Abstract
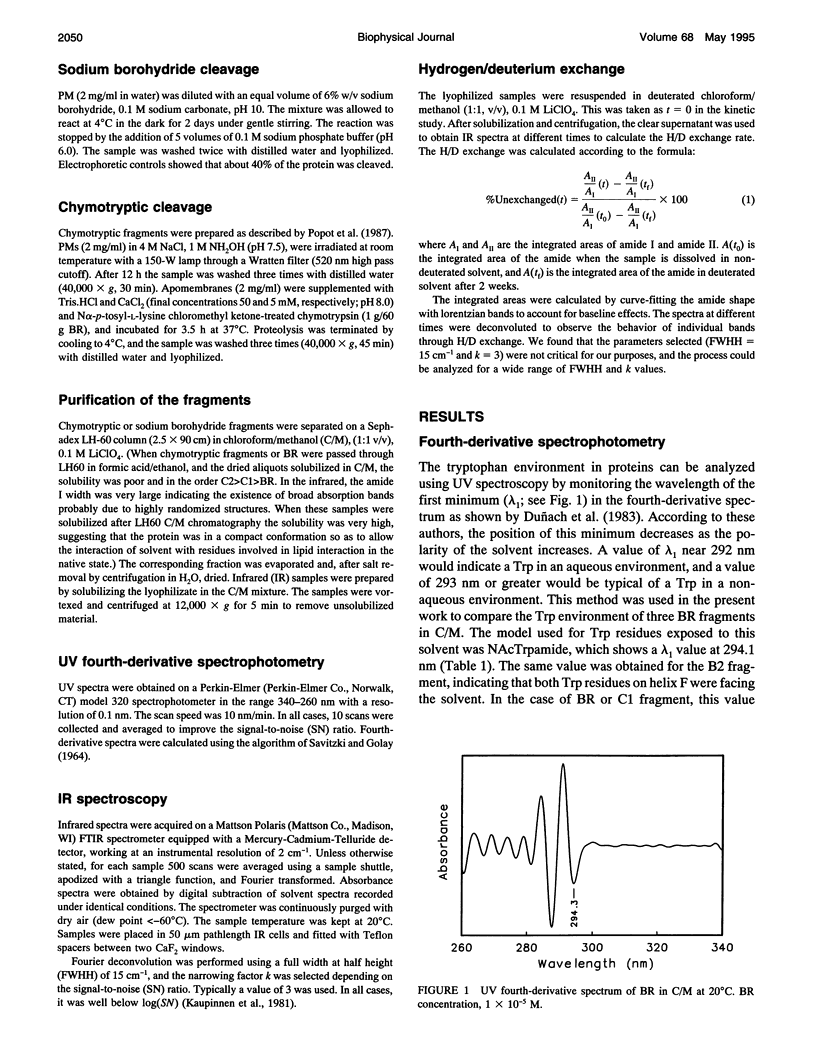
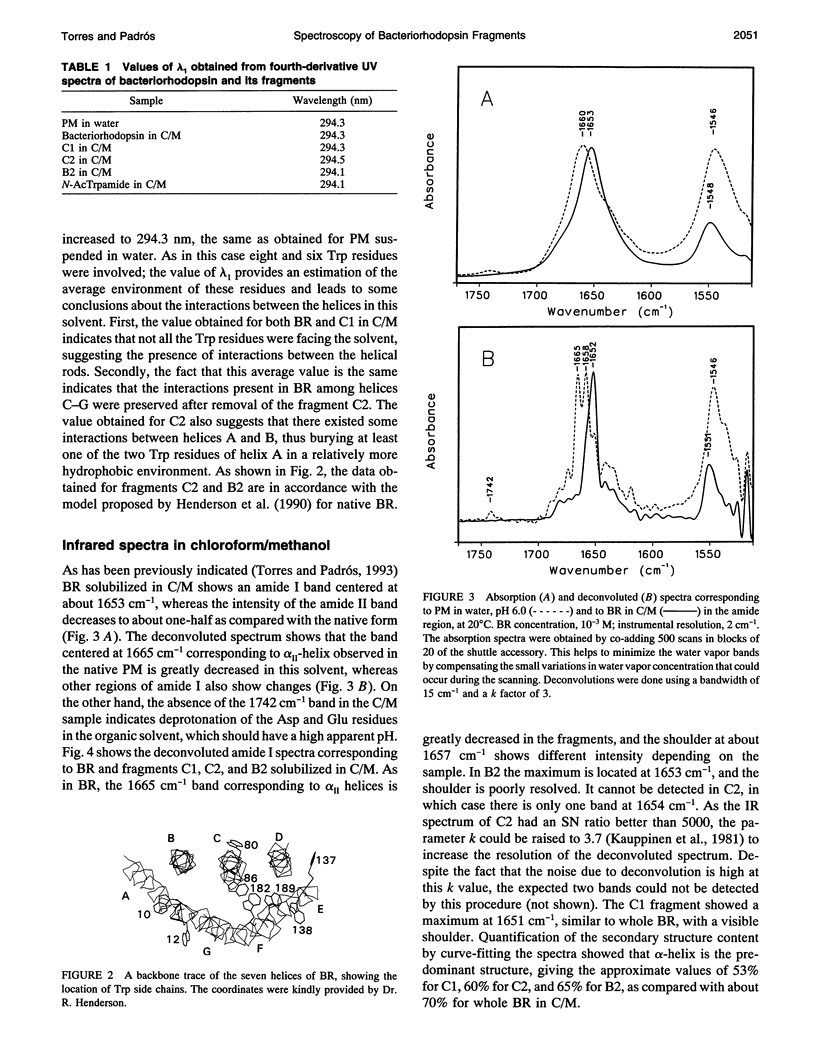
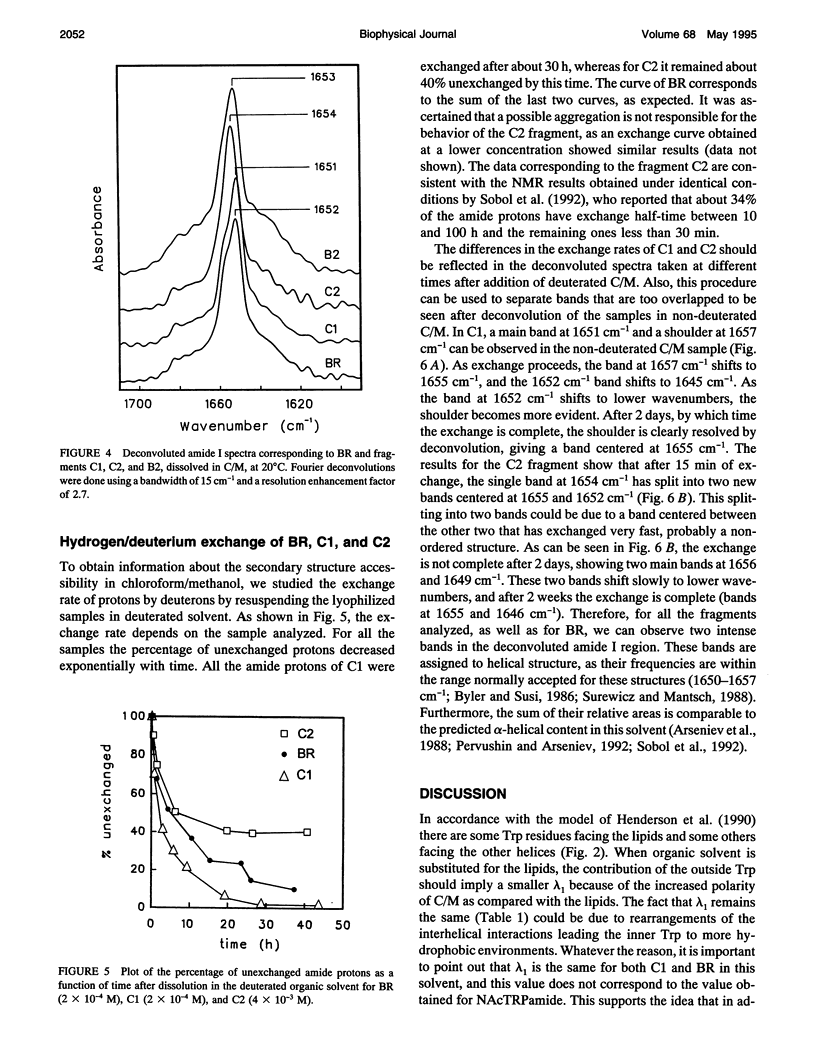
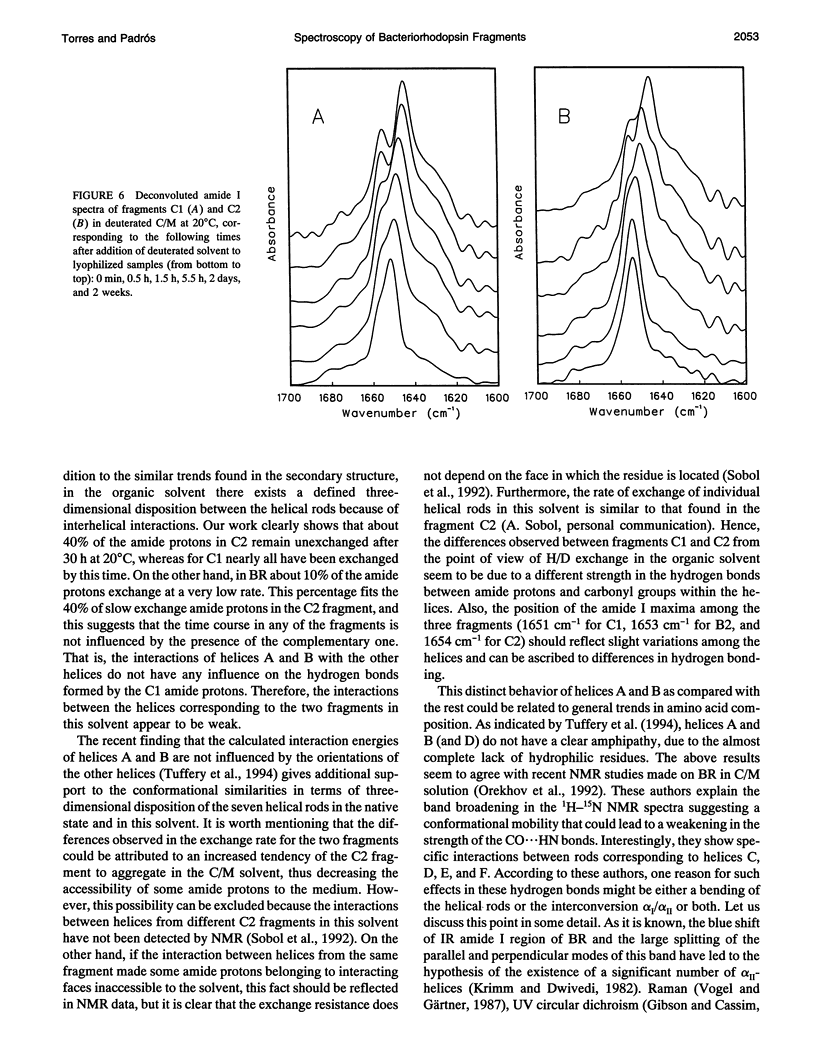
Fourier transform infrared and UV fourth-derivative spectroscopies were used to study the secondary structure of bacteriorhodopsin and its chymotryptic and one of the sodium borohydride fragments dissolved in chloroform-methanol (1:1, v/v), 0.1 M LiClO4. The C1 fragment (helices C, D, E, F, and G) showed an alpha-helical content of about 53%, whereas C2 (helices A and B) had about 60%, and B2 (helices F and G) about 65% alpha-helix. The infrared main band indicated differences in alpha-helical properties between these fragments. These techniques were also used to obtain information on the interactions among helices. According to the results obtained from the hydrogen/deuterium exchange kinetics, about 40% of the amide protons of C2 are particularly protected against exchange, whereas for the C1 fragment this process is unexpectedly fast. UV fourth-derivative spectra of these samples were used to obtain information about the environment of Trp side chains. The results showed that the Trp residues of C2 are more shielded from the solvent than those of C1 or B2. The results of this work indicate that the specific interactions existing between the transmembrane segments induce different types of helical conformations in native bacteriorhodopsin.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barsukov I. L., Abdulaeva G. V., Arseniev A. S., Bystrov V. F. Sequence-specific 1H-NMR assignment and conformation of proteolytic fragment 163-231 of bacterioopsin. Eur J Biochem. 1990 Sep 11;192(2):321–327. doi: 10.1111/j.1432-1033.1990.tb19230.x. [DOI] [PubMed] [Google Scholar]
