Abstract
Sequence-dependent bending of the helical axes in 112 oligonucleotide duplex crystal structures resident in the Nucleic Acid Database have been analyzed and compared with the use of bending dials, a computer graphics tool. Our analysis includes structures of both A and B forms of DNA and considers both uncomplexed forms of the double helix as well as those bound to drugs and proteins. The patterns in bending preferences in the crystal structures are analyzed by base pair steps, and emerging trends are noted. Analysis of the 66 B-form structures in the Nucleic Acid Database indicates that uniform trends within all pyrimidine-purine and purine-pyrimidine steps are not necessarily observed but are found particularly at CG and GC steps of dodecamers. The results support the idea that AA steps are relatively straight and that larger roll bends occur at or near the junctions of these A-tracts with their flanking sequences. The data on 16 available crystal structures of protein-DNA complexes indicate that the majority of the DNA bends induced via protein binding are sharp localized kinks. The analysis of the 30 available A-form DNA structures indicates that these structures are also bent and show a definitive preference for bending into the deep major groove over the shallow minor groove.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Aymami J., Coll M., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Molecular structure of nicked DNA: a substrate for DNA repair enzymes. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2526–2530. doi: 10.1073/pnas.87.7.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikalov I., Grzeskowiak K., Yanagi K., Quintana J., Dickerson R. E. The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn. J Mol Biol. 1993 Jun 5;231(3):768–784. doi: 10.1006/jmbi.1993.1325. [DOI] [PubMed] [Google Scholar]
- Barber A. M., Zhurkin V. B. CAP binding sites reveal pyrimidine-purine pattern characteristic of DNA bending. J Biomol Struct Dyn. 1990 Oct;8(2):213–232. doi: 10.1080/07391102.1990.10507803. [DOI] [PubMed] [Google Scholar]
- Beamer L. J., Pabo C. O. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992 Sep 5;227(1):177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D., Bansal M. Local variability and base sequence effects in DNA crystal structures. J Biomol Struct Dyn. 1990 Dec;8(3):539–572. doi: 10.1080/07391102.1990.10507828. [DOI] [PubMed] [Google Scholar]
- Bingman C. A., Zon G., Sundaralingam M. Crystal and molecular structure of the A-DNA dodecamer d(CCGTACGTACGG). Choice of fragment helical axis. J Mol Biol. 1992 Oct 5;227(3):738–756. doi: 10.1016/0022-2836(92)90221-5. [DOI] [PubMed] [Google Scholar]
- Bingman C., Jain S., Zon G., Sundaralingam M. Crystal and molecular structure of the alternating dodecamer d(GCGTACGTACGC) in the A-DNA form: comparison with the isomorphous non-alternating dodecamer d(CCGTACGTACGG). Nucleic Acids Res. 1992 Dec 25;20(24):6637–6647. doi: 10.1093/nar/20.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. G., Sanderson M. R., Garman E., Neidle S. Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features. J Mol Biol. 1992 Jul 20;226(2):481–490. doi: 10.1016/0022-2836(92)90962-j. [DOI] [PubMed] [Google Scholar]
