Abstract
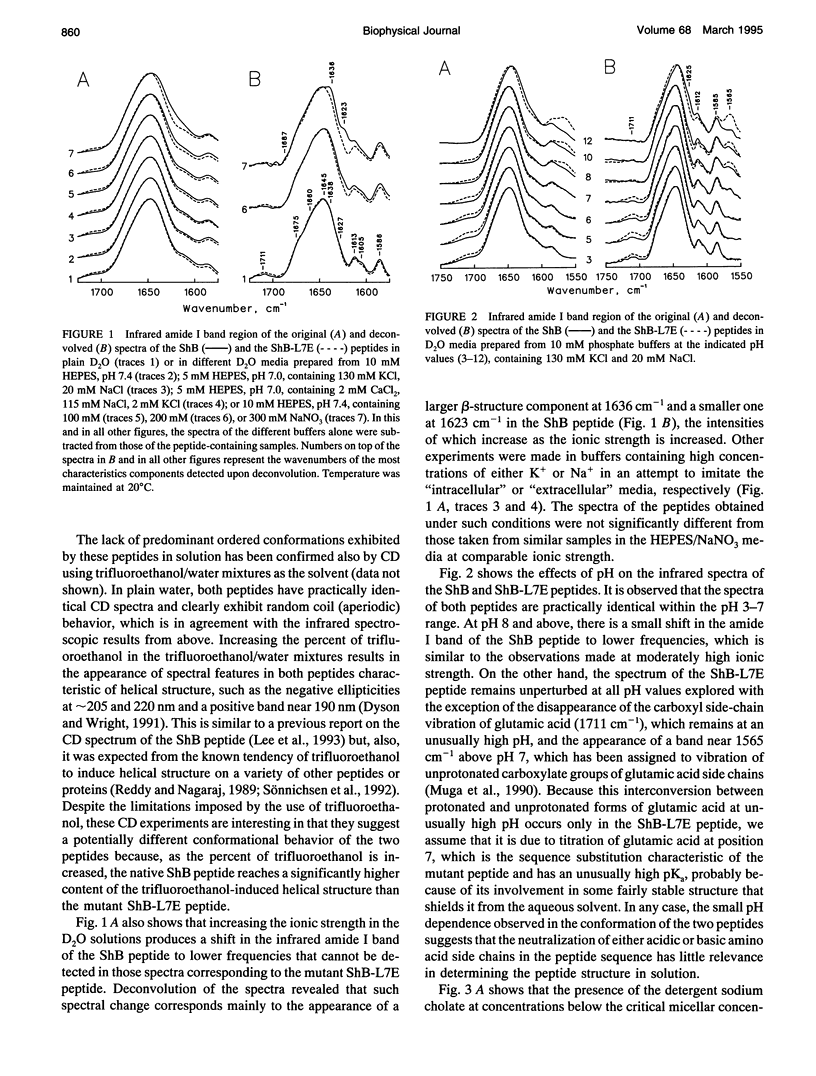
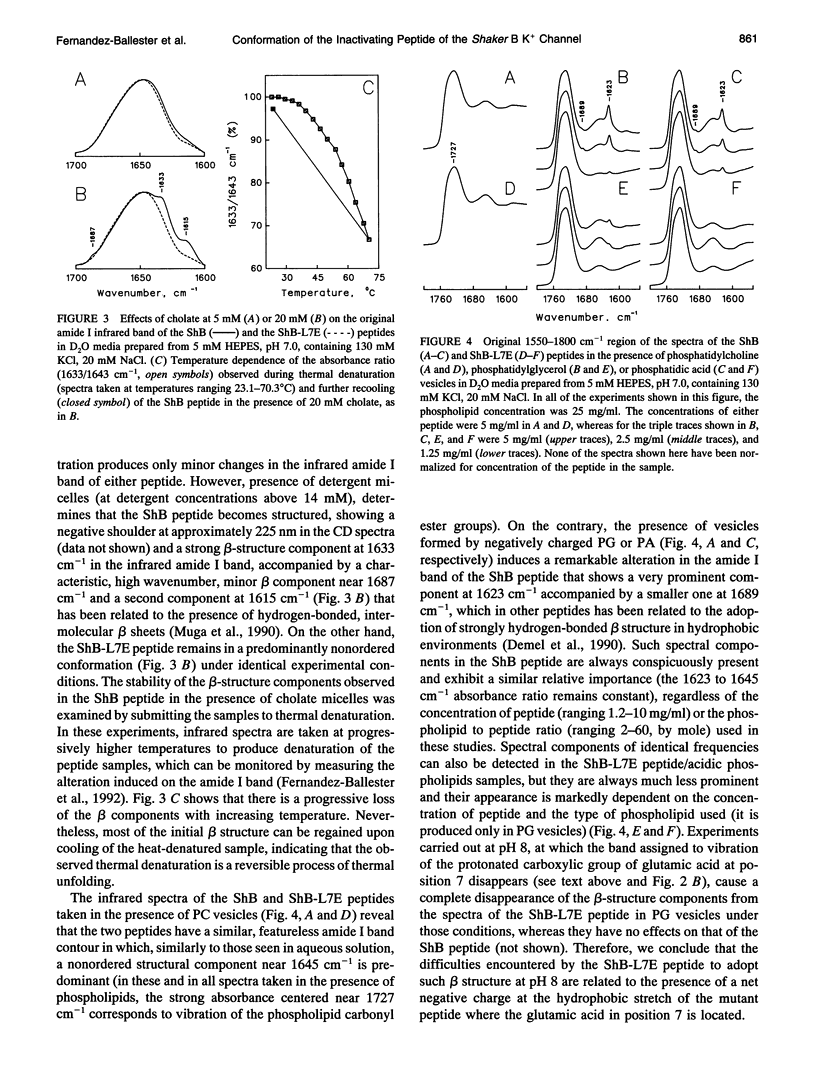
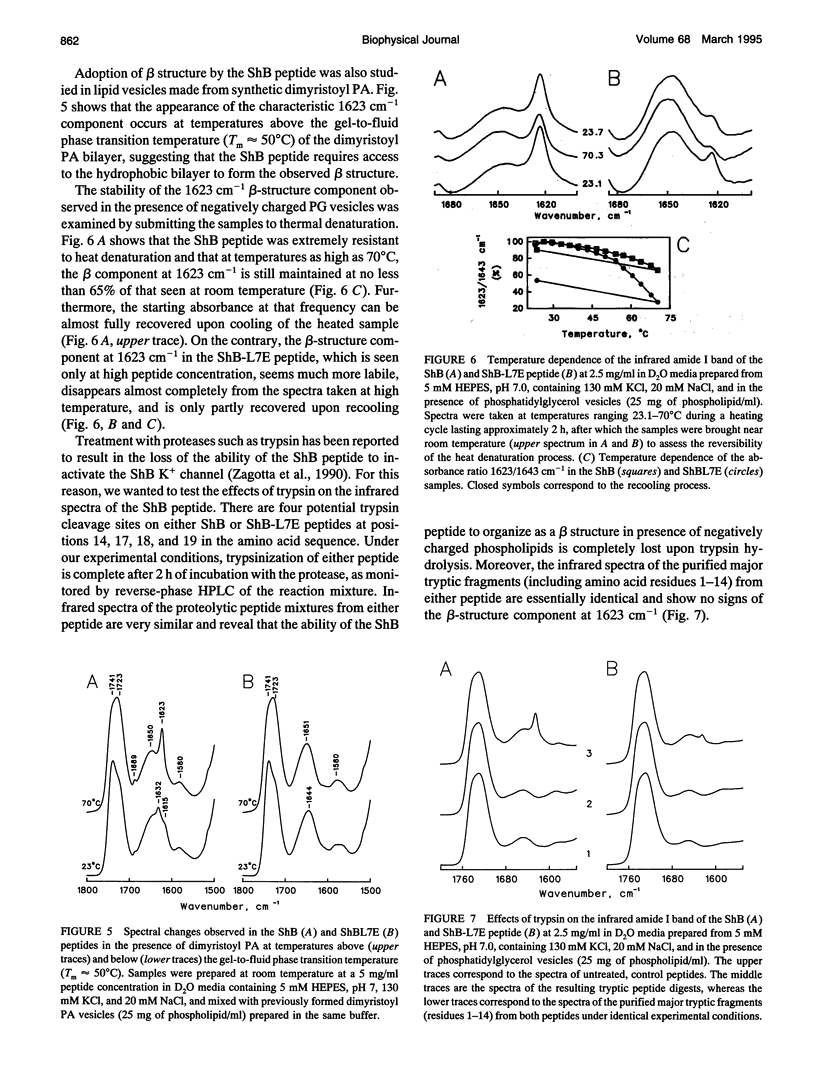
The conformation of the inactivating peptide of the Shaker B K+ channel (ShB peptide) and that of a noninactivating mutant (ShBL7E peptide) have been studied. Under all experimental conditions explored, the mutant peptide remains in a predominantly nonordered conformation. On the contrary, the inactivating ShB peptide has a great tendency to adopt a highly stable beta structure, particularly when challenged "in vitro" by anionic phospholipid vesicles. Because the putative peptide binding elements at the inner mouth of the channel comprise a ring of anionic residues and a hydrophobic pocket, we hypothesize that the conformational restrictions imposed on the ShB peptide by its interaction with the anionic lipid vesicles could partly imitate those imposed by the above ion channel elements. Thus, we propose that adoption of beta structure by the inactivating peptide may also occur during channel inactivation. Moreover, the difficulties encountered by the noninactivating ShBL7E peptide mutant to adopt beta structure and the observation that trypsin hydrolysis of the ShB peptide prevent both structure formation and channel inactivation lend further support to the hypothesis that adoption of beta structure by the inactivating peptide in a hydrophobic environment is important in determining channel blockade.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong C. M., Bezanilla F. Inactivation of the sodium channel. II. Gating current experiments. J Gen Physiol. 1977 Nov;70(5):567–590. doi: 10.1085/jgp.70.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrondo J. L., Mantsch H. H., Mullner N., Pikula S., Martonosi A. Infrared spectroscopic characterization of the structural changes connected with the E1----E2 transition in the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1987 Jul 5;262(19):9037–9043. [PubMed] [Google Scholar]
- Bandekar J. Amide modes and protein conformation. Biochim Biophys Acta. 1992 Apr 8;1120(2):123–143. doi: 10.1016/0167-4838(92)90261-b. [DOI] [PubMed] [Google Scholar]
