Abstract
Haemophilus influenzae is a commensal and opportunistic pathogen of the human airways. A number of surface molecules contribute to colonization of the airways by H. influenzae, such as adhesins, including structures found in the lipooligosaccharide (LOS). A human bronchiolar xenograft model was employed to investigate the host-bacterial interactions involved in the colonization of the airway by H. influenzae. Differential display was used to identify H. influenzae mRNA that reflect genes which were preferentially expressed in the xenograft compared to growth. Eleven mRNA fragments had consistent increased expression when the bacteria grew in xenografts. On sequencing these fragments, eight open reading frames were identified. Three of these had no match in the NCBI or the TIGR database, while an additional three were homologous to genes involved in heme or iron acquisition and utilization: two of the mRNAs encoded proteins homologous to enzymes involved in LOS biosynthesis: a heptosyl transferase (rfaF) involved in the synthesis of the LOS core and a ketodeoxyoctonate phosphate-dependent acyltransferase (htrB) that performs one of the late acylation reactions in lipid A synthesis. Inoculation of human bronchiolar xenografts revealed a significant reduction in colonization capacity by htrB mutants. In vitro, htrB mutants elicited lesser degrees of cytoskeletal rearrangement and less stimulation of host cell signaling with 16HBE14o− cells and decreased intracellular survival. These results implicate acylation of H. influenzae lipid A as playing a key role in the organisms' colonization of the normal airway.
Haemophilus influenzae is a nearly ubiquitous commensal of the human upper airways, especially among children (12). Encapsulated strains, of which there are six capsular serotypes, cause invasive disease such as meningitis and cellulitis. The majority of strains isolated from asymptomatic patients and those with localized airway infections are strains lacking capsular polysaccharide, i.e., nontypeable H. influenzae (NTHi) (28). The colonization of the airways is facilitated by a number of adhesive factors used by NTHi to circumvent mucociliary clearance. These include long-thin pili (or fimbriae), surface fibrils, and two high-molecular-weight adhesins (34). The receptors for these adhesins are unknown, although considerable data indicate that H. influenzae bind to mucins and other glycoproteins at the airway surface (22, 42).
Lipooligosaccharide (LOS) is the major immunogen on the H. influenzae surface and features an assortment of short (<15 saccharide units) oligosaccharides extending from all three heptoses of a triheptose core region (33). These oligosaccharides contain a number of molecules which mimic host structures, such as human blood-group antigens containing sialic acid and phosphorylcholine (ChoP) (25, 26, 35-37). The expression of host structures within the LOS has been proposed to be a means for utilizing host receptors to facilitate colonization (25). Studies with the gonococcus have shown that the expression of a terminal lactosamine unit upon the LOS allows for adherence of the organism to the asialoglycoprotein receptor on human sperm (19). Work by Tuomanen and colleagues established that Streptococcus pneumoniae utilizes ChoP within the cell wall teichoic acid to bind to the platelet-activating factor (PAF) receptor on host cells (10). Similarly, the expression of ChoP on the LOS of NTHi allows the organism to bind to the PAF receptor on human airway epithelial cells (42). More-recent data have indicated that the NTHi LOS can act as a PAF receptor agonist and that receptor activation after NTHi infection initiates a multifactorial signal cascade that is involved in bacterial entry (43).
As with lipopolysaccharide (LPS), most of the endotoxic activity of LOS is ascribed to lipid A. Much of the toxicity of enteric lipid A is conferred by the late acylation reactions, encoded by htrB and msbB (7). The lipid A of H. influenzae is hexa-acylated, and H. influenzae htrB mutants produce hyperphosphorylated LOS with a mixture of penta- and tetra-acylated lipid A (23). Monocytes and epithelial cells challenged with LOS isolated from an htrB mutant produce significantly less tumor necrosis factor alpha and interleukin-6 than those challenged with LOS from the parental strain (31). An htrB mutant of NTHi was also significantly attenuated in infection studies with an infant rat model (31).
A number of possible factors appear to be involved in the colonization of respiratory epithelium by H. influenzae. We sought to identify H. influenzae genes expressed during the colonization of normal human respiratory epithelium. A differential display approach was employed to identify H. influenzae mRNA representative of genes with increased expression in human airway xenografts compared to growth in vitro. The results indicate that the expression of genes involved in the acquisition and utilization of heme and LOS biosynthesis are increased during infection. Further experiments revealed that htrB mutants with lesser acylation of lipid A have decreased ability to colonize human airway xenografts in comparison to the parental strains. An htrB mutant also elicited lesser cytoskeletal rearrangements and cellular activation after inoculation of immortalized 16HBE14o− airway cells. These results indicate that the late acylation of the lipid A is important in the colonization of respiratory epithelium by H. influenzae and represents a key step in LOS biosynthesis.
MATERIALS AND METHODS
Bacteria.
Descriptions of the H. influenzae strains used in this study are provided in Table 1. Escherichia coli strains DH5α and HB101 were used as recipients in the cloning experiments. All H. influenzae strains were propagated on brain heart infusion agar or broth. (Difco-Becton Dickinson, Franklin Lakes, N.J.) supplemented with 10 μg of hemin (ICN Biomedicals [Aurora, Ohio] or Sigma [St. Louis, Mo.])/ml and 10 μg of NAD (Sigma)/ml (sBHI). Xenograft flushes (see below) were plated on the same media containing bacitracin at 100 μg/ml. Kanamycin-resistant transformants were selected on sBHI-bacitracin agar containing kanamycin at 15 μg/ml. Plasmid pGB19 was obtained from G. J. Barcak (University of Maryland) and introduced in to H. influenzae by electroporation (3).
TABLE 1.