- Byler D. M., Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986 Mar;25(3):469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- Cladera J., Sabés M., Padrós E. Fourier transform infrared analysis of bacteriorhodopsin secondary structure. Biochemistry. 1992 Dec 15;31(49):12363–12368. doi: 10.1021/bi00164a010. [DOI] [PubMed] [Google Scholar]
- Duñach M., Sabés M., Padrós E. Fourth-derivative spectrophotometry analysis of tryptophan environment in proteins. Application to melittin, cytochrome c and bacteriorhodopsin. Eur J Biochem. 1983 Jul 15;134(1):123–128. doi: 10.1111/j.1432-1033.1983.tb07540.x. [DOI] [PubMed] [Google Scholar]
- Fillingame R. H., Girvin M. E., Fraga D., Zhang Y. Correlations of structure and function in H+ translocating subunit c of F1F0 ATP synthase. Ann N Y Acad Sci. 1992 Nov 30;671:323–334. doi: 10.1111/j.1749-6632.1992.tb43806.x. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Krimm S., Dwivedi A. M. Infrared spectrum of the purple membrane: clue to a proton conduction mechanism? Science. 1982 Apr 23;216(4544):407–408. doi: 10.1126/science.6280277. [DOI] [PubMed] [Google Scholar]
- Liao M. J., London E., Khorana H. G. Regeneration of the native bacteriorhodopsin structure from two chymotryptic fragments. J Biol Chem. 1983 Aug 25;258(16):9949–9955. [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Tittor J., Bamberg E. A unifying concept for ion translocation by retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- Orekhov VYu, Abdulaeva G. V., Musina LYu, Arseniev A. S. 1H-15N-NMR studies of bacteriorhodopsin Halobacterium halobium. Conformational dynamics of the four-helical bundle. Eur J Biochem. 1992 Nov 15;210(1):223–229. doi: 10.1111/j.1432-1033.1992.tb17412.x. [DOI] [PubMed] [Google Scholar]
- Pervushin K. V., Arseniev A. S. Three-dimensional structure of (1-36)bacterioopsin in methanol-chloroform mixture and SDS micelles determined by 2D 1H-NMR spectroscopy. FEBS Lett. 1992 Aug 17;308(2):190–196. doi: 10.1016/0014-5793(92)81272-n. [DOI] [PubMed] [Google Scholar]
- Popot J. L., Gerchman S. E., Engelman D. M. Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J Mol Biol. 1987 Dec 20;198(4):655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- Rothschild K. J. FTIR difference spectroscopy of bacteriorhodopsin: toward a molecular model. J Bioenerg Biomembr. 1992 Apr;24(2):147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- Seigneuret M., Neumann J. M., Rigaud J. L. Detergent delipidation and solubilization strategies for high-resolution NMR of the membrane protein bacteriorhodopsin. J Biol Chem. 1991 Jun 5;266(16):10066–10069. [PubMed] [Google Scholar]
- Sigrist H., Wenger R. H., Kislig E., Wüthrich M. Refolding of bacteriorhodopsin. Protease V8 fragmentation and chromophore reconstitution from proteolytic V8 fragments. Eur J Biochem. 1988 Oct 15;177(1):125–133. doi: 10.1111/j.1432-1033.1988.tb14352.x. [DOI] [PubMed] [Google Scholar]
