Abstract
A computationally efficient method to describe the organization of water around solvated biomolecules is presented. It is based on a statistical mechanical expression for the water-density distribution in terms of particle correlation functions. The method is applied to analyze the hydration of small nucleic acid molecules in the crystal environment, for which high-resolution x-ray crystal structures have been reported. Results for RNA [r(ApU).r(ApU)] and DNA [d(CpG).d(CpG) in Z form and with parallel strand orientation] and for DNA-drug complexes [d(CpG).d(CpG) with the drug proflavine intercalated] are described. A detailed comparison of theoretical and experimental data shows positional agreement for the experimentally observed water sites. The presented method can be used for refinement of the water structure in x-ray crystallography, hydration analysis of nuclear magnetic resonance structures, and theoretical modeling of biological macromolecules such as molecular docking studies. The speed of the computations allows hydration analyses of molecules of almost arbitrary size (tRNA, protein-nucleic acid complexes, etc.) in the crystal environment and in aqueous solution.
Full text
PDF





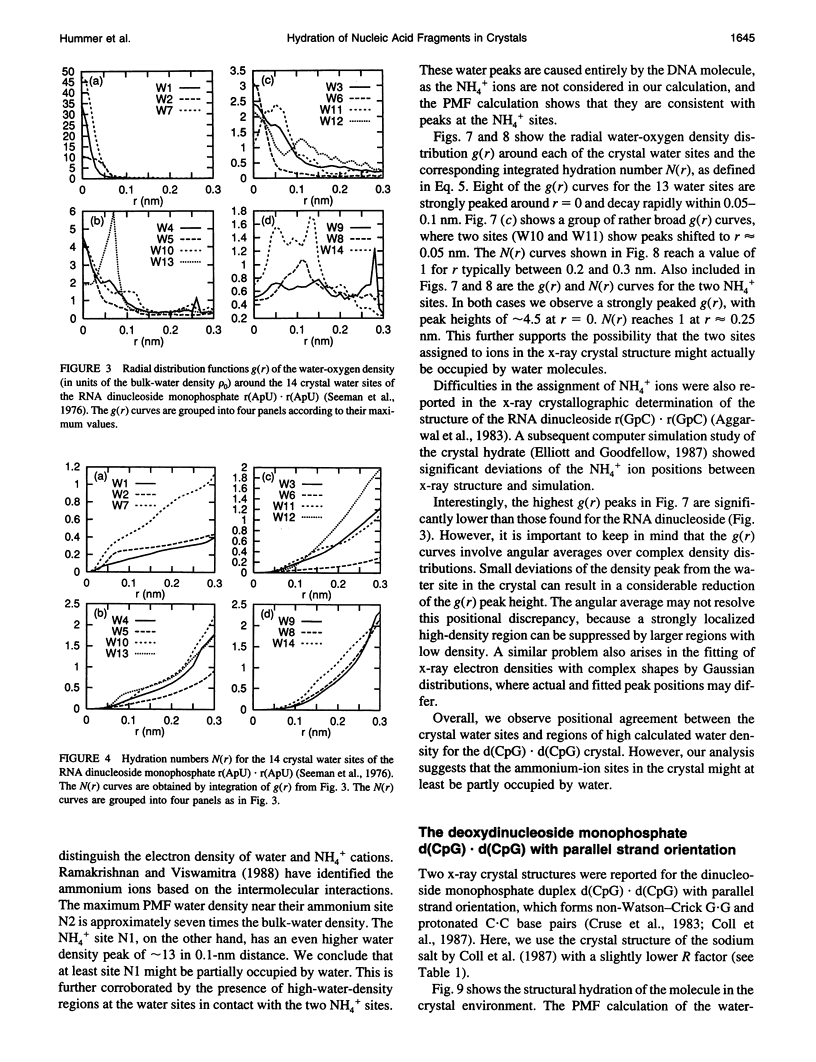
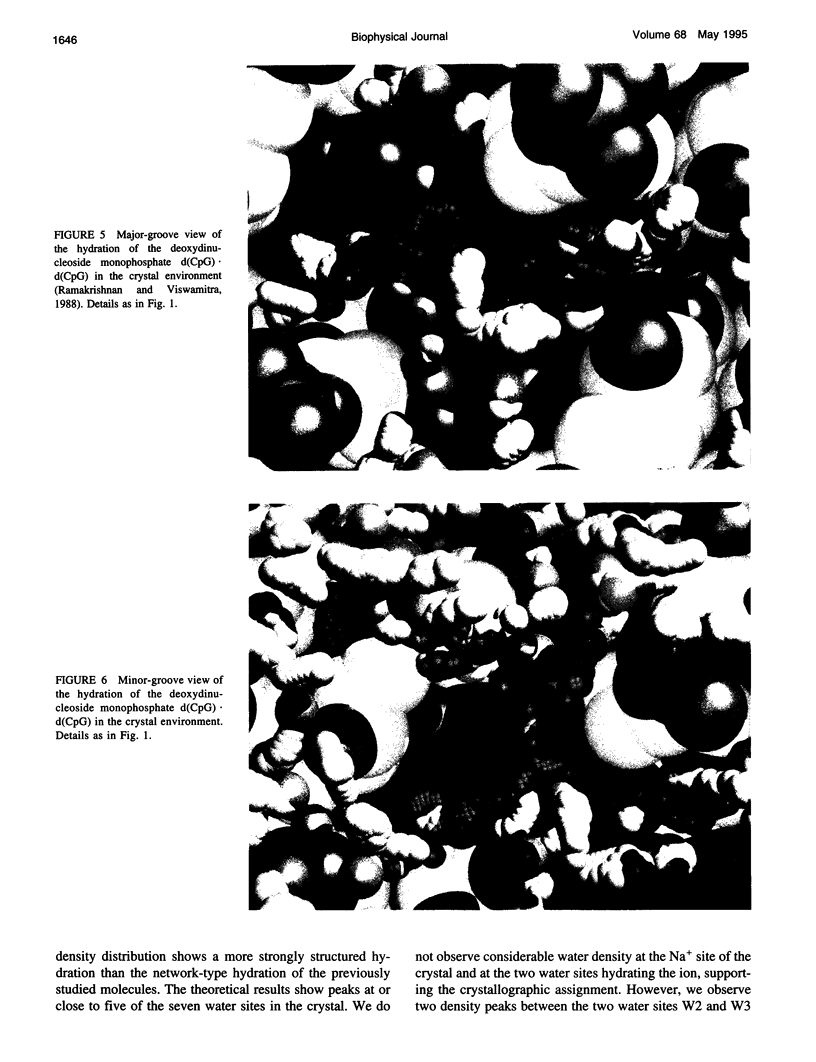
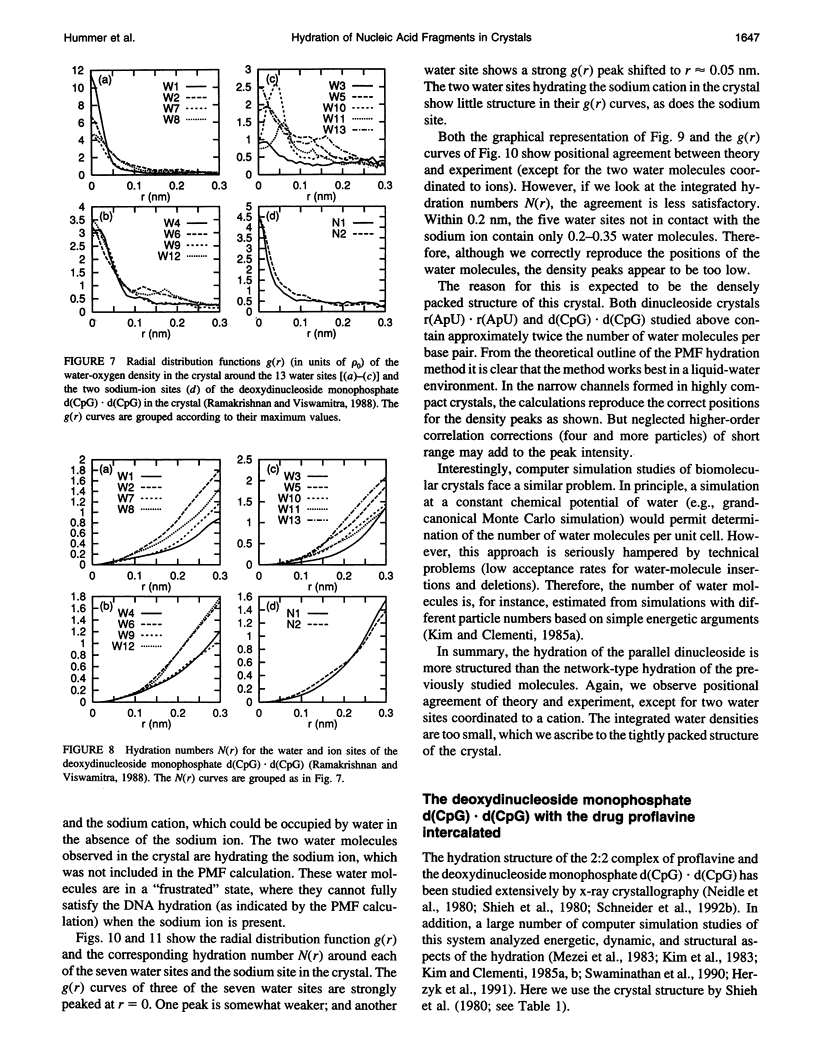



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman H. M. Hydration of nucleic acid crystals. Ann N Y Acad Sci. 1986;482:166–178. doi: 10.1111/j.1749-6632.1986.tb20948.x. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
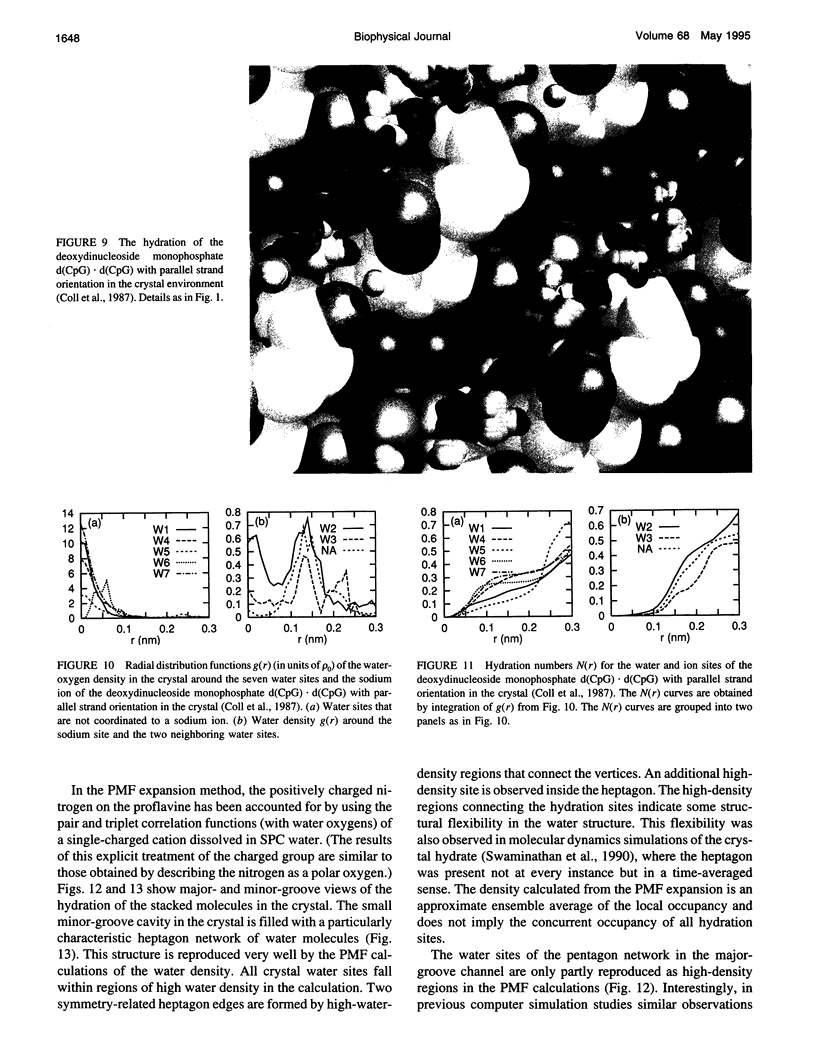