H. influenzae strains
| Strain | Relevant genotype | Capsular type | Description | Source or reference |
|---|---|---|---|---|
| R3001 | Wild type | NTHi | Clinical isolate from CF patient | This study |
| R3001a | htrB mutant | NTHi | Tn5 mutant of R3001 | This study |
| E1a | Wild type | b | Serotype b strain | 41 |
| E1a∗ | htrB mutant | b | Tn5 mutant of E1a | This study |
| 2019 | Wild type | NTHi | Clinical isolate from COPD patient | 6, 30 |
| B29 | htrB mutant | NTHi | Tn3 htrB mutant of 2019 (cam) | 23 |
| 2019 pgmB::erm | htrB+ | NTHi | Erythromycin-resistant phosphoglucomutase mutant | 42 |
| 2019 licD::kan | htrB+ | NTHi | licD with kanamycin cassette from pUC4K | 24 |
| B29 (KKH1) | ΔhtrB(htrB+) | NTHi | Wild-type htrB in shuttle vector | This study |
| B29 (KKH2) | ΔhtrB(htrB+) | NTHi | Wild-type htrB in shuttle vector | This study |
Human airway xenografts.
Primary human airway epithelial cells were cultured on a devitalized rat tracheal matrix as xenografts implanted in nude mice as described previously (8). This tissue is repopulated with pseudostratified respiratory epithelium and is open at both ends (8). For differential display experiments, six xenografts were inoculated with 2 × 103 to 5 × 103 CFU of H. influenzae R3001. To assess xenograft colonization by wild-type and mutant strains of H. influenzae, xenografts were inoculated with 5 × 102 to 6 × 103 CFU of bacteria: 5 days after inoculation the xenografts were flushed with 0.2 ml of phosphate-buffered saline (PBS) containing 0.1% gelatin (PBS-G) and plated on sBHI-bacitracin agar. After 36 h of incubation at 37°C, the plates were inspected. If there was no growth, the xenograft was removed and homogenized in 5 ml of PBS-G in a standard clearance Potter-Elvehjem homogenizer,and the numbers of viable H. influenzae were determined by quantitative plate counting on sBHI-bacitracin agar. Colonization was defined as occurring if any inoculated H. influenzae organisms were recovered from a xenograft effulent or homogenate. A typical xenograft flush and homogenate of the entire xenograft contain 3.2 × 105 to 6.4 × 107 CFU 5 days after inoculation with 102 to 103 CFU of strain R3001.
Cryosections of the xenografts were prepared as follows: the central portions of the airway xenografts were excised and fixed in 2% paraformaldehyde in PBS and then stored at 4°C. They were then infiltrated with 30% sucrose and embedded in TBS tissue freezing medium from American Master Tech Scientific (Lodi, Calif.). Sections (6 to 8 nm) were cut, placed onto microscope slides, and stored at −20°C. Prior to the studies with labeled monoclonal antibodies to H. influenzae OMP6 or ChoP, the sections were placed at room temperature for 1 h.
Immortalized human bronchial epithelial cells.
Infection studies were performed with the simian virus 40-derived human bronchial epithelial line 16HBE14o−, which contains a diverse population of airway cells (16). For the adherence and invasion experiments, the cells were seeded into 24-well dishes (105 cells/well) in Eagle modified essential medium (Difco) supplemented with 10% fetal calf serum and 1% l-glutamine. For the electron microscopy experiments, the cells were seeded onto BioCoat membranes (1 μm [pore size], 105 cells/membrane; Becton Dickinson, Franklin Lakes, N.J.) in supplemented Eagle modified essential medium and incubated in submerged culture for 48 h. The medium overlying the monolayer was then aspirated to establish an air-liquid interface, and the cells were incubated for an additional 48 h with Widdicombe's medium in the basolateral compartment to permit maximum differentiation (49). Prior to inoculation, the transepithelial resistance (TER) of the monolayers was measured with a MilliCell (MilliPore, Bedford, Mass.) to confirm the establishment of polarized monolayers with tight junctions. The TER was between 1,000 and 2,500 Ω/cm2.
Differential display.
Xenografts were inoculated with 2 × 103 to 5 × 103 CFU of NTHi strain R3001 3 weeks after seeding with normal human tracheal epithelial cells and flushed or harvested as previously described (8). The xenograft was flushed with 0.2 ml of PBS-G, which was serially diluted and quantitatively cultured by plate count. Those xenografts whose effluent contained >106 CFU/ml were then flushed every other day with 0.2 ml of PBS with the effluent directed into 1 ml of Trizol (BRL, Bethesda, Md.) heated to 60°C. The tube was vortexed and the centrifuged (12,000 × g) for 10 min, and the supernatant was added to 0.1 ml of chloroform and again centrifuged. The supernatant was aspirated, and a 1/10 volume of 3 M sodium acetate (pH 5.2) was added, followed by 2 volumes of isopropanol; the mixture was then cooled to −20°C for 2 h. The nucleic acid was next pelleted by centrifugation (12,000 × g) for 10 min, the supernatant was aspirated, and the pellet was dried in a Speedy-Vac. The pellet was dissolved in 0.15 ml of diethylpyrocarbonate (DEPC)-treated water and treated with RNase-free DNase I by using the Message Clean Kit (GeneHunter, Brookline, Mass.). Eight flushes of the same xenograft yielded ca. 0.05 μg of RNA quantitated on its absorbance at 260 and 280 nm. The material from six separate xenografts flushed eight times was used as a source of in vivo RNA. For comparison, 25 ml of stationary-phase culture of H. influenzae strain R3001 grown in sBHI broth was centrifuged (10,000 × g) for 15 min at 0 to 4°C, and the pellet was suspended in 0.05 M sodium acetate (pH 5.2) and 5 ml of Trizol (at 60°C) added, and the suspension vigorously vortexed for 30 s. After centrifugation for 5 min at 10,000 × g the supernatant was aspirated, and a 1/10 volume of 3 M sodium acetate (pH 5.2) was added, followed by 2 volumes of isopropanol. After the mixture was cooled to −20°C for 2 h, the preparation was centrifuged (10,000 × g) for 30 min and the supernatant was aspirated and discarded; the pellet was dried in a Speedy-Vac. The pellet was dissolved in DEPC-treated water and DNase I treated as described above. The total RNA concentration was 1.1 μg/ml.