- Sobol A. G., Arseniev A. S., Abdulaeva G. V., Musina LYu, Bystrov V. F. Sequence-specific resonance assignment and secondary structure of (1-71) bacterioopsin. J Biomol NMR. 1992 Mar;2(2):161–171. doi: 10.1007/BF01875527. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K., Mantsch H. H. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim Biophys Acta. 1988 Jan 29;952(2):115–130. doi: 10.1016/0167-4838(88)90107-0. [DOI] [PubMed] [Google Scholar]
- Torres J., Padrós E. The secondary structure of bacteriorhodopsin in organic solution. A Fourier transform infrared study. FEBS Lett. 1993 Feb 22;318(1):77–79. doi: 10.1016/0014-5793(93)81331-s. [DOI] [PubMed] [Google Scholar]
- Tuffery P., Etchebest C., Popot J. L., Lavery R. Prediction of the positioning of the seven transmembrane alpha-helices of bacteriorhodopsin. A molecular simulation study. J Mol Biol. 1994 Mar 4;236(4):1105–1122. doi: 10.1016/0022-2836(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Tuzi S., Naito A., Saitô H. A high-resolution solid-state 13C-NMR study on [1-13C]Ala and [3-13C]Ala and [1-13C]Leu and Val-labelled bacteriorhodopsin. Conformation and dynamics of transmembrane helices, loops and termini, and hydration-induced conformational change. Eur J Biochem. 1993 Dec 15;218(3):837–844. doi: 10.1111/j.1432-1033.1993.tb18439.x. [DOI] [PubMed] [Google Scholar]
- Vogel H., Gärtner W. The secondary structure of bacteriorhodopsin determined by Raman and circular dichroism spectroscopy. J Biol Chem. 1987 Aug 25;262(24):11464–11469. [PubMed] [Google Scholar]