- Brown D. G., Sanderson M. R., Skelly J. V., Jenkins T. C., Brown T., Garman E., Stuart D. I., Neidle S. Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. EMBO J. 1990 Apr;9(4):1329–1334. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner B. N., Yoon C., Dickerson J. L., Dickerson R. E. Helix geometry and hydration in an A-DNA tetramer: IC-C-G-G. J Mol Biol. 1984 Apr 25;174(4):663–695. doi: 10.1016/0022-2836(84)90089-5. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Cruse W. B., Aymani J., Kennard O., Brown T., Jack A. G., Leonard G. A. Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs. Nucleic Acids Res. 1989 Jan 11;17(1):55–72. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Salisbury S. A., Brown T., Cosstick R., Eckstein F., Kennard O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J Mol Biol. 1986 Dec 20;192(4):891–905. doi: 10.1016/0022-2836(86)90035-5. [DOI] [PubMed] [Google Scholar]
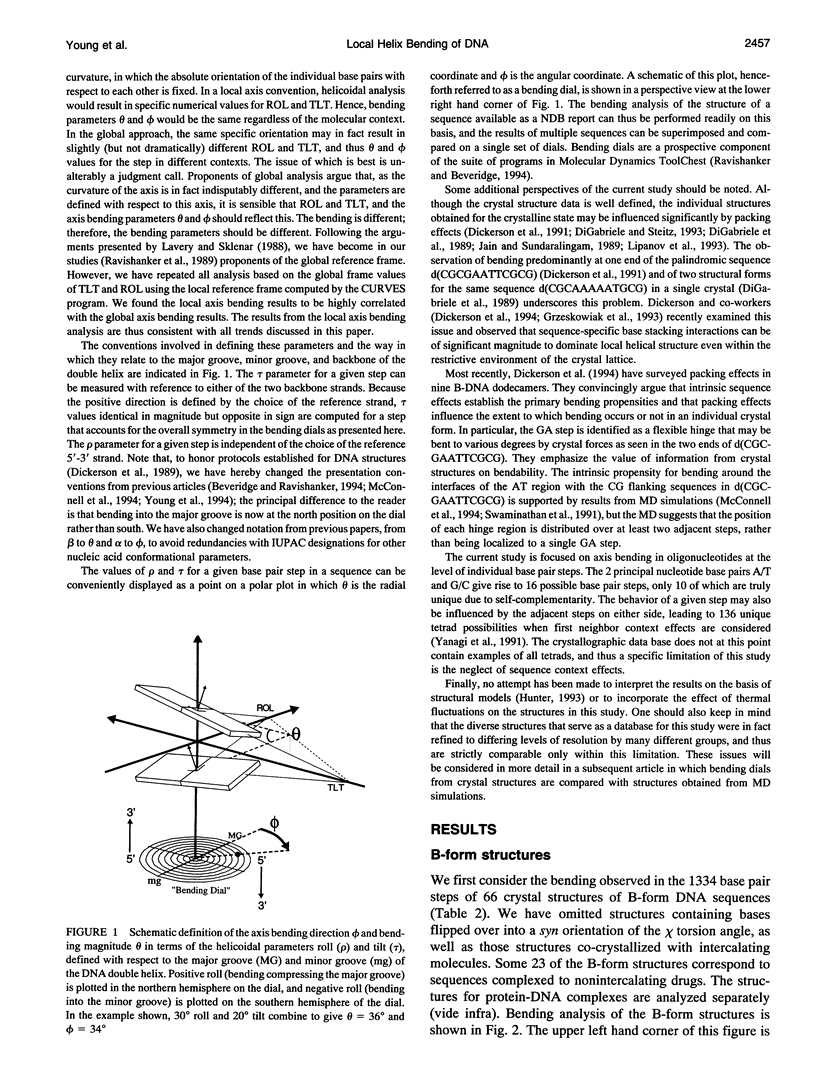
- Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989 Jan;8(1):1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Steitz T. A. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J Mol Biol. 1993 Jun 20;231(4):1024–1039. doi: 10.1006/jmbi.1993.1349. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. DNA structure from A to Z. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Kopka M. L., Pjura P. E. The effect of crystal packing on oligonucleotide double helix structure. J Biomol Struct Dyn. 1987 Dec;5(3):557–579. doi: 10.1080/07391102.1987.10506413. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Neidle S. "...the tyranny of the lattice...". Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Samson S., Dickerson R. E. Structure of a B-DNA dodecamer at 16 K. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4040–4044. doi: 10.1073/pnas.79.13.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Brown D. G., Spink N., Skelly J. V., Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992 Aug 20;226(4):1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Jenkins T. C., Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992 Aug 11;31(31):7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
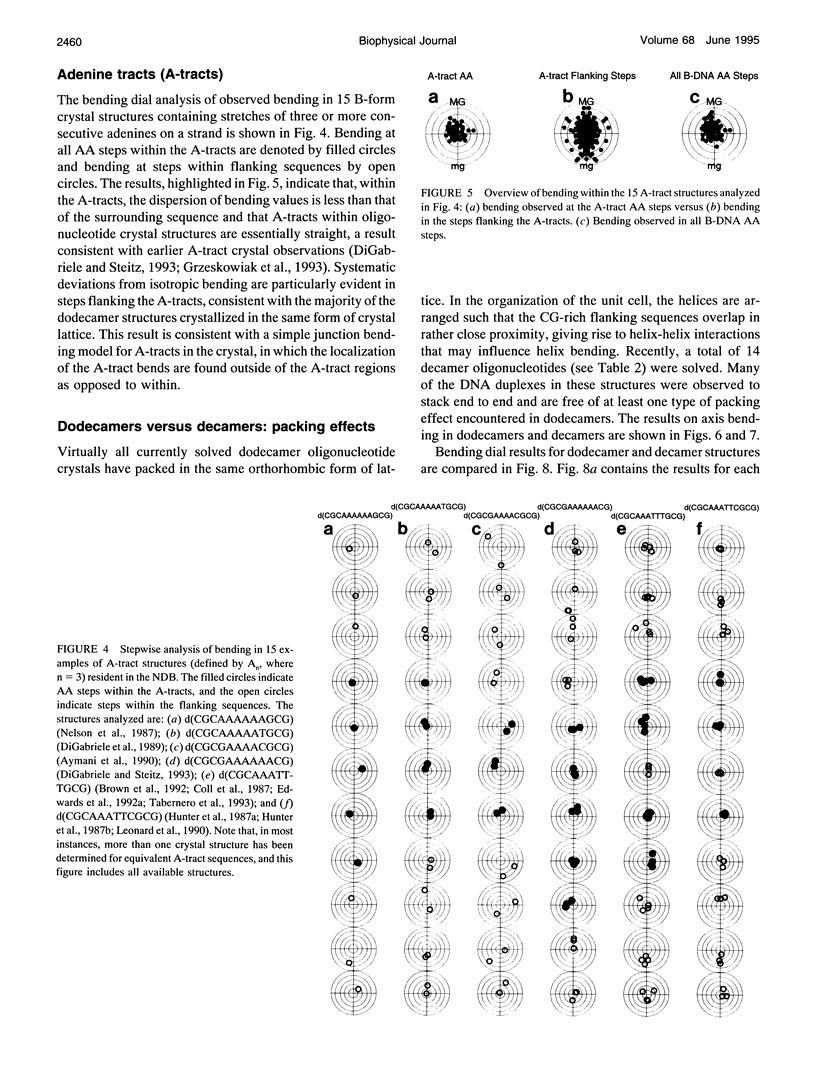
- Egli M., Usman N., Rich A. Conformational influence of the ribose 2'-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993 Apr 6;32(13):3221–3237. [PubMed] [Google Scholar]