To verify that mRNA was present in the xenograft flush, we initially used the primers and conditions described by Wong and McClelland (48). We then designed 10 10-mers essentially by the technique of Fislage et al. (13). After we determined the 8-mer frequency in the genome of H. influenzae strain Rd KW20, we randomly added an A and a T to the 5′ and 3′ ends, respectively, of the 8-mer sequence. For the reverse transcription reaction, 10 ng of RNA in 10 μl of DEPC-treated water was heated to 65°C and added to 10 μl of first-strand synthesis buffer (Gibco-BRL) containing 0.01 μM concentrations of each of the 10-mer primers, 10 μl of 10 mM deoxynucleoside triphosphate mix (Gibco-BRL), and 50 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL). The tube containing the first-strand reaction mix was incubated at 37°C for 1 h, and then an equal volume (20 μl) of polymerase buffer (2 mM Tris-HCl, pH 8.3; 5 mM KCl; 4 mM MgCl2), 1 μM concentrations of the same primers, 1 μCi of [α-32P]dCTP, and 0.5 U of AmpliTaq DNA polymerase (Perkin-Elmer-Roche, Branchburg, N.J.) were added. An initial low-stringency PCR cycle started at 65°C, followed by 94°C for 1 min, 37°C for 2 min, and 72°C for 2 min, and in turn followed by 30 high-stringency cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min. Four volumes of loading solution (80% formamide with bromphenol blue and xylene cyanol) were added to each 50 μl of PCR mixture, and the samples were heated to 65°C and loaded onto a 5% acrylamide-50% urea sequencing gel. After electrophoresis, the gel was dried onto Whatman 3MM paper and autoradiographed. Lanes containing dilutions of the reverse transcription-PCR from broth-grown and xenograft-grown bacteria were loaded side by side.
The autoradiographs were scanned with an Alpha Inotech image processor, and each band in the in vitro lane was assigned relative value based on its density. We then compared the ratio of that band to the adjacent band found from RNA isolated from xenograft effluent. Our initial goal was to investigate in vivo bands whose density was twice that of the adjacent in vitro band. However, the run-to-run variation in each band intensity was two- to threefold. Eleven bands were 1.5- to 5.0-fold greater in intensity in the in vivo RNA prep. One of us (A.L.S.) independently selected the same 11 in vivo bands on the basis of their density being consistently greater than the adjacent in vitro band. These bands were excised with a scalpel and heated in 50 μl of Tris-EDTA buffer at 65°C for 1 h. The fragments were cloned into the PCR-Trap vector (Gene Hunter) and sequenced by using PCR-Trap primers (Gene Hunter).
Construction of mutants.
Once htrB was chosen as a target for further study, PCR primers directed against regions flanking the gene were identified in the H. influenzae Rd KW20 genome. The primers were htr-F (AGCTTTACGCCACGAAACAAA; coordinates 1597846 to 1597861) and htr-R (CGCAAAATTCACGAATAGCA; coordinates 1599155 to 1599171). After amplification by using PCR, a 1.3-kb fragment was gel purified, blunt-ended, and cloned into pUC19, and the clone was then transformed into E. coli HB101. htrB was mutagenized by using λTn5 according to the method of de Bruijn and Lupski (11). The resulting plasmid was linearized and used to transform H. influenzae R3001 and E1a after the induction of competence (20), with transformants selected on sBHI agar containing kanamycin at 15 μg/ml. The allelic exchange of htrB was confirmed by Southern analysis. H. influenzae strain B29 is an htrB mutant of NTHi 2019 that has been described previously (23).
Cloning of htrB.
The htrB gene in plasmid pCRII (Invitrogen, Carlsbad, Calif.) (23) was excised with EcoRI and cloned into pGB19 (3), a derivative of the E. coli-H. influenzae shuttle vector pHVT1, with an increased copy number. After electroporation (23) into strain B29, two clones were retained: B29/pKKH1 and B29/pKKH2.
Bacterial adherence and invasion experiments.
The invasion of 16HBE14o− cells by wild-type H. influenzae and htrB mutants was assessed by using a gentamicin-survival assay, as described elsewhere (42, 43). Bronchial cells were infected with NTHi (multiplicity of infection, 100:1) for 4 h, washed thoroughly, and incubated with 50 μg of gentamicin/ml for 1.5 h to kill extracellular bacteria. Control experiments revealed no significant permeation of the bronchial cells by gentamicin and no significant differences in the growth of extracellular bacteria during the course of the initial infection (data not shown). Cellular association (adherence plus invasion) was defined as the percentage of the inoculum ± the standard error of the mean recovered as viable CFU after a vigorous washing and no gentamicin treatment, whereas invasion was defined as the percentage of the inoculum recovered after gentamicin treatment. Significance was evaluated with a paired t test by using the interactive version of SAS software (SAS Institute).
Confocal laser scanning microscopy.
Human bronchial epithelial cells (16HBE14o−) were cultured at an air-fluid interface on BioCoat units inoculated with strain 2019 and isogenic mutants as described above and incubated for 4 h at 37°C. The membranes were washed twice in Dulbecco PBS (D-PBS; Gibco, Invitrogen, Carlsbad, Calif.) and fixed in 2% paraformaldehyde for 15 min. Immunohistochemical labeling was performed essentially as described previously (42). The PAF receptor was visualized by using a rabbit polyclonal antibody directed against an N-terminal peptide as described by Palade and coworkers (32). H. influenzae were visualized by using the monoclonal antibody 3B9, which recognizes the outer membrane protein P6 (5). After they were processed, the membranes were excised from the inserts and mounted under coverslips by using the ProLong Antifade mounting resin (Molecular Probes, Eugene, Oreg.). Confocal analysis was performed by using a Zeiss 510 Axiovert confocal laser scanning microscope (Carl Zeiss, Galveston, Tex.).
Electron microscopy.
After infection, the BioCoat membranes were washed twice in D-PBS and fixed in 4% paraformaldehyde for 30 min. The membranes were then divided and processed for scanning electron microscopy (SEM) and transmission electron microscopy (TEM). For SEM analysis, the membranes were incubated in 1% tannic acid in D-PBS, washed, and fixed in 2% glutaraldehyde for 1 h at room temperature. The membranes were treated with osmium tetraoxide, dehydrated with a graded ethanol series and hexamethyldisilazine, and mounted with adhesive tabs and colloidal silver. After sputter coating with gold-palladium, the stubs were viewed on a Hitachi S-4000 scanning electron microscope (Hitachi Plaza, San Francisco, Calif.).
