Abstract
Grand canonical Monte Carlo (GCMC) simulations are reported for aqueous solutions containing excess univalent salt (activities a +/- = 1.76-12.3 mM) and one of the following species: an octacationic rod-like ligand, L8+; a B-DNA oligomer with N phosphate charges (8 < or = N < or = 100); or a complex resulting from the binding of L8+ at the center of an N-mer (24 < or = N < or = 250). Simplified models of these multiply charged species are used in the GCMC simulations to predict the fundamental coulombic contributions to the following experimentally relevant properties: 1) the axial distance over which ligand binding affects local counterion concentrations at the surface of the N-mer; 2) the dependence on N of GCMC preferential interaction coefficients, gamma 32MC identical to delta C3/delta C2l a +/-, T, where C3 and C2 are, respectively, the molar concentrations of salt and the multiply charged species (ligand, N-mer or complex); and 3) the dependence on N of SaKobs identical to d in Kobs/d in a +/- = delta (magnitude of ZJ + 2 gamma 32J), where Kobs is the equilibrium concentration quotient for the binding of L8+ to the center of an N-mer and delta denotes the stoichiometric combination of terms, each of which pertains to a reactant or product J having magnitude of ZJ charges. The participation of electrolyte ions in the ligand binding interaction is quantified by the magnitude of SaKobs, which reflects the net (stoichiometrically weighted) difference in the extent of thermodynamic binding of salt ions to the products and reactants. Results obtained here from GCMC simulations yield a picture of the salient molecular consequences of binding a cationic ligand, as well as thermodynamic predictions whose