- Egli M., Usman N., Zhang S. G., Rich A. Crystal structure of an Okazaki fragment at 2-A resolution. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):534–538. doi: 10.1073/pnas.89.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M., Frolow F., Shakked Z., Rabinovich D. The structure and hydration of the A-DNA fragment d(GGGTACCC) at room temperature and low temperature. Nucleic Acids Res. 1990 Jun 11;18(11):3185–3194. doi: 10.1093/nar/18.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Frederick C. A., Quigley G. J., Teng M. K., Coll M., Van der Marel G. A., Van Boom J. H., Rich A., Wang A. H. Molecular structure of an A-DNA decamer d(ACCGGCCGGT). Eur J Biochem. 1989 May 1;181(2):295–307. doi: 10.1111/j.1432-1033.1989.tb14724.x. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution. J Biol Chem. 1988 Nov 25;263(33):17872–17879. doi: 10.2210/pdb4dnb/pdb. [DOI] [PubMed] [Google Scholar]
- Goodsell D. S., Dickerson R. E. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994 Dec 11;22(24):5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Kopka M. L., Cascio D., Dickerson R. E. Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeskowiak K., Goodsell D. S., Kaczor-Grzeskowiak M., Cascio D., Dickerson R. E. Crystallographic analysis of C-C-A-A-G-C-T-T-G-G and its implications for bending in B-DNA. Biochemistry. 1993 Aug 31;32(34):8923–8931. doi: 10.1021/bi00085a025. [DOI] [PubMed] [Google Scholar]
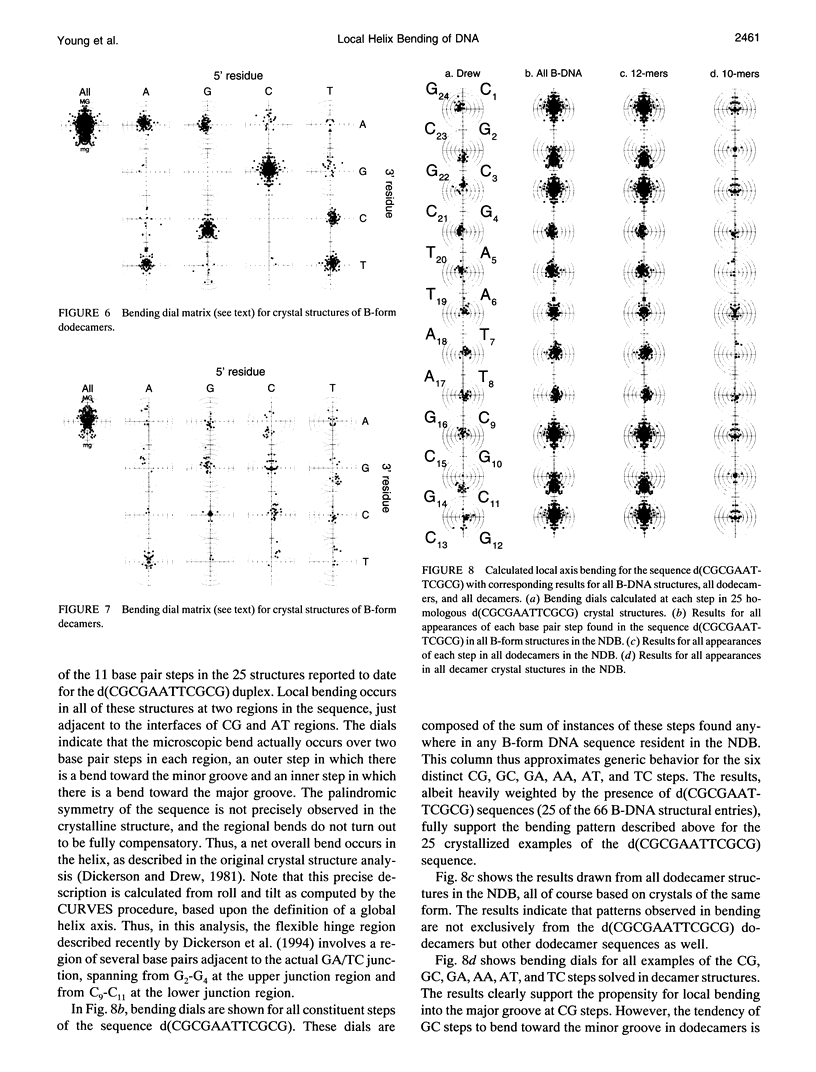
- Grzeskowiak K., Yanagi K., Privé G. G., Dickerson R. E. The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. The effect of base pair reversals. J Biol Chem. 1991 May 15;266(14):8861–8883. doi: 10.2210/pdb1d23/pdb. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Straightening out the bends in curved DNA. Biochim Biophys Acta. 1992 Jun 15;1131(2):125–132. doi: 10.1016/0167-4781(92)90066-9. [DOI] [PubMed] [Google Scholar]
- Hahn M., Heinemann U. DNA helix structure and refinement algorithm: comparison of models for d(CCAGGCm5CTGG) derived from NUCLSQ, TNT and X-PLOR. Acta Crystallogr D Biol Crystallogr. 1993 Sep 1;49(Pt 5):468–477. doi: 10.1107/S0907444993004858. [DOI] [PubMed] [Google Scholar]
- Han F., Watt W., Duchamp D. J., Callahan L., Kézdy F. J., Agarwal K. Molecular structure of deoxycytidyl-3'-methylphosphonate (RP) 5'-deoxyguanidine, d[Cp(CH3)G]. A neutral dinucleotide with Watson-Crick base pairing and a right handed helical twist. Nucleic Acids Res. 1990 May 11;18(9):2759–2767. doi: 10.1093/nar/18.9.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran T. E., Crothers D. M. Cooperativity in A-tract structure and bending properties of composite TnAn blocks. Biochemistry. 1989 Apr 4;28(7):2763–2767. doi: 10.1021/bi00433a003. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Grossman S. R., Laimins L. A., Sigler P. B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992 Oct 8;359(6395):505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C., Bansal M. Double helix conformation, groove dimensions and ligand binding potential of a G/C stretch in B-DNA. EMBO J. 1992 May;11(5):1931–1939. doi: 10.1002/j.1460-2075.1992.tb05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Hahn M. C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J Biol Chem. 1992 Apr 15;267(11):7332–7341. [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Rudolph L. N., Alings C., Morr M., Heikens W., Frank R., Blöcker H. Effect of a single 3'-methylene phosphonate linkage on the conformation of an A-DNA octamer double helix. Nucleic Acids Res. 1991 Feb 11;19(3):427–433. doi: 10.1093/nar/19.