For TEM analysis, 2% gluteraldehyde fixed membranes were treated with osmium tetraoxide and dehydrated with a graded acetone series prior to embedding within Epon resin. Sections (0.1 μm) were cut with a diamond knife, and the sections were stained with uranyl acetate and lead prior to viewing them on a Hitachi T-7000 or a JEOL 1200X transmission electron microscope (JEOL, Inc., Peabody, Mass.).
Immunoblot analysis.
The expression of ChoP on the H. influenzae LOS was assessed by colony immunoblot analysis as described previously (47). Monoclonal antibody TEPC-15 (immunoglobulin A [IgA]; Sigma-Aldrich, St. Louis, Mo.) was used at a 1:10,000 dilution. A goat anti-mouse IgA alkaline phosphatase conjugate (Bio-Rad, Hercules, Calif.) was used at a 1:10,000 dilution, and positive colonies were detected by using BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitro blue tetrazolium.
IP assay.
To gain insight into cell signaling by htrB mutants, cytosolic inositol phosphate (IP) levels were assessed by using a well-described radioassay (14). Immortalized 16HBE14o− airway cells were labeled with 3H-labeled myoinositol, infected with H. influenzae 2019 and derivatives with mutations in htrB pgmB licD (42), and cytosolic fractions prepared by methanol-chloroform extraction. IPs were isolated by ion-exchange chromatography as described previously (43). The statistical significance of the differences was calculated by t test by using SAS software.
RESULTS
Identification of genes with increased expression in vivo by differential display.
Differential display analysis was performed on pooled total RNA isolated from nontypeable H. influenzae strain R3001 grown in vitro and from the effulent of infected human airway epithelial xenografts. Eleven fragments with increased expression within the infected xenografts were identified and confirmed by Southern blotting to be found within the H. influenzae R3001 genome (data not shown). On sequencing, these fragments were portions of eight open reading frames (ORFs) (Table 2). These ORFs were compared against the TIGR and GenBank databases by using BLASTP, BLASTN, BLASTX, and TBLASTN. In addition, a motif search was performed at Swiss-Prot (www.expasy.ch). Three of these ORFs had no significant homology with any genes in the NCBI GenBank, Swiss-Prot, or TIGR database. Three of the ORFs were homologous to H. influenzae genes involved in the acquisition or utilization of heme. Two other ORFs were homologous to two other genes, rfaF and htrB, and are involved in the biosynthesis of H. influenzae LOS: rfaF encodes a heptosyltransferase that is required for the synthesis of the LOS core (30), and htrB encodes one of two putative ketodeoxyoctonate (KDO)-dependent acyltransferases responsible for the late acylation of the lipid A region of LOS (23).
TABLE 2.
In vivo-expressed genes identified by differential cDNA display analysis
| Databasea homology | Gene | Description | Source or reference |
|---|---|---|---|
| No match | ORF1 | No known homology | NA |
| No match | ORF2 | No known homology | NA |
| No match | ORF3 | No known homology | NA |
| HI0097-0099 | hitABC | Iron transport | 1 |
| HI0263-0264 | hxuAB | Heme utilization | 9 |
| HI0853 | hbpA | Heme-binding lipoprotein | 18 |
| HI1527 | htrB | Acyltransferase | 23 |
| HI1105 | rfaF | Heptosyl transferase | 30 |
The NCBI, TIGR, and Swiss-Prot databases were searched.
Effect of htrB mutation on adherence and invasion.
To assess the role of H. influenzae lipid A acylation in the adherence to and invasion of human bronchial epithelial cells, studies were performed by using the simian virus 40-immortalized airway cell line 16HBE14o− and H. influenzae with an intact htrB allele (strain 2019), an insertionally inactivated htrB allele (strain B29), or a null allele complemented with a plasmid-borne htrB gene as described previously (23). The cell association (adherence plus invasion) of strain 2019 was unaffected by the htrB mutation (Table 3). However, the results of gentamicin-survival invasion experiments revealed a significant decrease in the number of intracellular bacteria (P = 0.008). In strains in which the htrB mutation was complemented with an episomal copy of the gene the recovery of viable intracellular bacteria was partially restored; however, the invasion of these strains was still significantly less than that of the parental strain (Table 3).
TABLE 3.
Adherence and invasion by htrB strains of bronchial epithelial cells
| Strain | Genotype | Cell associationa (%) | SEMb | Invasiona (%) | SEM | Pc |
|---|---|---|---|---|---|---|
| 2019 | htrB+ | 29 | 4 | 0.79 | 0.13 | NA |
| B29 | htrB mutant | 22 | 8 | 0.04 | 0.018 | 0.008 |
| B29(pKKH1) | ΔhtrB (htrB+) | 19 | 7 | 0.18 | 0.004 | 0.004 |
| B29(pKKH2) | ΔhtrB (htrB+) | 33 | 11 | 0.24 | 0.021 | 0.002 |
Percentage of the initial inoculum.
n = 4.
P values were calculated by using a paired t test.
The effect of the htrB mutation on invasion by strain 2019 was examined qualitatively in cryosections of bronchial xenografts to determine the presence and location (intracellular versus extracellular) of bacteria. The cryosections were immunostained with a H. influenzae-specific monoclonal antibody directed against outer membrane protein P6 and examined by confocal microscopy (Fig. 1). Colocalization of the immunostaining indicated intracellular and extracellular bacteria in xenografts inoculated with H. influenzae B29. Similar observations were made with strain 2019 (data not shown). These data suggest that intracellular survival, as opposed to invasion, may be the primary defect in NTHi htrB mutants. SEM of 16HBE14o− bronchial cells inoculated with strain 2019 and the isogenic htrB mutant B29 revealed nonuniform distribution of the bacteria on the cell surface for both strains (Fig. 2A and B). For cells infected with 2019, there was a tendency toward host cell microvilli elongation, whereas no such pattern was seen with the htrB mutant. Attachment and internalization assessed by TEM were comparable for the two strains (Fig. 2C to H). While the heterogeneity of attachment of the organisms across the cell surface precluded accurate quantitation of adherence and internalization by TEM, a survey of ca. 200 surface epithelial cells per strain was performed. The average number organisms attached extracellularly per epithelial cell was greater for the htrB mutant than the parent (0.85 ± 0.7 for strain B29 per epithelial cell versus 0.50 ± 0.57 for strain 2019 per epithelial cell; P = 0.014). The number of intracellular organisms represented ca. 8% of the total organisms detected per NTHi strain (8.1% for the htrB mutant B29 and 8.2% for the parent 2019). These data support the hypothesis derived from the gentamicin survival assay (above), indicating that intracellular survival rather than epithelial cell attachment or invasion are affected by changes in LOS acylation.