applicability can be tested experimentally. Formation of the central complex is predicted to cause a dramatic reduction in the surface counterion (e.g., Na+) concentration over a region including but extending well beyond the location of the ligand binding site. For binding a cationic ligand, SaKobs is predicted to be negative, indicating net electrolyte ion release in the binding process. At small enough N, -SaKobs is predicted to decrease strongly toward zero with decreasing N. At intermediate N, -SaKobs appears to exceed its limiting value as N-->infinity.
Full text
PDF




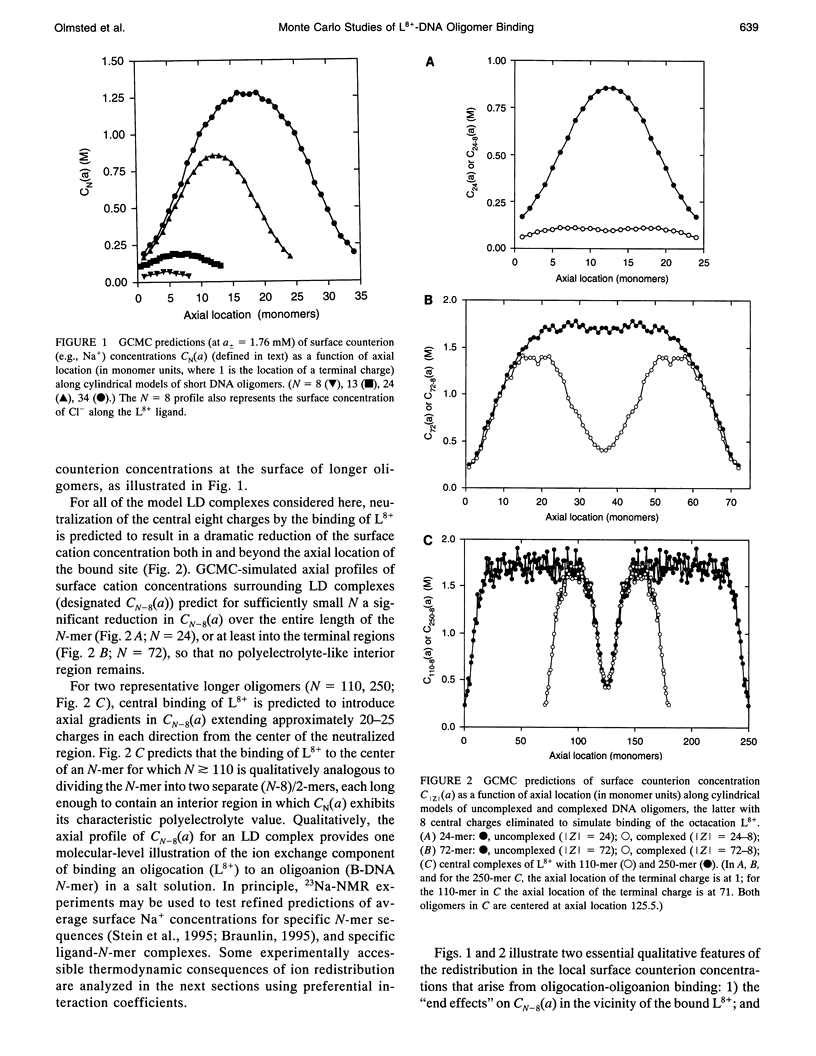
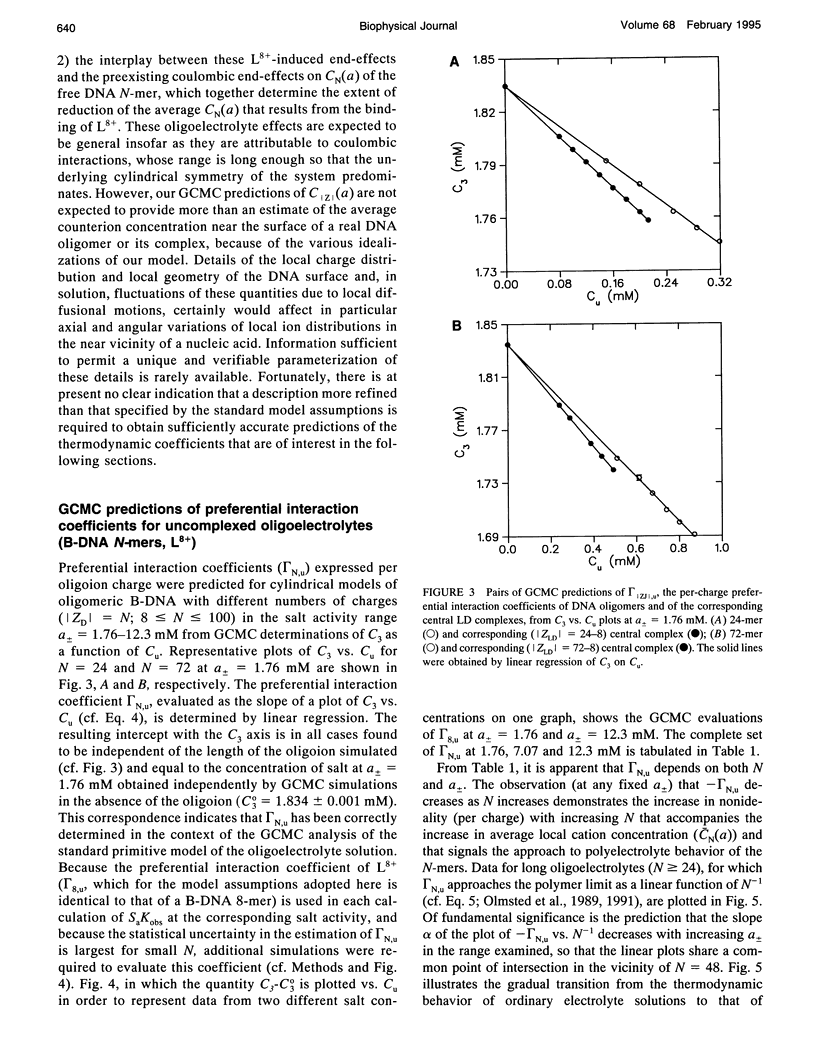
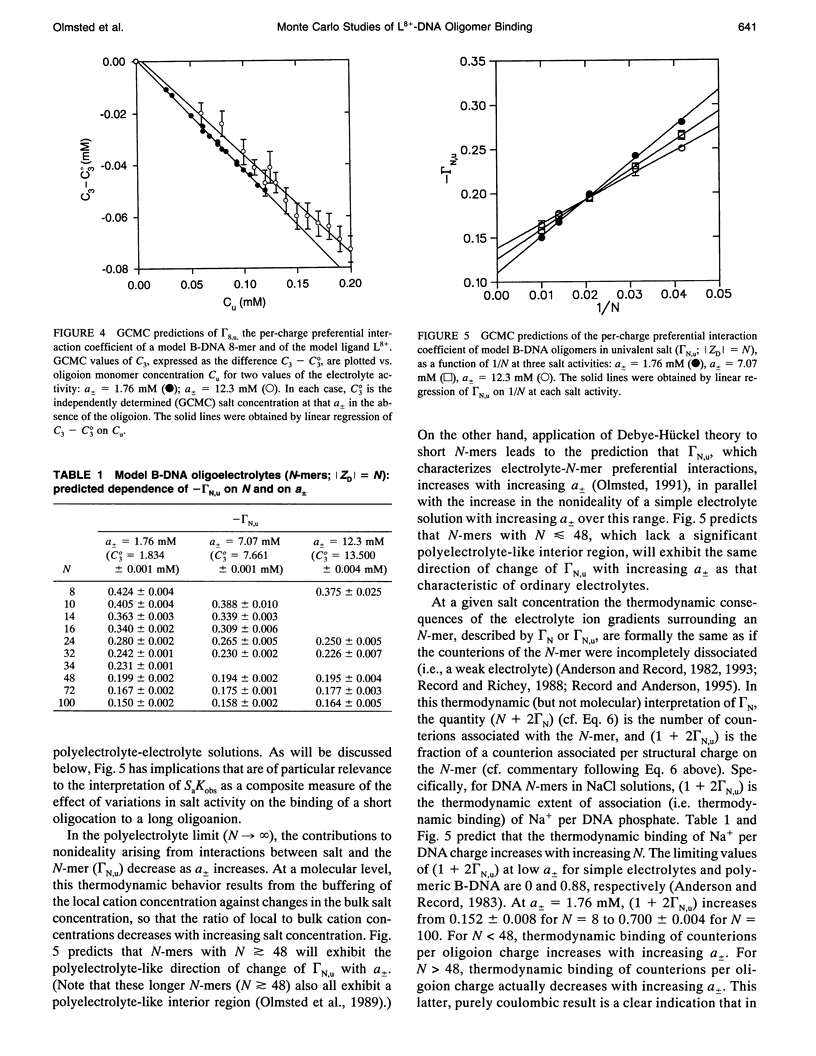

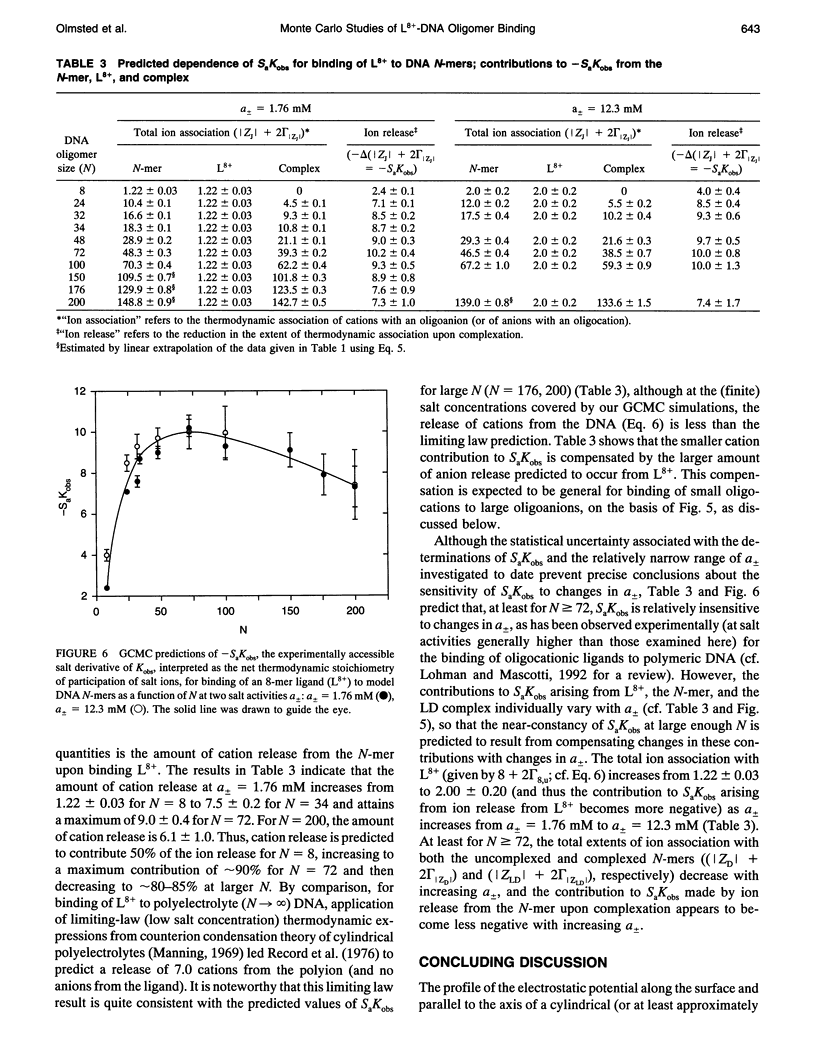




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. F., Record M. T., Jr Ion distributions around DNA and other cylindrical polyions: theoretical descriptions and physical implications. Annu Rev Biophys Biophys Chem. 1990;19:423–465. doi: 10.1146/annurev.bb.19.060190.002231. [DOI] [PubMed] [Google Scholar]
- Anderson C. F., Record M. T., Jr The relationship between the poisson-boltzmann model and the condensation hypothesis: an analysis based on the low salt form of the Donnan coefficient. Biophys Chem. 1980 Jun;11(3-4):353–360. doi: 10.1016/0301-4622(80)87008-6. [DOI] [PubMed] [Google Scholar]
- Bond J. P., Anderson C. F., Record M. T., Jr Conformational transitions of duplex and triplex nucleic acid helices: thermodynamic analysis of effects of salt concentration on stability using preferential interaction coefficients. Biophys J. 1994 Aug;67(2):825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson E. L., Scheffler I. E., Baldwin R. L. Helix formation by d(TA) oligomers. 3. Electrostatic effects. J Mol Biol. 1970 Dec 28;54(3):401–415. doi: 10.1016/0022-2836(70)90118-x. [DOI] [PubMed] [Google Scholar]