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C. A. Sequence-dependent DNA structure. The role of base stacking interactions. J Mol Biol. 1993 Apr 5;230(3):1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kennard O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987 Aug 25;15(16):6589–6606. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kneale G., Anand N. N., Rabinovich D., Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J Biol Chem. 1987 Jul 25;262(21):9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., D'Estaintot B. L., Kennard O. Structural variation in d(CTCTAGAG). Implications for protein-DNA interactions. Biochemistry. 1989 Mar 21;28(6):2444–2451. doi: 10.1021/bi00432a015. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Kneale G., Brown T., Rabinovich D., Kennard O. Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs. J Mol Biol. 1986 Aug 20;190(4):605–618. doi: 10.1016/0022-2836(86)90246-9. [DOI] [PubMed] [Google Scholar]
- Jain S., Sundaralingam M. Effect of crystal packing environment on conformation of the DNA duplex. Molecular structure of the A-DNA octamer d(G-T-G-T-A-C-A-C) in two crystal forms. J Biol Chem. 1989 Aug 5;264(22):12780–12784. [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry. 1989 Mar 21;28(6):2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kennard O., Cruse W. B., Nachman J., Prange T., Shakked Z., Rabinovich D. Ordered water structure in an A-DNA octamer at 1.7 A resolution. J Biomol Struct Dyn. 1986 Feb;3(4):623–647. doi: 10.1080/07391102.1986.10508452. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Goodsell D. S., Cascio D., Grzeskowiak K., Dickerson R. E. The structure of DAPI bound to DNA. J Biomol Struct Dyn. 1989 Dec;7(3):477–491. doi: 10.1080/07391102.1989.10508505. [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Kopka M. L., Dickerson R. E. Crystal structure analysis of the B-DNA dodecamer CGTGAATTCACG. Biochemistry. 1991 May 7;30(18):4443–4449. doi: 10.1021/bi00232a010. [DOI] [PubMed] [Google Scholar]
- Lauble H., Frank R., Blöcker H., Heinemann U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988 Aug 25;16(16):7799–7816. doi: 10.1093/nar/16.16.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., Hunter W. N. Crystal and molecular structure of d(CGTAGATCTACG) at 2.25 A resolution. J Mol Biol. 1993 Nov 5;234(1):198–208. doi: 10.1006/jmbi.1993.1574. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., Thomson J., Watson W. P., Brown T. High-resolution structure of a mutagenic lesion in DNA. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9573–9576. doi: 10.1073/pnas.87.24.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Wu H. M., Crothers D. M. Bending and flexibility of kinetoplast DNA. Biochemistry. 1986 Jul 15;25(14):3988–3995. doi: 10.1021/bi00362a003. [DOI] [PubMed] [Google Scholar]