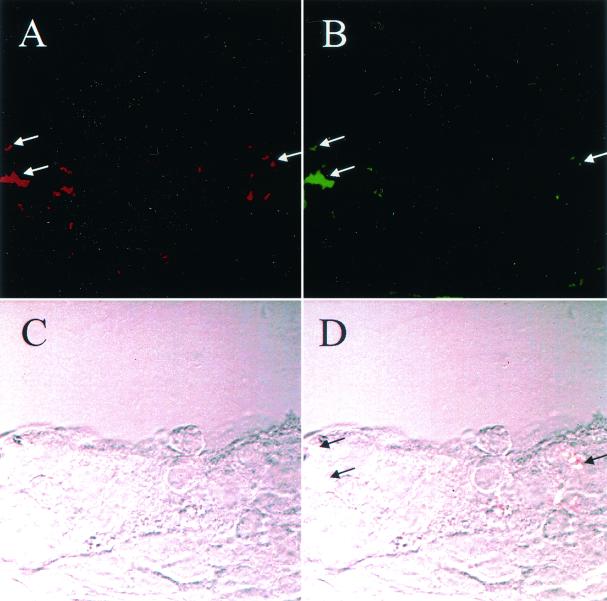
FIG. 1.
Cryosection of human bronchial epithelial cell xenograft infected with the htrB mutant B29. Bacteria were visualized by immunohistochemical staining with monoclonal antibody 3B9 (IgG), which recognizes the H. influenzae outer membrane protein P6 (A) and monoclonal antibody TEPC-15 (IgA), which recognizes ChoP (B). Goat anti-mouse IgG-FITC (Molecular Probes) and anti-mouse IgA-Texas red conjugate (Cortex Biochemical) secondary antibodies were used to identify bacteria (arrows). (C) Epithelial cell monolayer as visualized by Nomarski differential-interference contrast microscopy. (D) Image in which panels A and B are merged, with the resultant color (faint yellow) photographing as white (arrows). Comparable results were obtained in similar analyses of cells infected with strain 2019 (data not shown).
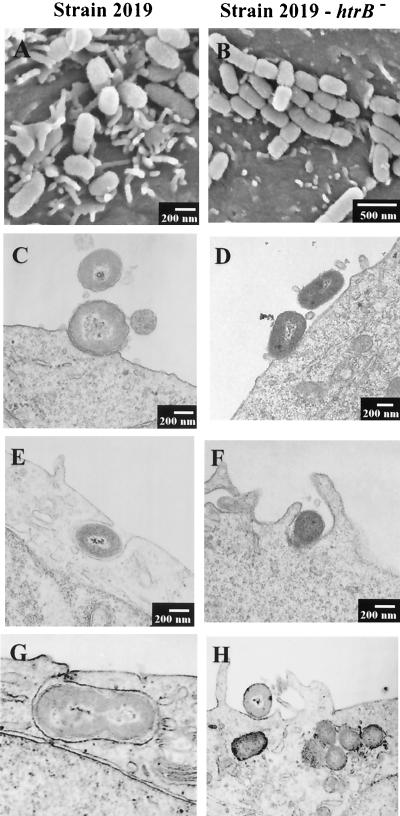
FIG. 2.
Analysis of immortalized 16HBE14o− human airway epithelial cells infected with NTHi 2019 (A, C, E, and G) and the isogenic NTHi 2019 htrB mutant B29 (B, D, F, and H). The airway cells were cultured at an air-fluid interface on BioCoat semipermeable membrane inserts (1-μm pore size), infected for 4 h as indicated, and processed for SEM analysis (A and B) or embedded in Epon resin, sectioned, and stained for TEM analysis (C to H). Note the difference in microvillus extension in airway cells infected with strain 2019 (A) compared to B29 (B). No apparent differences in the numbers or morphology of intracellular bacteria were observed in TEM analysis of cells infected with strain 2019 (C, E, and G) or B29 (D, F, and H).
Involvement of htrB in host cell signaling.
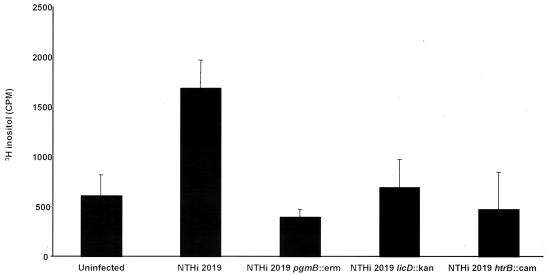
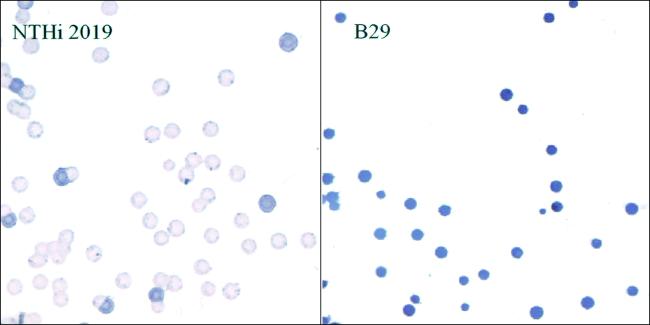
The effect of the htrB mutation on host cell signaling was evaluated by using a radioassay to measure cytosolic IP concentrations (Fig. 3). The inoculation of 16HBE14o− cells with strain 2019 resulted in a significant increase in cytosolic IP levels over those observed in uninfected cells (P = 0.012), as has been described elsewhere (43). In parallel experiments, no changes in IP levels were observed in cells infected with the isogenic htrB mutant (P = 0.29). These results are similar to those obtained in experiments with the poorly invasive 2019 pgmB and 2019 licD mutants (43). Since the loss of ChoP on the LOS may result in an inability of the mutants to activate PAF receptor signaling, the presence of ChoP on the htrB mutant B29 was assessed. Of particular interest, the 2019 htrB mutant had an increased, rather than decreased, reactivity on colony immunoblots with a T15 idiotype anti-ChoP monoclonal antibody compared to the parental strain (Fig. 4). Analysis of 16HBE14o− cells inoculated with 2019 and isogenic htrB mutants revealed no apparent difference in bacterial colocalization with the PAF receptor (data not shown). These results demonstrate that although htrB mutants express ChoP and bind to the PAF receptor, they fail to initiate cellular signaling, suggesting that the structural context of ChoP in the NTHi LOS is important to PAF receptor activation. These findings are consistent with previous data (42, 43).
