Abstract
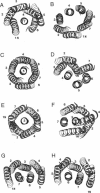
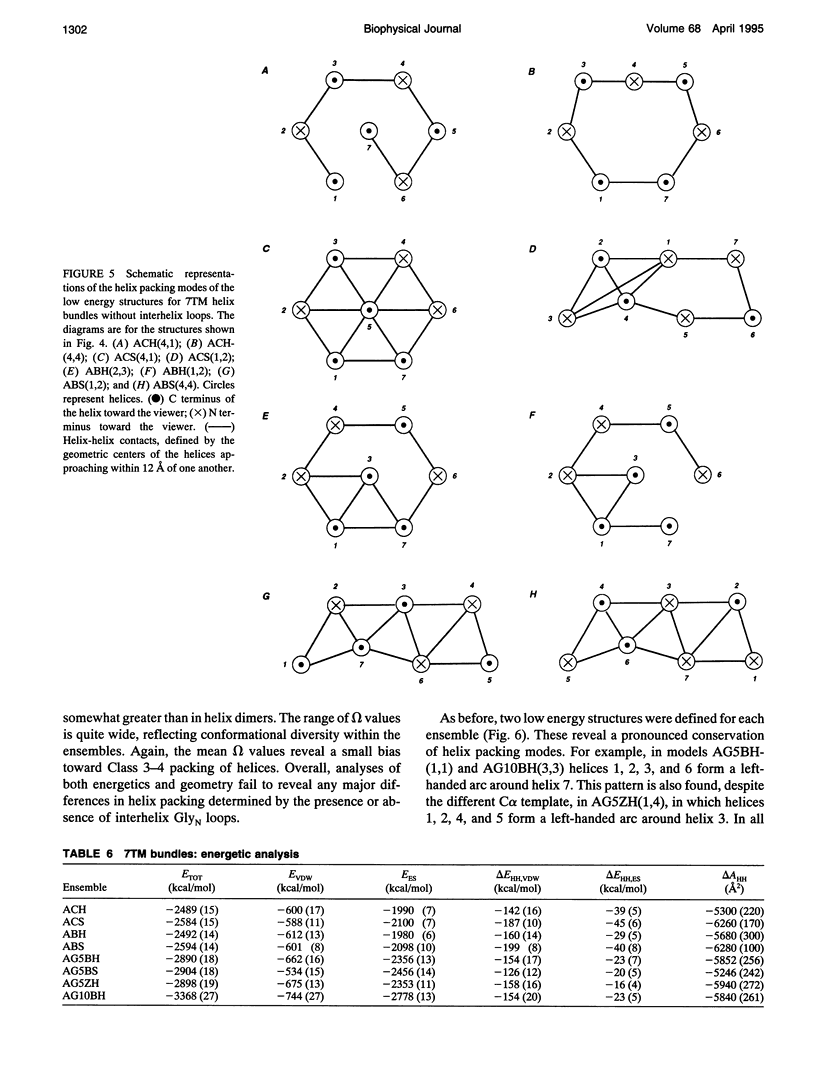
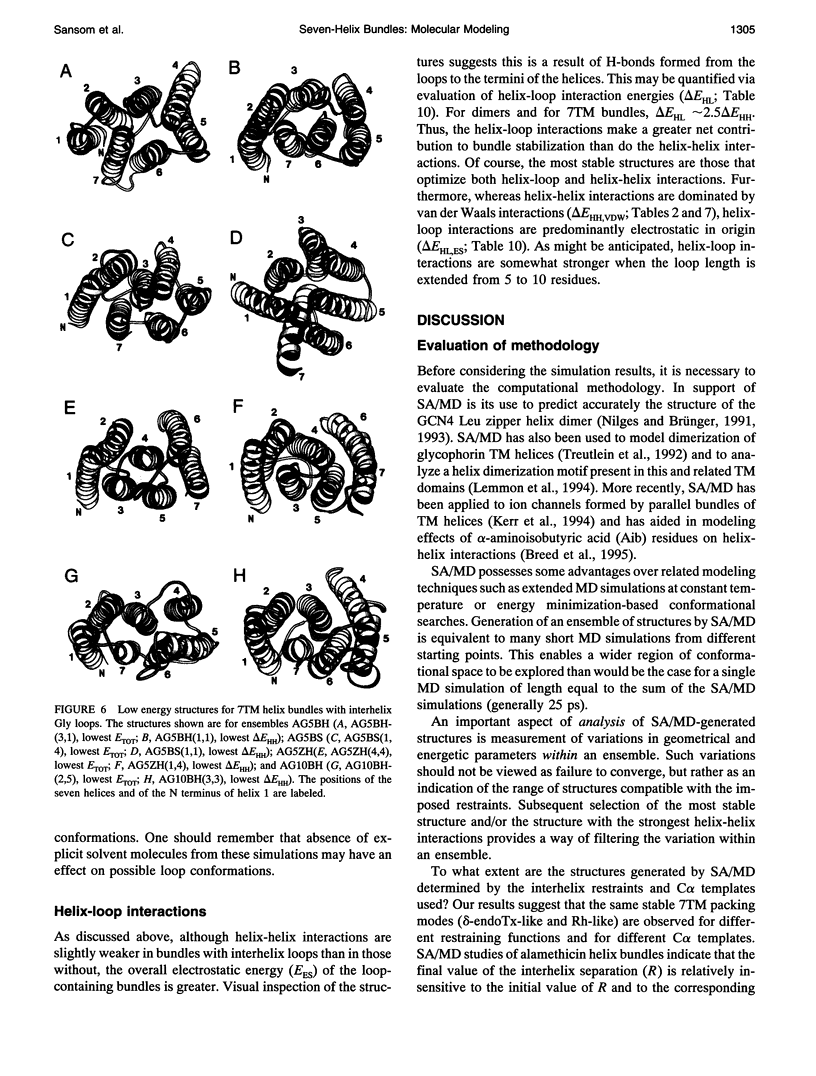
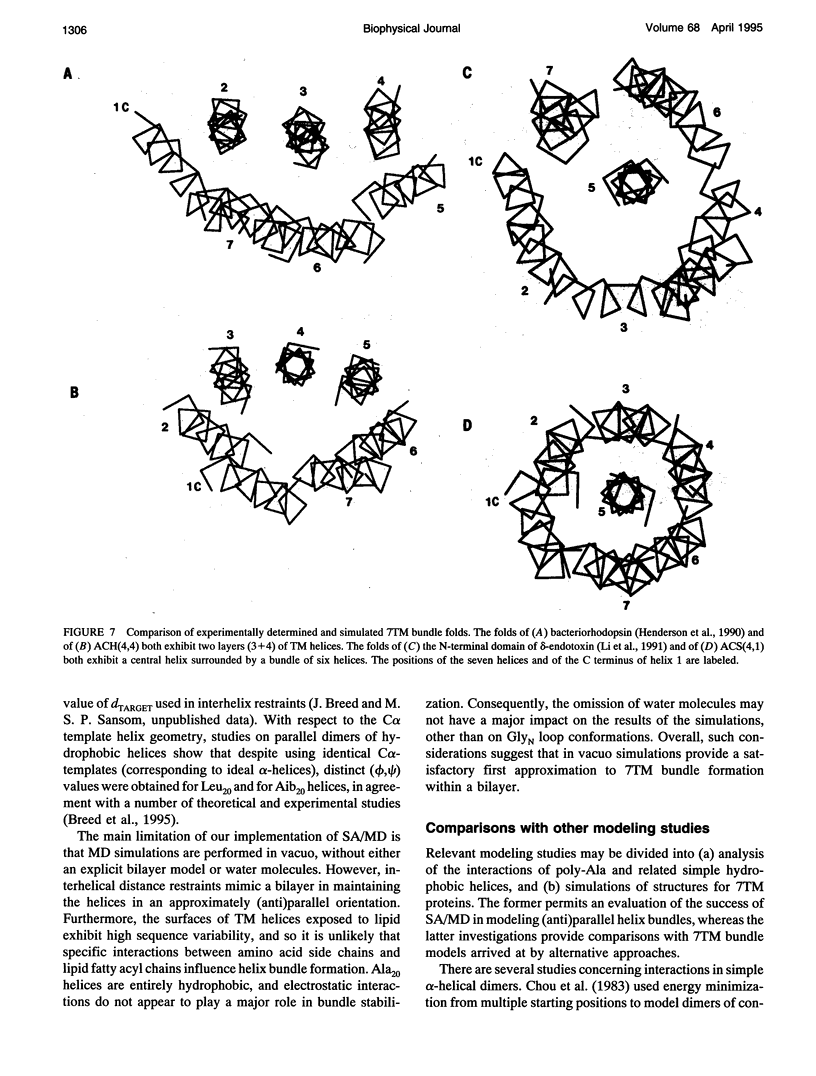
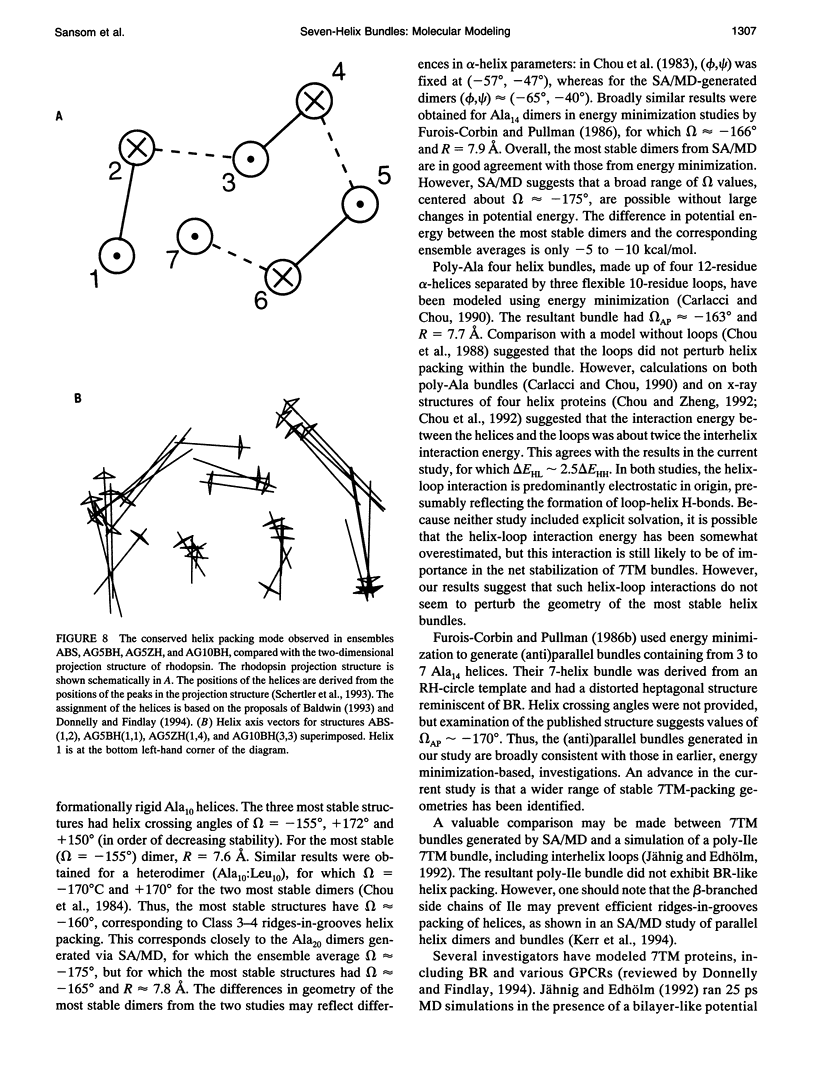
Simulated annealing via restrained molecular dynamics (SA/MD) has been used to model compact bundles of seven approximately (anti)parallel alpha-helices. Seven such helix bundles occur, e.g., in bacteriorhodopsin, in rhodopsin, and in the channel-forming N-terminal domain of Bacillus thuringiensis delta-endotoxin. Two classes of model are considered: (a) those consisting of seven Ala20 peptide chains; and (b) those containing a single polypeptide chain, made up of seven Ala20 helices linked by GlyN interhelix loops (where N = 5 or 10). Three different starting C alpha templates for SA/MD are used, in which the seven helices are arranged (a) on a left-handed circular template, (b) on a bacteriorhodopsin-like template, or (c) on a zig-zag template. The ensembles of models generated by SA/MD are analyzed in terms of their geometry and energetics, and the most stable structures from each ensemble are examined in greater detail. Structures resembling bacteriorhodopsin and structures resembling delta-endotoxin are both represented among the most stable structures. delta-Endotoxin-like structures arise from both circular and bacteriorhodopsin-like C alpha templates. A third helix-packing mode occurs several times among the stable structures, regardless of the C alpha template and of the presence or absence of interhelix loops. It is characterized by a "4 + 1" core, in which four helices form a distorted left-handed supercoil around a central, buried helix. The remaining two helices pack onto the outside of the core. This packing mode is comparable with that proposed for rhodopsin on the basis of two-dimensional electron crystallographic and sequence analysis studies.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. M. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993 Apr;12(4):1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. Helix geometry in proteins. J Mol Biol. 1988 Jun 5;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Barsukov I. L., Nolde D. E., Lomize A. L., Arseniev A. S. Three-dimensional structure of proteolytic fragment 163-231 of bacterioopsin determined from nuclear magnetic resonance data in solution. Eur J Biochem. 1992 Jun 15;206(3):665–672. doi: 10.1111/j.1432-1033.1992.tb16972.x. [DOI] [PubMed] [Google Scholar]