- Coll M., Solans X., Font-Altaba M., Subirana J. A. Crystal and molecular structure of the sodium salt of the dinucleotide duplex d(CpG). J Biomol Struct Dyn. 1987 Apr;4(5):797–811. doi: 10.1080/07391102.1987.10507679. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Egert E., Kennard O., Sala G. B., Salisbury S. A., Viswamitra M. A. Self base pairing in a complementary deoxydinucleoside monophosphate duplex: crystal and molecular structure of deoxycytidylyl-(3'-5')-deoxyguanosine. Biochemistry. 1983 Apr 12;22(8):1833–1839. doi: 10.1021/bi00277a014. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. DNA structure from A to Z. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- Elliott R. J., Goodfellow J. M. Monte Carlo simulations of nucleotide crystal hydrates and their counter-ions. J Theor Biol. 1987 Aug 21;127(4):403–412. doi: 10.1016/s0022-5193(87)80138-8. [DOI] [PubMed] [Google Scholar]
- Herzyk P., Goodfellow J. M., Neidle S. Molecular dynamics simulations of dinucleoside and dinucleoside-drug crystal hydrates. J Biomol Struct Dyn. 1991 Oct;9(2):363–386. doi: 10.1080/07391102.1991.10507918. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Corongiu G., Clementi E. Networks of water molecules in a proflavine deoxydinucleoside phosphate complex. J Biomol Struct Dyn. 1983 Oct;1(1):263–285. doi: 10.1080/07391102.1983.10507439. [DOI] [PubMed] [Google Scholar]
- Klement R., Soumpasis D. M., Jovin T. M. Computation of ionic distributions around charged biomolecular structures: results for right-handed and left-handed DNA. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4631–4635. doi: 10.1073/pnas.88.11.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M., Park B. H. Water: now you see it, now you don't. Structure. 1993 Dec 15;1(4):223–226. doi: 10.1016/0969-2126(93)90011-5. [DOI] [PubMed] [Google Scholar]