FIG. 3.
Effect of inoculation of NTHi 2019 and isogenic mutants on cytosolic IP levels in immortalized 16HBE14o− human airway epithelial cells. Wild-type strain 2019, in comparison with the poorly invasive strain with mutations in pgmB and licD results in the increased cytosolic IP levels by means of PAF receptor activation; mutations affecting the addition of ChoP onto the oligosaccharides of the strain 2019 LOS reduce this effect (43). The 16HBE14o− cells were labeled with 3H-labeled myoinositol and inoculated with the strain indicated, and the IP levels in the cytosolic fractions were assessed.
FIG. 4.
Effect of htrB mutation on ChoP expression by NTHi 2019. Colonies of strain 2019 and the isogenic htrB mutant (strain B29) were lifted onto nitrocellulose, and immunoblot analyses were performed as described in Materials and Methods and in prior studies (42, 47).
Effect of htrB mutation on the colonization of human airway xenografts.
Human airway xenografts were inoculated with the H. influenzae serotype b strain E1a, and NTHi strains R3001 and 2019, as well as isogenic htrB mutants (Table 4). Each parental strain colonized the xenografts; however, the htrB mutants were all markedly decreased in their ability to survive and colonize the xenografts. Considering the in vitro adherence and invasion data, these data strongly suggest that the acylation of the lipid A is required for the colonization of respiratory epithelium by H. influenzae.
TABLE 4.
Colonization capacity of H. influenzae and isogenic htrB mutants
| Strain | htrB | Colonization of human respiratory xenograftsa
|
||
|---|---|---|---|---|
| Type | Inoculum | No. colonized/total no. | ||
| E1a | + | b | 103 | 10/10 |
| E1a∗ | − | b | 103 | 1/10 |
| R3001 | + | NTHi | 102 | 10/10 |
| R3001a | − | NTHi | 102 | 2/10 |
| 2019 | + | NTHi | 103 | 10/10 |
| B29 | − | NTHi | 103 | 0/10 |
Colonization is defined as present if the xenograft flush or homogenate culture yielded H. influenzae 5 days (flush) or 7 days (homogenate) after inoculation.
DISCUSSION
Innate immune defenses such as defensins, C reactive protein, and lactoferrin are found at the airway surface and are beginning to be recognized as major contributors to the clearance and containment of bacteria within the airways (15, 38, 39). The uptake of bacteria within airway epithelial cells may serve to protect the bacteria from killing by these factors, or the uptake of bacteria by epithelial cells may be itself a component of the innate immunity (50). Mutations affecting lipid A acylation are known to destabilize the gram-negative bacterial cell envelope and could easily confer increased sensitivity to killing by host cells. Key insights have been provided by prior work with Salmonella enterica serovar Typhimurium, in which mutations that eliminate the addition of long-chain fatty acids to lipid A increase the sensitivity to killing by antimicrobial peptides and polymyxin B and lead to attenuation of virulence in animal models (17).
The acylation of the lipid A has long been recognized as a key determinant of the toxicity of LPS or LOS. In particular, the late acylation reactions mediated by the products of htrB and msbB are responsible for much of the toxicity of enterobacterial lipid A. Previous data from our laboratories have shown that mutation of htrB results in a largely detoxified LOS and significantly reduces the virulence of H. influenzae in an infant rat model of infection (31). The current data show that htrB expression is increased during colonization of human respiratory epithelium and that the loss of htrB largely attenuates the ability of NTHi to colonize and persist in a bronchiolar xenograft. In an independent series of experiments, we have found that as few as 10 CFU of H. influenzae strain R3001 can colonize human bronchiolar xenografts. Within 24 h of inoculation PBS-G flushed through the xenograft contains 2 × 106 to 8 × 106 CFU/ml, which then increases to 2 × 107 to 5 × 107 CFU/ml over the next 24 h. This density is maintained for 3 weeks, the maximum time that the lumen of the xenograft is patent. Our data suggest transcriptional regulation of LOS biosynthesis and late acylation. Given the importance of the late acylation reactions to the endotoxic nature of LOS or LPS, it is interesting that previous work has shown that there is heterogeneity among lipid A produced by different NTHi strains (2). It is tempting to speculate that NTHi may differentially acylate its LOS during commensal and disease states, resulting not only in differences in LOS toxicity but also differences in persistence or clearance. Differential lipid A acylation may therefore be a widely shared adaptive change in bacterial populations during long-term colonization of the airway.
The results of the in vitro infection studies also suggest that htrB mutants are unable to survive within airway epithelial cells. These and prior data clearly demonstrate that NTHi colonize airway cells in a nonuniform fashion that may preclude a representative quantitative analysis by TEM (21, 43). However, present analyses of immortalized airway cells inoculated with NTHi and isogenic htrB mutants by TEM and thick cryosection revealed comparable numbers of intracellular bacteria. Since bacterial viability cannot be assessed by TEM, we relied on the gentamicin kill experiments to indicate the numbers of viable, intracellular bacteria. These experiments indicated that the majority of the intracellular htrB mutants are not viable. Furthermore, htrB mutants express ChoP and bind to the PAF receptor and yet are deficient in the ability to activate PAF receptor-mediated signaling. Although the role of ChoP in persistent host colonization is not yet fully understood, data from a number of different studies with animal models and patient samples have indicated that host nasopharyngeal carriage enriches for ChoP+ H. influenzae variants (44-47). ChoP may be expressed within a variety of structural contexts upon the oligosaccharide portion of the LOS (24, 26, 37), and the nature of the presentation of ChoP affects binding to the PAF receptor (42) and C reactive protein (24). The present data suggest that ChoP must be present within a fully acylated LOS structure for activation of the PAF receptor by NTHi. These findings are reminiscent of biochemical work that has demonstrated that the removal of the acetyl side group from PAF, the host ligand for the PAF receptor, diminishes its ability to activate receptor-mediated signaling (4).