- Byler D. M., Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986 Mar;25(3):469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- Castresana J., Fernandez-Ballester G., Fernandez A. M., Laynez J. L., Arrondo J. L., Ferragut J. A., Gonzalez-Ros J. M. Protein structural effects of agonist binding to the nicotinic acetylcholine receptor. FEBS Lett. 1992 Dec 14;314(2):171–175. doi: 10.1016/0014-5793(92)80967-l. [DOI] [PubMed] [Google Scholar]
- Chirgadze Y. N., Fedorov O. V., Trushina N. P. Estimation of amino acid residue side-chain absorption in the infrared spectra of protein solutions in heavy water. Biopolymers. 1975 Apr;14(4):679–694. doi: 10.1002/bip.1975.360140402. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Demel R. A., Goormaghtigh E., de Kruijff B. Lipid and peptide specificities in signal peptide--lipid interactions in model membranes. Biochim Biophys Acta. 1990 Aug 24;1027(2):155–162. doi: 10.1016/0005-2736(90)90079-4. [DOI] [PubMed] [Google Scholar]
- Durell S. R., Guy H. R. Atomic scale structure and functional models of voltage-gated potassium channels. Biophys J. 1992 Apr;62(1):238–250. doi: 10.1016/S0006-3495(92)81809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. Defining solution conformations of small linear peptides. Annu Rev Biophys Biophys Chem. 1991;20:519–538. doi: 10.1146/annurev.bb.20.060191.002511. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ballester G., Castresana J., Arrondo J. L., Ferragut J. A., Gonzalez-Ros J. M. Protein stability and interaction of the nicotinic acetylcholine receptor with cholinergic ligands studied by Fourier-transform infrared spectroscopy. Biochem J. 1992 Dec 1;288(Pt 2):421–426. doi: 10.1042/bj2880421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ballester G., Castresana J., Fernandez A. M., Arrondo J. L., Ferragut J. A., Gonzalez-Ros J. M. A role for cholesterol as a structural effector of the nicotinic acetylcholine receptor. Biochemistry. 1994 Apr 5;33(13):4065–4071. doi: 10.1021/bi00179a035. [DOI] [PubMed] [Google Scholar]