- Carlacci L., Chou K. C. Electrostatic interactions between loops and alpha-helices in four-helix bundle proteins. Protein Eng. 1990 Dec;4(2):225–227. doi: 10.1093/protein/4.2.225. [DOI] [PubMed] [Google Scholar]
- Carlacci L., Chou K. C. Energetic approach to the folding of four alpha-helices connected sequentially. Protein Eng. 1990 May;3(6):509–514. doi: 10.1093/protein/3.6.509. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981 Jan 5;145(1):215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Chou K. C., Carlacci L., Maggiora G. M., Parodi L. A., Schulz M. W. An energy-based approach to packing the 7-helix bundle of bacteriorhodopsin. Protein Sci. 1992 Jun;1(6):810–827. doi: 10.1002/pro.5560010613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Maggiora G. M., Némethy G., Scheraga H. A. Energetics of the structure of the four-alpha-helix bundle in proteins. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4295–4299. doi: 10.1073/pnas.85.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Maggiora G. M., Scheraga H. A. Role of loop-helix interactions in stabilizing four-helix bundle proteins. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7315–7319. doi: 10.1073/pnas.89.16.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K. C., Zheng C. Strong electrostatic loop-helix interactions in bundle motif protein structures. Biophys J. 1992 Sep;63(3):682–688. doi: 10.1016/S0006-3495(92)81653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronet P., Sander C., Vriend G. Modeling of transmembrane seven helix bundles. Protein Eng. 1993 Jan;6(1):59–64. doi: 10.1093/protein/6.1.59. [DOI] [PubMed] [Google Scholar]
- Donnelly D., Overington J. P., Ruffle S. V., Nugent J. H., Blundell T. L. Modeling alpha-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 1993 Jan;2(1):55–70. doi: 10.1002/pro.5560020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm O., Jähnig F. The structure of a membrane-spanning polypeptide studied by molecular dynamics. Biophys Chem. 1988 Jul 15;30(3):279–292. doi: 10.1016/0301-4622(88)85023-3. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Henderson R., McLachlan A. D., Wallace B. A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hibert M. F., Trumpp-Kallmeyer S., Bruinvels A., Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol Pharmacol. 1991 Jul;40(1):8–15. [PubMed] [Google Scholar]