Previous data have shown an increase in the transcription of rfaE in an htrB mutant (23), and structural analysis of LOS isolated from H. influenzae htrB mutants revealed hyperphosphorylation of the oligosaccharide portion of the LOS (23). The means whereby htrB expression is controlled and how it affects the expression of other LOS biosynthetic genes is not defined. The data also show that the expression of rfaF is increased during host colonization. rfaF encodes a heptosyl transferase that carries out one of the early steps in the synthesis of the LOS core region, and thus its increased transcription may be an index of increased LOS biosynthesis. Data presented here therefore provide support for the hypothesis that htrB expression and LOS biosynthesis are controlled at the transcriptional level and are increased during colonization of human respiratory epithelial cells. Future studies will address the means of transcriptional regulation of htrB and its role in the regulation of the expression of other genes.
Acknowledgments
We acknowledge the staff of the University of Iowa Central Microscopy Research Facility, including Randy Nessler and Tom Moninger, as well as the staff of the University of Missouri-Columbia Electron Microscopy Core Facility, including Cheryl Jensen and Randy Tindall, for their assistance. We thank Allan Weber for assistance with differential display.
The Experimental Station Chemical Laboratories of the University of Missouri provided financial assistance. This work was also supported by research grants from the Cystic Fibrosis Foundation (A.L.S.), the Allen P. and Josephine B. Green Foundation (D.L.C.), and the USPHS (AI10053, D.L.C.; AI44002, A.L.S.; AI165298 and AI24616, M.A.A.). Assistance in xenograft studies at the University of Iowa was provided by Yulong Zhang and John Engelhardt of the Animal Models Core of the Iowa Center for Gene Therapy of Cystic Fibrosis and Other Genetic Diseases, which is cofunded by the National Institutes of Health and the Cystic Fibrosis Foundation (P30 DK54759). W.E.S. was supported by a training grant from the NIAID (AI07511). Work in W.E.S.'s laboratory is supported in part by USPHS grant AI50108.
Editor: B. B. Finlay
REFERENCES
- 1.Adhikari, P., S. D. Kirby, A. J. Nowalk, K. L. Veraldi, A. B. Schryvers, and T. A. Mietzner. 1995. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J. Biol. Chem. 270:25142-25149. [DOI] [PubMed] [Google Scholar]
- 2.Apicella, M. A., K. C. Dudas, A. Campagnari, P. Rice, J. M. Mylotte, and T. F. Murphy. 1985. Antigenic heterogeneity of lipid A of Haemophilus influenzae. Infect. Immun. 50:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcak, C. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 4.Blank, M. L., T. Lee, V. Fitzgerald, and F. Snyder. 1981. A specific acetylhydrolase for 1-alkyl-2-acetyl-sn-glycero-3-phosphocholine (a hypotensive and platelet-activating lipid). J. Biol. Chem. 256:175-178. [PubMed] [Google Scholar]
- 5.Bogdan, J. A., Jr., and M. A. Apicella. 1995. Mapping of a surface-exposed, conformational epitope of the P6 protein of Haemophilus influenzae. Infect. Immun. 63:4395-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campagnari, A. A., M. R. Gupta, K. C. Dudas, T. F. Murphy, and M. A. Apicella. 1987. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect. Immun. 55:882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz, T., Z. Zhou, and C. R. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A: acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 8.Cohn, L. A., A. Weber, T. Phillips, S. Lory, M. Kaplan, and A. Smith. 2001. Pseudomonas aeruginosa infection of respiratory epithelium in a cystic fibrosis xenograft model. J. Infect. Dis. 183:919-927. [DOI] [PubMed] [Google Scholar]
- 9.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 11.de Bruijn, F. J., and J. R. Lupski. 1984. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene 27:131-149. [DOI] [PubMed] [Google Scholar]
- 12.Faden, H., M. J. Waz, J. M. Bernstein, L. Brodsky, J. Stanievich, and P. L. Ogra. 1991. Nasopharyngeal flora in the first three years of life in normal and otitis-prone children. Ann. Otol. Rhinol. Laryngol. 100:612-615. [DOI] [PubMed] [Google Scholar]
- 13.Fislage, R., M. Berceanu, Y. Humboldt, M. Wendt, and H. Oberender. 1997. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 25:1830-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foubister, V., I. Rosenshine, and B. B. Finlay. 1994. A diarrheal pathogen, enteropathogenic Escherichia coli (EPEC), triggers a flux of inositol phosphates in infected epithelial cells. J. Exp. Med. 179:993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould, J. M., and J. N. Weiser. 2001. Expression of C-reactive protein in the human respiratory tract. Infect. Immun. 69:1747-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruenert, D. C., W. E. Finkbeiner, and J. H. Widdicombe. 1995. Culture and transformation of human airway epithelial cells. Am. J. Physiol. 269:L347-L360. [DOI] [PubMed] [Google Scholar]
- 17.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson, M. S., C. Slaughter, and E. J. Hansen. 1992. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect. Immun. 60:2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey, H. A., N. Porat, C. A. Campbell, M. Jennings, B. W. Gibson, N. J. Phillips, M. A. Apicella, and M. S. Blake. 2000. Gonococcal lipooligosaccharide is a ligand for the asialoglycoprotein receptor on human sperm. Mol. Microbiol. 36:1059-1070. [DOI] [PubMed] [Google Scholar]
- 20.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketterer, M. R., J. Q. Shao, D. B. Hornick, B. Buscher, V. K. Bandi, and M. A. Apicella. 1999. Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect. Immun. 67:4161-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubiet, M., and R. Ramphal. 1995. Adhesion of nontypeable Haemophilus influenzae from blood and sputum to human tracheobronchial mucins and lactoferrin. Infect. Immun. 63:899-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, N.-G., M. G. Sunshine, J. J. Engstrom, B. W. Gibson, and M. A. Apicella. 1995. Mutation of the htrB locus of Haemophilus influenzae nontypable strain 2019 is associated with modifications of lipid A and phosphorylation of the lipo-oligosaccharide. J. Biol. Chem. 270:27151-27159. [PubMed] [Google Scholar]