- Fraser P. E., Nguyen J. T., Surewicz W. K., Kirschner D. A. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys J. 1991 Nov;60(5):1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T., Zagotta W. N., Aldrich R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990 Oct 26;250(4980):533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Isacoff E. Y., Jan Y. N., Jan L. Y. Putative receptor for the cytoplasmic inactivation gate in the Shaker K+ channel. Nature. 1991 Sep 5;353(6339):86–90. doi: 10.1038/353086a0. [DOI] [PubMed] [Google Scholar]
- Jackson M., Mantsch H. H., Spencer J. H. Conformation of magainin-2 and related peptides in aqueous solution and membrane environments probed by Fourier transform infrared spectroscopy. Biochemistry. 1992 Aug 18;31(32):7289–7293. doi: 10.1021/bi00147a012. [DOI] [PubMed] [Google Scholar]
- Jan L. Y., Jan Y. N. Structural elements involved in specific K+ channel functions. Annu Rev Physiol. 1992;54:537–555. doi: 10.1146/annurev.ph.54.030192.002541. [DOI] [PubMed] [Google Scholar]
- Kamb A., Tseng-Crank J., Tanouye M. A. Multiple products of the Drosophila Shaker gene may contribute to potassium channel diversity. Neuron. 1988 Jul;1(5):421–430. doi: 10.1016/0896-6273(88)90192-4. [DOI] [PubMed] [Google Scholar]
- King D. S., Fields C. G., Fields G. B. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Pept Protein Res. 1990 Sep;36(3):255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Muga A., Surewicz W. K., Wong P. T., Mantsch H. H. Structural studies with the uveopathogenic peptide M derived from retinal S-antigen. Biochemistry. 1990 Mar 27;29(12):2925–2930. doi: 10.1021/bi00464a006. [DOI] [PubMed] [Google Scholar]
- Murrell-Lagnado R. D., Aldrich R. W. Interactions of amino terminal domains of Shaker K channels with a pore blocking site studied with synthetic peptides. J Gen Physiol. 1993 Dec;102(6):949–975. doi: 10.1085/jgp.102.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian D. M., Schwarz T. L., Tempel B. L., Jan Y. N., Jan L. Y. