Abstract
Caenorhabditis elegans is currently introduced as a new, facile, and cheap model organism to study the pathogenesis of gram-negative bacteria such as Pseudomonas aeruginosa and Salmonella enterica serovar Typhimurium. The mechanisms of killing involve either diffusible exotoxins or infection-like processes. Recently, it was shown that also some gram-positive bacteria kill C. elegans, although the precise mechanisms of killing remained open. We examined C. elegans as a pathogenesis model for the gram-positive bacterium Streptococcus pyogenes, a major human pathogen capable of causing a wide spectrum of diseases. We demonstrate that S. pyogenes kills C. elegans, both on solid and in liquid medium. Unlike P. aeruginosa and S. enterica serovar Typhimurium, the killing by S. pyogenes is solely mediated by hydrogen peroxide. Killing required live streptococci; the killing capacity depends on the amount of hydrogen peroxide produced, and killing can be inhibited by catalase. Major exotoxins of S. pyogenes are not involved in the killing process as confirmed by using specific toxin inhibitors and knockout mutants. Moreover, no accumulation of S. pyogenes in C. elegans is observed, which excludes the involvement of infection-like processes. Preliminary results show that S. pneumoniae can also kill C. elegans by hydrogen peroxide production. Hydrogen peroxide-mediated killing might represent a common mechanism by which gram-positive, catalase-negative pathogens kill C. elegans.
Streptococcus pyogenes is a major human pathogen, causing a wide spectrum of pyogenic infections such as tonsillitis, pharyngitis, scarlet fever, and skin inflammation. The organism can also invade tissues and cells and cause life-threatening diseases such as necrotizing fasciitis and toxic shock syndrome. Untreated infections can lead to serious postinfection complications in the form of rheumatic heart disease and glomerulonephritis in predisposed individuals (22). The pathogenesis of streptococci is so complex that the infections caused by these organisms and their sequelae have not been completely understood. Bacterial factors, host factors, and an abnormal immune response determine the outcome of the infection. One of the difficulties in understanding the streptococcus-host interaction is the lack of a suitable animal model. Recently a mouse model of invasive streptococcal infections has been developed (16), but there remains a great need for new animal models to understand streptococcal pathogenesis.
Ausubel and coworkers have introduced Caenorhabditis elegans as a new, facile, and cheap model organism to study the pathogenesis of the gram-negative bacteria Pseudomonas aeruginosa (5, 15, 23-25, 27) and Salmonella enterica serovar Typhimurium (1, 2, 13), and Garsin et al. showed that the gram-positive bacteria Enterococcus faecalis and Streptococcus pneumoniae kill C. elegans (8). Hodgkin et al. demonstrated that the genetically amenable nematode C. elegans is ideally suited to identify host factors (12).
We examined C. elegans as a pathogenesis model for S. pyogenes. We show that S. pyogenes kills C. elegans, both on plates and in culture medium. Killing is exclusively mediated by hydrogen peroxide. Preliminary results indicate that the same mechanism of killing is used by S. pneumoniae. These data imply that besides infection-like processes and diffusible proteins, hydrogen peroxide release might be a third common mechanism by which bacteria can kill C. elegans.
MATERIALS AND METHODS
The general methods for handling C. elegans are described in reference 26, and those for S. pyogenes are described in reference 4.
Strains.
Killing assays were performed with C. elegans N2 (Bristol) or glp-1 (e2144), a temperature-sensitive mutant strain that grows permissively at 15°C but produces no progeny at 25°C (18). S. pyogenes strains used in this study are depicted in Table 1. A streptococcal pyrogenic exotoxin B (SPE-B)-negative S. pyogenes mutant 1579 was derived from strain 634 (M49 CS101) (14, 21), and a streptolysin O (SLO)-negative mutant A468 (T17) was derived from strain 3413 (M1 38541) (20). As control strains E. faecalis (H1A, Minneapolis), Staphylococcus aureus (SA1, H Mi 1, Giessen), and Escherichia coli OP50 were used.
TABLE 1.
S. pyogenes strain source and killing capacity in the solid assay
| Strain | Code | Source | Killing in solid assaya |
|---|---|---|---|
| A20 | DSM 2071 | Giessen, Germany | + |
| A21 | DSM 2072 | Giessen, Germany | ± |
| A23 | DSM 2074 | Giessen, Germany | ± |
| A29 | BS 06 | Braunschweig, Germany | ± |
| A40 | BS 20 | Braunschweig, Germany | ± |
| A115 | PR-791 | Minneapolis, Minn. | + |
| A153 | BSB 19 | Australia | + |
| A154 | DRY 6 | Australia | ± |
| A158 | NS 342 | Australia | ± |
| A162 | NS 345 | Australia | ± |
| A171 | NS 297 | Australia | + |
| A194 | NS 300 | Australia | + |
| A195 | NS 343 | Australia | + |
| A198 | NS 727 | Australia | ± |
| A216 | CK 470 | Australia | + |
| A241 | #273 | Australia | + |
| A248 | NS 226 | Australia | ± |
| A249 | NS 210 | Australia | + |
| A294 | Xy 283 | Hannover, Germany | ± |
| A296 | Xy 395 | Hannover, Germany | + |
| A302, M1 | SF 370 | Reference strain | ± |
| A306 | NS 244 | Australia | ± |
| A467 | M1-38541 | Jena, Germany | ± |
| A468 | SLO mutant T17 | Jena, Germany | ± |
Since killing efficiency of intermediate killing strains varied between experiments, strains are subdivided into good versus intermediate killers, rather than presenting actual percentages of killing. Symbols: +, strain killed 100% of worms within 24 h; ±, strain caused delayed and/or incomplete killing.