- Liepinsh E., Otting G., Wüthrich K. NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Res. 1992 Dec 25;20(24):6549–6553. doi: 10.1093/nar/20.24.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezei M., Beveridge D. L., Berman H. M., Goodfellow J. M., Finney J. L., Neidle S. Monte Carlo studies on water in the dCpG/proflavin crystal hydrate. J Biomol Struct Dyn. 1983 Oct;1(1):287–297. doi: 10.1080/07391102.1983.10507440. [DOI] [PubMed] [Google Scholar]
- Neidle S., Berman H. M., Shieh H. S. Highly structured water network in crystals of a deoxydinucleoside---drug complex. Nature. 1980 Nov 13;288(5787):129–133. doi: 10.1038/288129a0. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Wüthrich K. Protein hydration in aqueous solution. Science. 1991 Nov 15;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Viswamitra M. A. Crystal and molecular structure of the ammonium salt of the dinucleoside monophosphate d(CpG). J Biomol Struct Dyn. 1988 Dec;6(3):511–523. doi: 10.1080/07391102.1988.10506504. [DOI] [PubMed] [Google Scholar]
- Saenger W. Structure and dynamics of water surrounding biomolecules. Annu Rev Biophys Biophys Chem. 1987;16:93–114. doi: 10.1146/annurev.bb.16.060187.000521. [DOI] [PubMed] [Google Scholar]
- Savage H., Wlodawer A. Determination of water structure around biomolecules using X-ray and neutron diffraction methods. Methods Enzymol. 1986;127:162–183. doi: 10.1016/0076-6879(86)27014-7. [DOI] [PubMed] [Google Scholar]