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987 Aug 14;237(4816):749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- Pongs O., Kecskemethy N., Müller R., Krah-Jentgens I., Baumann A., Kiltz H. H., Canal I., Llamazares S., Ferrus A. Shaker encodes a family of putative potassium channel proteins in the nervous system of Drosophila. EMBO J. 1988 Apr;7(4):1087–1096. doi: 10.1002/j.1460-2075.1988.tb02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G. L., Nagara R. Circular dichroism studies on synthetic signal peptides indicate beta-conformation as a common structural feature in highly hydrophobic environment. J Biol Chem. 1989 Oct 5;264(28):16591–16597. [PubMed] [Google Scholar]
- Rothschild K. J. FTIR difference spectroscopy of bacteriorhodopsin: toward a molecular model. J Bioenerg Biomembr. 1992 Apr;24(2):147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K., Mantsch H. H., Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993 Jan 19;32(2):389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K., Mantsch H. H. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim Biophys Acta. 1988 Jan 29;952(2):115–130. doi: 10.1016/0167-4838(88)90107-0. [DOI] [PubMed] [Google Scholar]
- Sönnichsen F. D., Van Eyk J. E., Hodges R. S., Sykes B. D. Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry. 1992 Sep 22;31(37):8790–8798. doi: 10.1021/bi00152a015. [DOI] [PubMed] [Google Scholar]
- Toro L., Ottolia M., Stefani E., Latorre R. Structural determinants in the interaction of Shaker inactivating peptide and a Ca(2+)-activated K+ channel. Biochemistry. 1994 Jun 14;33(23):7220–7228. doi: 10.1021/bi00189a026. [DOI] [PubMed] [Google Scholar]
- Toro L., Stefani E., Latorre R. Internal blockade of a Ca(2+)-activated K+ channel by Shaker B inactivating "ball" peptide. Neuron. 1992 Aug;9(2):237–245. doi: 10.1016/0896-6273(92)90163-8. [DOI] [PubMed] [Google Scholar]
- Zagotta W. N., Hoshi T., Aldrich R. W. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990 Oct 26;250(4980):568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P., Lewis R. N., Hodges R. S., McElhaney R. N. FTIR spectroscopic studies of the conformation and amide hydrogen exchange of a peptide model of the hydrophobic transmembrane alpha-helices of membrane proteins. Biochemistry. 1992 Nov 24;31(46):11572–11578. doi: 10.1021/bi00161a041. [DOI] [PubMed] [Google Scholar]