C. elegans killing assays. (i) Solid assay.
The solid assay was adapted from the plate assay described by Tan et al. (24). An overnight culture of bacteria in Todd-Hewitt Broth supplemented with 0.5% yeast extract (THY) was placed (100 μl) on 35-mm-diameter nematode growth medium agar plates. The plates were incubated aerobically at room temperature (on the work bench) for 24 h, to enable bacterial toxins to diffuse through the plate. C. elegans nematodes were bleached to obtain sterile eggs by standard methods (26), and approximately 200 eggs were added to each plate. Plates were incubated at 25°C, and the viability of the nematodes was scored every 24 h. Worms were considered dead when they showed no detectable response upon firmly tapping the plate on the microscope platform.
(ii) Liquid assay.
From a bacterial overnight culture in THY a 1:25-diluted culture was started and grown to early log phase (optical density at 660 nm between 0.2 and 0.3). Bacteria were washed and taken up in THY, diluted 1:4 in M9. Since OP50 grown in rich medium can be lethal to worms (8), it is necessary to dilute the THY medium. Cultures were transferred to 24-well plates, and 200 sterile L1 larvae, obtained from bleached eggs, were added. Plates were incubated at 25°C under shaking (150 rpm [orbital shaker]), and the viability of worms was scored at different time points.
(iii) Hydrogen peroxide production assay.
Hydrogen peroxide production was measured as described in reference 17. Bacterial cultures taken from wells with and without worms were diluted 2:5 in M9 and filtered through 0.2-μm-pore-size membranes. Immediately prior to the assay, phenol red and horseradish peroxidase were added to peroxidase buffer (5.0 mM K2HPO4, 1.0 mM KH2PO4, 140 mM NaCl, 0.5 mM glucose; pH 7,4) at final concentrations of 0.46 mM and 0.046 U/ml, respectively. Aliquots of filtered supernatant were added to the assay mixture at a ratio of 1 to 4 and incubated for 30 min at 37°C in duplicate. After the reactions were stopped by the addition of NaOH (4 mM final concentration), the absorbance was recorded at a wavelength of 610 nm. Concentrations were calculated in comparison to a standard curve with known amounts of hydrogen peroxide added to control supernatant from wells containing catalase, which had been heated to 100°C for 20 min to eliminate catalase activity.
Statistical analysis.
Killing of nematodes with and without addition of catalase was analyzed by the Mann-Whitney test (Prism software). Correlation between hydrogen peroxide production and the killing capacity in different bacterial strains was assessed by Pearson linear regression analysis (Prism software).
RESULTS
S. pyogenes kills C. elegans nematodes on plates and in culture medium.
Since our killing assays extended over the generation time of C. elegans (2.7 days at 25°C) and new progeny would interfere with a quantitative evaluation of the experiments, we prevented the reproduction of the worms in our experiments using the conditionally sterile mutant (e2144) with a mutation in the Notch receptor glp-1 of the worm. This mutation specifically prevents the propagation of the germ cells (18). Initially, wild-type and mutant nematodes were evaluated in parallel in the solid and liquid killing assays. Since both worm strains gave similar killing patterns (data not shown), all subsequent experiments were performed with the e2144 mutant strain.
S. pyogenes strains (n = 24) were tested for their capability to kill C. elegans larvae in the solid assay. Thirty-eight percent of S. pyogenes strains tested (n = 9) killed C. elegans larvae within 24 h, whereas the other 62% (n = 15) showed delayed and/or incomplete killing (Table 1). E. faecalis, S. aureus, and E. coli OP50 did not kill (Fig. 1) within the time interval measured (5 days). However, some variation in killing occurred depending on the uniformity of the bacterial lawn and mobility of the worms. Therefore, a second setup was developed (liquid assay) in which worms were added to bacterial cultures. Killing was more pronounced in the liquid assay. All S. pyogenes strains killed C. elegans within 24 h in the liquid set up (Fig. 2). L1 larvae and adult nematodes gave similar killing patterns (e.g., strain A296 killed 100% ± 0% [mean ± standard deviation {SD}; n = 2] of C. elegans adults and 100% ± 0% larvae in the liquid assay at 24 h). In all further experiments, killing results from the liquid assay were confirmed by using the solid assay.