- Lipanov A., Kopka M. L., Kaczor-Grzeskowiak M., Quintana J., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA. Biochemistry. 1993 Feb 9;32(5):1373–1389. doi: 10.1021/bi00056a024. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroun R. C., Olson W. K. Base sequence effects in double-helical DNA. II. Configurational statistics of rodlike chains. Biopolymers. 1988 Apr;27(4):561–584. doi: 10.1002/bip.360270403. [DOI] [PubMed] [Google Scholar]
- Maroun R. C., Olson W. K. Base sequence effects in double-helical DNA. III. Average properties of curved DNA. Biopolymers. 1988 Apr;27(4):585–603. doi: 10.1002/bip.360270404. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Kennard O. The crystal structure of d(G-G-G-G-C-C-C-C). A model for poly(dG).poly(dC). J Mol Biol. 1985 Jun 5;183(3):385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich A. K., Bhattacharyya D., Brahmachari S. K., Bansal M. CA/TG sequence at the 5' end of oligo(A)-tracts strongly modulates DNA curvature. J Biol Chem. 1994 Mar 11;269(10):7824–7833. [PubMed] [Google Scholar]
- Narayana N., Ginell S. L., Russu I. M., Berman H. M. Crystal and molecular structure of a DNA fragment: d(CGTGAATTCACG). Biochemistry. 1991 May 7;30(18):4449–4455. doi: 10.1021/bi00232a011. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nunn C. M., Jenkins T. C., Neidle S. Crystal structure of d(CGCGAATTCGCG) complexed with propamidine, a short-chain homologue of the drug pentamidine. Biochemistry. 1993 Dec 21;32(50):13838–13843. doi: 10.1021/bi00213a012. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Marky N. L., Jernigan R. L., Zhurkin V. B. Influence of fluctuations on DNA curvature. A comparison of flexible and static wedge models of intrinsically bent DNA. J Mol Biol. 1993 Jul 20;232(2):530–554. doi: 10.1006/jmbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Price M. A., Tullius T. D. Using hydroxyl radical to probe DNA structure. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Prévost C., Louise-May S., Ravishanker G., Lavery R., Beveridge D. L. Persistence analysis of the static and dynamical helix deformations of DNA oligonucleotides: application to the crystal structure and molecular dynamics simulation of d(CGCGAATTCGCG)2. Biopolymers. 1993 Mar;33(3):335–350. doi: 10.1002/bip.360330303. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Grzeskowiak K., Yanagi K., Dickerson R. E. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. J Mol Biol. 1992 May 20;225(2):379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Lipanov A. A., Dickerson R. E. Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry. 1991 Oct 22;30(42):10294–10306. doi: 10.1021/bi00106a030. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Sundaralingam M. Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures. Biochemistry. 1993 Oct 26;32(42):11458–11468. doi: 10.1021/bi00093a025. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Sundaralingam M. High resolution crystal structure of the A-DNA decamer d(CCCGGCCGGG). Novel intermolecular base-paired G*(G.C) triplets. J Mol Biol. 1993 May 20;231(2):431–444. doi: 10.1006/jmbi.1993.1292. [DOI] [PubMed] [Google Scholar]