- Fritsch V., Ravishanker G., Beveridge D. L., Westhof E. Molecular dynamics simulations of poly(dA).poly(dT): comparisons between implicit and explicit solvent representations. Biopolymers. 1993 Oct;33(10):1537–1552. doi: 10.1002/bip.360331005. [DOI] [PubMed] [Google Scholar]
- Klein B. J., Pack G. R. Calculations of the spatial distribution of charge density in the environment of DNA. Biopolymers. 1983 Nov;22(11):2331–2352. doi: 10.1002/bip.360221103. [DOI] [PubMed] [Google Scholar]
- Latt S. A., Sober H. A. Protein-nucleic acid interactions. II. Oligopeptide-polyribonucleotide binding studies. Biochemistry. 1967 Oct;6(10):3293–3306. doi: 10.1021/bi00862a040. [DOI] [PubMed] [Google Scholar]
- Laughton C. A., Neidle S. Molecular dynamics simulation of the DNA triplex d(TC)5.d(GA)5.d(C+T)5. J Mol Biol. 1992 Jan 20;223(2):519–529. doi: 10.1016/0022-2836(92)90667-9. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Mascotti D. P. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 1992;212:400–424. doi: 10.1016/0076-6879(92)12026-m. [DOI] [PubMed] [Google Scholar]
- Mascotti D. P., Lohman T. M. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3142–3146. doi: 10.1073/pnas.87.8.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascotti D. P., Lohman T. M. Thermodynamics of single-stranded RNA binding to oligolysines containing tryptophan. Biochemistry. 1992 Sep 22;31(37):8932–8946. doi: 10.1021/bi00152a033. [DOI] [PubMed] [Google Scholar]
- Misra V. K., Sharp K. A., Friedman R. A., Honig B. Salt effects on ligand-DNA binding. Minor groove binding antibiotics. J Mol Biol. 1994 Apr 29;238(2):245–263. doi: 10.1006/jmbi.1994.1285. [DOI] [PubMed] [Google Scholar]