- Schneider B., Cohen D. M., Schleifer L., Srinivasan A. R., Olson W. K., Berman H. M. A systematic method for studying the spatial distribution of water molecules around nucleic acid bases. Biophys J. 1993 Dec;65(6):2291–2303. doi: 10.1016/S0006-3495(93)81306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B., Cohen D., Berman H. M. Hydration of DNA bases: analysis of crystallographic data. Biopolymers. 1992 Jul;32(7):725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- Schneider B., Ginell S. L., Berman H. M. Low temperature structures of dCpG-proflavine. Conformational and hydration effects. Biophys J. 1992 Dec;63(6):1572–1578. doi: 10.1016/S0006-3495(92)81755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Suddath F. L., Kim J. J., Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3',5'-uridine hexahydrate. J Mol Biol. 1976 Jun 14;104(1):109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- Shieh H. S., Berman H. M., Dabrow M., Neidle S. The structure of drug-deoxydinucleoside phosphate complex; generalized conformational behavior of intercalation complexes with RNA and DNA fragments. Nucleic Acids Res. 1980 Jan 11;8(1):85–97. doi: 10.1093/nar/8.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texter J. Nucleic acid-water interactions. Prog Biophys Mol Biol. 1978;33(1):83–97. doi: 10.1016/0079-6107(79)90026-9. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Matthews B. W. Conservation of solvent-binding sites in 10 crystal forms of T4 lysozyme. Protein Sci. 1994 Jul;3(7):1031–1039. doi: 10.1002/pro.5560030705. [DOI] [PMC free article] [PubMed] [Google Scholar]