- Higgins C. F. The multidrug resistance P-glycoprotein. Curr Opin Cell Biol. 1993 Aug;5(4):684–687. doi: 10.1016/0955-0674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Hol W. G., Halie L. M., Sander C. Dipoles of the alpha-helix and beta-sheet: their role in protein folding. Nature. 1981 Dec 10;294(5841):532–536. doi: 10.1038/294532a0. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994 Mar 15;33(10):3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Jähnig F., Edholm O. Modeling of the structure of bacteriorhodopsin. A molecular dynamics study. J Mol Biol. 1992 Aug 5;226(3):837–850. doi: 10.1016/0022-2836(92)90635-w. [DOI] [PubMed] [Google Scholar]
- Kahn T. W., Engelman D. M. Bacteriorhodopsin can be refolded from two independently stable transmembrane helices and the complementary five-helix fragment. Biochemistry. 1992 Jul 7;31(26):6144–6151. doi: 10.1021/bi00141a027. [DOI] [PubMed] [Google Scholar]
- Kerr I. D., Sankararamakrishnan R., Smart O. S., Sansom M. S. Parallel helix bundles and ion channels: molecular modeling via simulated annealing and restrained molecular dynamics. Biophys J. 1994 Oct;67(4):1501–1515. doi: 10.1016/S0006-3495(94)80624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W., Wang D. N., Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994 Feb 17;367(6464):614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Engelman D. M. Specificity and promiscuity in membrane helix interactions. Q Rev Biophys. 1994 May;27(2):157–218. doi: 10.1017/s0033583500004522. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Treutlein H. R., Adams P. D., Brünger A. T., Engelman D. M. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994 Mar;1(3):157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- Li J. D., Carroll J., Ellar D. J. Crystal structure of insecticidal delta-endotoxin from Bacillus thuringiensis at 2.5 A resolution. Nature. 