavage site. Biophys J. 1994 Feb;66(2 Pt 1):335–344. doi: 10.1016/s0006-3495(94)80782-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E. S., Hendrick J. P., Kraus J. P., Wall D., Kalousek F., Rosenberg L. E. Import of rat ornithine transcarbamylase precursor into mitochondria: two-step processing of the leader peptide. J Cell Biol. 1987 Dec;105(6 Pt 1):2631–2639. doi: 10.1083/jcb.105.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassarotti A., Chen W. J., Smagula C., Douglas M. G. Sequences distal to the mitochondrial targeting sequences are necessary for the maturation of the F1-ATPase beta-subunit precursor in mitochondria. J Biol Chem. 1987 Jan 5;262(1):411–418. [PubMed] [Google Scholar]
- Zamyatnin A. A. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]