- 24.Lysenko, E., J. C. Richards, A. D. Cox, A. Stewart, A. Martin, M. Kapoor, and J. N. Weiser. 2000. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35:234-245. [DOI] [PubMed] [Google Scholar]
- 25.Mandrell, R. E., and M. A. Apicella. 1993. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology 187:382-402. [DOI] [PubMed] [Google Scholar]
- 26.Mansson, M., S. H. Bauer, D. W. Hood, J. C. Richards, E. R. Moxon, E. K. Schweda. 2001. A new structural type for Haemophilus influenzae lipopolysaccharide: structural analysis of the lipopolysaccharide from nontypeable Haemophilus influenzae strain 486. Eur. J. Biochem. 268:2148-2159. [DOI] [PubMed] [Google Scholar]
- 27.Masoud, H., E. Moxon, A. Martin, D. Krajcarski, and J. Richards. 1997. Structure of the variable and conserved lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae serotype b strain eagan. Biochemistry 36:2091-2103. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, T. F., and M. A. Apicella. 1987. Nontypable Haemophilus influenzae: a review of clinical aspects, surface antigens and the human response to infection. Rev. Infect. Dis. 9:1-15. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, T. F., K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, W. A., B. W. Gibson, W. Melaugh, N. G. Lee, M. Sunshine, and M. A. Apicella. 1997. Identification of the ADP-l-glycero-d-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect. Immun. 65:1377-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols, W. A., C. R. H. Raetz, T. Clementz, A. L. Smith, J. A. Hanson, M. R. Ketterer, M. Sunshine, and M. A. Apicella. 1997. htrB of Haemophilus influenzae: determination of biochemical activity and effects on virulence and lipooligosaccharide toxicity. J. Endotoxin Res. 4:163-172. [Google Scholar]
- 32.Predescu, D., K. Ihida, S. Predescu, and G. E. Palade. 1996. The vascular distribution of the platelet-activating factor receptor. Eur. J. Cell Biol. 69:86-98. [PubMed] [Google Scholar]
- 33.Preston, A., R. E. Mandrell, B. W. Gibson, and M. A. Apicella. 1996. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 22:139-180. [DOI] [PubMed] [Google Scholar]
- 34.Rao, V. K., G. P. Krasan, D. R. Hendrixson, S. Dawid, and J. W. St. Geme III. 1999. Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 23:99-129. [DOI] [PubMed] [Google Scholar]
- 35.Risberg, A., G. Alvelius, and E. K. Schweda. 1999. Structural analysis of the lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae strain RM.118-26. Eur. J. Biochem. 265:1067-1074. [DOI] [PubMed] [Google Scholar]
- 36.Risberg, A., H. Masoud, A. Martin, J. C. Richards, E. R. Moxon, and E. K. Schweda. 1999. Structural analysis of the lipopolysaccharide oligosaccharide epitopes expressed by a capsule-deficient strain of Haemophilus influenzae Rd. Eur. J. Biochem. 261:171-180. [DOI] [PubMed] [Google Scholar]
- 37.Schweda, E. K., J. R. Brisson, G. Alvelius, A. Martin, J. N. Weiser, D. W. Hood, E. R. Moxon, and J. C. Richards. 2000. Characterization of the phosphocholine-substituted oligosaccharide in lipopolysaccharides of type b Haemophilus influenzae. Eur. J. Biochem. 267:3902-3913. [DOI] [PubMed] [Google Scholar]
- 38.Singh, P. K., H. P. Jia, K. Wiles, J. Hesselberth, L. Liu, B. A. Conway, E. P. Greenberg, E. V. Valore, M. J. Welsh, T. Ganz, B. F. Tack, and P. B. McCray, Jr. 1998. Production of beta-defensins by human airway epithelia. Proc. Natl. Acad. Sci. USA 95:14961-14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, P. K., B. F. Tack, P. B. McCray, Jr., and M. J. Welsh. 2000. Synergistic and additive killing by antimicrobial factors found in human airway surface liquid. Am. J. Physiol. Lung Cell Mol. Physiol. 279:L799-L805. [DOI] [PubMed] [Google Scholar]
- 40.St. Geme, J. W. 3rd, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stull, T. L. 1987. Protein sources of heme for Haemophilus influenzae. Infect. Immun. 55:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swords, W. E., B. Buscher, K. Ver Steeg, W. Nichols, A. Preston, J. N. Weiser, B. Gibson, and M. A. Apicella. 2000. Nontypeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells by an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 43.Swords, W. E., M. R. Ketterer, J. Shao, C. A. Campbell, J. N. Weiser, and M. A. Apicella. 2001. Binding of the nontypeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signaling. Cell Microbiol. 8:525-536. [DOI] [PubMed] [Google Scholar]
- 44.Tong, H. H., L. E. Blue, M. A. James, Y. P. Chen, and T. F. DeMaria. 2000. Evaluation of phase variation of nontypeable Haemophilus influenzae lipooligosaccharide during nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:4593-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 46.Weiser, J. N., Pan, N., McGowan, K. L., Musher, D., Martin, A., Richards, J. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong, K. K., and M. McClelland. 1994. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc. Natl. Acad. Sci. USA 91:639-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaya, M., W. E. Finkbeiner, S. Y. Chun, and J. H. Widdicombe. 1992. Differentiated structure and function of cultures from human tracheal epithelium. Am. J. Physiol. 262:L713-L724. [DOI] [PubMed] [Google Scholar]
- 50.Zaidi, T. S., J. Lyczak, M. Preston, and G. B. Pier. 1999. Cystic fibrosis transmembrane conductance regulator-mediated corneal epithelial cell ingestion of Pseudomonas aeruginosa is a key component in the pathogenesis of experimental murine keratitis. Infect. Immun. 67:1481-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]