1991 Oct 31;353(6347):815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- Livingstone C. D., Strange P. G., Naylor L. H. Molecular modelling of D2-like dopamine receptors. Biochem J. 1992 Oct 1;287(Pt 1):277–282. doi: 10.1042/bj2870277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize A. L., Pervushin K. V., Arseniev A. S. Spatial structure of (34-65)bacterioopsin polypeptide in SDS micelles determined from nuclear magnetic resonance data. J Biomol NMR. 1992 Jul;2(4):361–372. doi: 10.1007/BF01874814. [DOI] [PubMed] [Google Scholar]
- MaloneyHuss K., Lybrand T. P. Three-dimensional structure for the beta 2 adrenergic receptor protein based on computer modeling studies. J Mol Biol. 1992 Jun 5;225(3):859–871. doi: 10.1016/0022-2836(92)90406-a. [DOI] [PubMed] [Google Scholar]
- Nilges M., Brünger A. T. Automated modeling of coiled coils: application to the GCN4 dimerization region. Protein Eng. 1991 Aug;4(6):649–659. doi: 10.1093/protein/4.6.649. [DOI] [PubMed] [Google Scholar]
- Nilges M., Brünger A. T. Successful prediction of the coiled coil geometry of the GCN4 leucine zipper domain by simulated annealing: comparison to the X-ray structure. Proteins. 1993 Feb;15(2):133–146. doi: 10.1002/prot.340150205. [DOI] [PubMed] [Google Scholar]
- Persson B., Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994 Mar 25;237(2):182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Pervushin K. V., Arseniev A. S. Three-dimensional structure of (1-36)bacterioopsin in methanol-chloroform mixture and SDS micelles determined by 2D 1H-NMR spectroscopy. FEBS Lett. 1992 Aug 17;308(2):190–196. doi: 10.1016/0014-5793(92)81272-n. [DOI] [PubMed] [Google Scholar]
- Popot J. L., Engelman D. M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990 May 1;29(17):4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- Reddy B. V., Blundell T. L. Packing of secondary structural elements in proteins. Analysis and prediction of inter-helix distances. J Mol Biol. 1993 Oct 5;233(3):464–479. doi: 10.1006/jmbi.1993.1524. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Getzoff E. D., Richardson D. C. The beta bulge: a common small unit of nonrepetitive protein structure. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2574–2578. doi: 10.1073/pnas.75.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom M. S. Structure and function of channel-forming peptaibols. Q Rev Biophys. 1993 Nov;26(4):365–421. doi: 10.1017/s0033583500002833. [DOI] [PubMed] [Google Scholar]