- Ravishanker G., Swaminathan S., Beveridge D. L., Lavery R., Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: "curves", dials and windows. J Biomol Struct Dyn. 1989 Feb;6(4):669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R. L. Origin of DNA helical structure and its sequence dependence. Biochemistry. 1988 Nov 1;27(22):8498–8502. doi: 10.1021/bi00422a030. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R. L. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989 Sep 19;28(19):7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Shimon L. J., Harrison S. C. The phage 434 OR2/R1-69 complex at 2.5 A resolution. J Mol Biol. 1993 Aug 5;232(3):826–838. doi: 10.1006/jmbi.1993.1434. [DOI] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Sriram M., van der Marel G. A., Roelen H. L., van Boom J. H., Wang A. H. Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342. EMBO J. 1992 Jan;11(1):225–232. doi: 10.1002/j.1460-2075.1992.tb05045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram M., van der Marel G. A., Roelen H. L., van Boom J. H., Wang A. H. Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex. Biochemistry. 1992 Dec 1;31(47):11823–11834. doi: 10.1021/bi00162a022. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Subramanian P. S., Beveridge D. L. A theoretical study of the aqueous hydration of canonical B d(CGCGAATTCGCG): Monte Carlo simulation and comparison with crystallographic ordered water sites. J Biomol Struct Dyn. 1989 Jun;6(6):1093–1122. doi: 10.1080/07391102.1989.10506539. [DOI] [PubMed] [Google Scholar]
- Tabernero L., Verdaguer N., Coll M., Fita I., van der Marel G. A., van Boom J. H., Rich A., Aymamí J. Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin. Biochemistry. 1993 Aug 24;32(33):8403–8410. doi: 10.1021/bi00084a004. [DOI] [PubMed] [Google Scholar]
- Takusagawa F. The crystal structure of d(GTACGTAC) at 2.25 A resolution: are the A-DNA's always unwound approximately 10 degrees at the C-G steps? J Biomol Struct Dyn. 1990 Feb;7(4):795–809. doi: 10.1080/07391102.1990.10508524. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota N., Li X. H., Bingman C., Sundaralingam M. High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A. Acta Crystallogr D Biol Crystallogr. 1993 Mar 1;49(Pt 2):282–291. doi: 10.1107/S0907444992007522. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Base sequence, local helix structure, and macroscopic curvature of A-DNA and B-DNA. J Biol Chem. 1986 Mar 15;261(8):3700–3709. [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulyanov N. B., Zhurkin V. B. Sequence-dependent anisotropic flexibility of B-DNA. A conformational study. J Biomol Struct Dyn. 1984 Oct;2(2):361–385. doi: 10.1080/07391102.1984.10507573. [DOI] [PubMed] [Google Scholar]