- Olmsted M. C., Anderson C. F., Record M. T., Jr Importance of oligoelectrolyte end effects for the thermodynamics of conformational transitions of nucleic acid oligomers: a grand canonical Monte Carlo analysis. Biopolymers. 1991 Nov;31(13):1593–1604. doi: 10.1002/bip.360311314. [DOI] [PubMed] [Google Scholar]
- Olmsted M. C., Anderson C. F., Record M. T., Jr Monte Carlo description of oligoelectrolyte properties of DNA oligomers: range of the end effect and the approach of molecular and thermodynamic properties to the polyelectrolyte limits. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7766–7770. doi: 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S., Brushaber V. M., Anderson C. F., Record M. T., Jr Relative affinities of divalent polyamines and of their N-methylated analogues for helical DNA determined by 23Na NMR. Biochemistry. 1991 Jul 30;30(30):7550–7559. doi: 10.1021/bi00244a026. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Ha J. H., Fisher M. A. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Seibel G. L., Singh U. C., Kollman P. A. A molecular dynamics simulation of double-helical B-DNA including counterions and water. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6537–6540. doi: 10.1073/pnas.82.19.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gunsteren W. F., Berendsen H. J., Geurtsen R. G., Zwinderman H. R. A molecular dynamics computer simulation of an eight-base-pair DNA fragment in aqueous solution: comparison with experimental two-dimensional NMR data. Ann N Y Acad Sci. 1986;482:287–303. doi: 10.1111/j.1749-6632.1986.tb20962.x. [DOI] [PubMed] [Google Scholar]