- Sansom M. S. The biophysics of peptide models of ion channels. Prog Biophys Mol Biol. 1991;55(3):139–235. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- Schertler G. F., Villa C., Henderson R. Projection structure of rhodopsin. Nature. 1993 Apr 22;362(6422):770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Sibanda B. L., Blundell T. L., Thornton J. M. Conformation of beta-hairpins in protein structures. A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J Mol Biol. 1989 Apr 20;206(4):759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- Sylte I., Edvardsen O., Dahl S. G. Molecular dynamics of the 5-HT1a receptor and ligands. Protein Eng. 1993 Sep;6(7):691–700. doi: 10.1093/protein/6.7.691. [DOI] [PubMed] [Google Scholar]
- Taylor W. R., Jones D. T., Green N. M. A method for alpha-helical integral membrane protein fold prediction. Proteins. 1994 Mar;18(3):281–294. doi: 10.1002/prot.340180309. [DOI] [PubMed] [Google Scholar]
- Treutlein H. R., Lemmon M. A., Engelman D. M., Brünger A. T. The glycophorin A transmembrane domain dimer: sequence-specific propensity for a right-handed supercoil of helices. Biochemistry. 1992 Dec 29;31(51):12726–12732. doi: 10.1021/bi00166a003. [DOI] [PubMed] [Google Scholar]
- Tuffery P., Etchebest C., Popot J. L., Lavery R. Prediction of the positioning of the seven transmembrane alpha-helices of bacteriorhodopsin. A molecular simulation study. J Mol Biol. 1994 Mar 4;236(4):1105–1122. doi: 10.1016/0022-2836(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Vlassi M., Steif C., Weber P., Tsernoglou D., Wilson K. S., Hinz H. J., Kokkinidis M. Restored heptad pattern continuity does not alter the folding of a four-alpha-helix bundle. Nat Struct Biol. 1994 Oct;1(10):706–716. doi: 10.1038/nsb1094-706. [DOI] [PubMed] [Google Scholar]
- Zhang D., Weinstein H. Polarity conserved positions in transmembrane domains of G-protein coupled receptors and bacteriorhodopsin. FEBS Lett. 1994 Jan 10;337(2):207–212. doi: 10.1016/0014-5793(94)80274-2. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992 May 20;225(2):487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]