- Verdaguer N., Aymami J., Fernández-Forner D., Fita I., Coll M., Huynh-Dinh T., Igolen J., Subirana J. A. Molecular structure of a complete turn of A-DNA. J Mol Biol. 1991 Sep 20;221(2):623–635. doi: 10.1016/0022-2836(91)80077-8. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Westhof E. Re-refinement of the B-dodecamer d(CGCGAATTCGCG) with a comparative analysis of the solvent in it and in the Z-hexamer d(5BrCG5BrCG5BrCG). J Biomol Struct Dyn. 1987 Dec;5(3):581–600. doi: 10.1080/07391102.1987.10506414. [DOI] [PubMed] [Google Scholar]
- Weston S. A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol. 1992 Aug 20;226(4):1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- Wing R. M., Pjura P., Drew H. R., Dickerson R. E. The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G. EMBO J. 1984 May;3(5):1201–1206. doi: 10.1002/j.1460-2075.1984.tb01951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Xuan J. C., Weber I. T. Crystal structure of a B-DNA dodecamer containing inosine, d(CGCIAATTCGCG), at 2.4 A resolution and its comparison with other B-DNA dodecamers. Nucleic Acids Res. 1992 Oct 25;20(20):5457–5464. doi: 10.1093/nar/20.20.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi K., Privé G. G., Dickerson R. E. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J Mol Biol. 1991 Jan 5;217(1):201–214. doi: 10.1016/0022-2836(91)90620-l. [DOI] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Quintana J., Dickerson R. E. Alternative structures for alternating poly(dA-dT) tracts: the structure of the B-DNA decamer C-G-A-T-A-T-A-T-C-G. Biochemistry. 1992 Sep 1;31(34):8009–8021. [PubMed] [Google Scholar]
- Zhurkin V. B., Lysov Y. P., Florentiev V. L., Ivanov V. I. Torsional flexibility of B-DNA as revealed by conformational analysis. Nucleic Acids Res. 1982 Mar 11;10(5):1811–1830. doi: 10.1093/nar/10.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurkin V. B., Lysov Y. P., Ivanov V. I. Anisotropic flexibility of DNA and the nucleosomal structure. Nucleic Acids Res. 1979 Mar;6(3):1081–1096. doi: 10.1093/nar/6.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurkin V. B. Sequence-dependent bending of DNA and phasing of nucleosomes. J Biomol Struct Dyn. 1985 Feb;2(4):785–804. doi: 10.1080/07391102.1985.10506324. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B. Specific alignment of nucleosomes on DNA correlates with periodic distribution of purine-pyrimidine and pyrimidine-purine dimers. FEBS Lett. 1983 Jul 25;158(2):293–297. doi: 10.1016/0014-5793(83)80598-5. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B., Ulyanov N. B., Gorin A. A., Jernigan R. L. Static and statistical bending of DNA evaluated by Monte Carlo simulations. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7046–7050. doi: 10.1073/pnas.88.16.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Comparative gel electrophoresis measurement of the DNA bend angle induced by the catabolite activator protein. Biopolymers. 1990 Jan;29(1):29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]