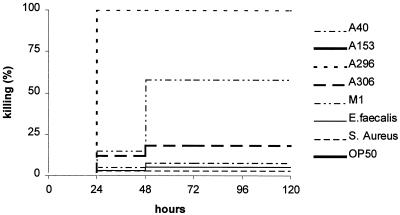
FIG. 1.
S. pyogenes kills C. elegans in the solid assay. Nematodes were fed on a representative set of S. pyogenes strains from Table 1: A40, A153, A296, A306, or M1. E. faecalis, S. aureus, and E. coli OP50 were used as control strains. S. pyogenes strains A153 and A296 killed all nematodes within 24 h, whereas A40, M1, and A306 showed intermediate or poor killing. Similar results were obtained in 10 independent experiments for a total of 24 different S. pyogenes strains, of which 9 strains killed within 24 h and 15 strains induced various degrees of killing in between 1 and 5 days (Table 1).
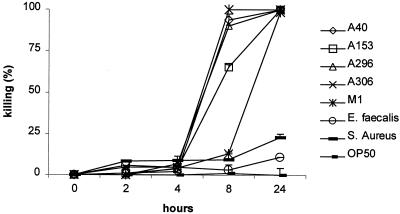
FIG. 2.
S. pyogenes kills C. elegans in the liquid assay. C. elegans nematodes were incubated in cultures of S. pyogenes strains A40, A153, A296, A306, or M1 or E. faecalis, S. aureus, or E. coli OP50. The experiment was performed in duplicate (results are presented as means ± SD [error bars]) and is representative of three independent experiments. All S. pyogenes strains tested in the liquid assay killed C. elegans within 24 h.
Killing of C. elegans by S. pyogenes is caused by neither infection-like processes nor streptococcal exotoxins SPE-B and SLO.
The killing of C. elegans required live bacteria, since heat-inactivated strains and antibiotic-treated strains did not kill in the liquid assay (Fig. 3A). To test whether killing is caused by infection-like processes or diffusible toxins, the worms were separated from streptococci by 0.4-μm-pore-size membranes, which did not interfere with the killing (Fig. 3A). Absence of bacteria in the worm compartment was confirmed by plating. Confocal microscopy confirmed the noninfectious killing of C. elegans by S. pyogenes. In contrast to E. faecalis, fluorescence-labeled S. pyogenes strains (Alexa; Molecular Probes) did not accumulate in the nematode intestines (Fig. 3B). Labeling did not interfere with the killing capacities of the strains (unlabeled and labeled S. pyogenes [strain A306] both killed 100% ± 0% [mean ± SD; n = 2] of the worms after, respectively, 6 and 9 h in the liquid assay).
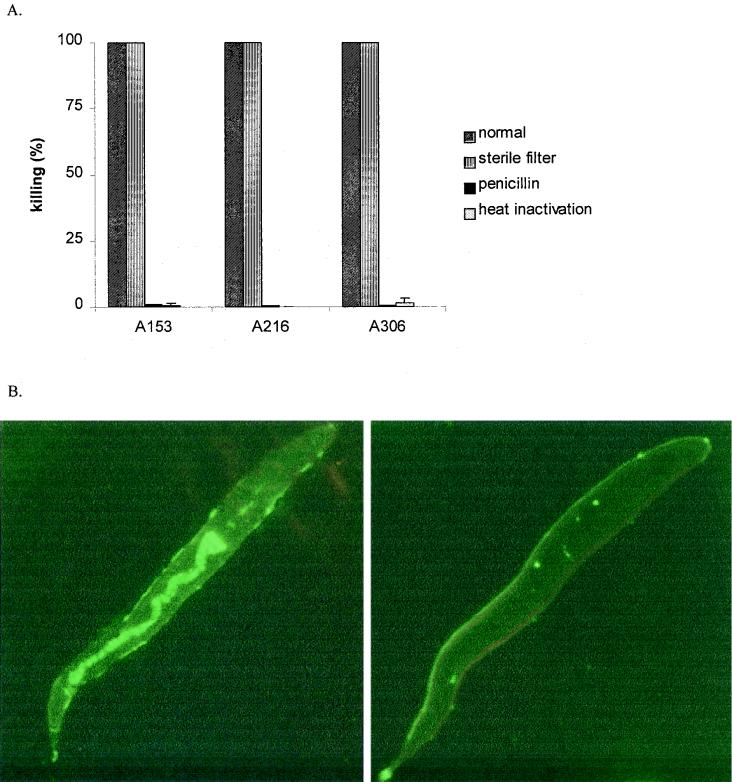
FIG. 3.
Killing of C. elegans by S. pyogenes requires live bacteria and is a noninfectious process. (A) A153, A216, and A306 killed in the liquid assay after 24 h under normal conditions (shaded bars), and killing was retained when nematodes were separated from bacteria by 0.4-μm-pore-size membranes (striped bars). In contrast, killing was abolished when bacteria were killed by heat (1 h, 90°C) (open bars) or penicillin (50 IU/ml; Gibco BRL) (solid bars). Killing was confirmed by plating (data not shown). Values represent the average of two independent experiments in duplicate. (B) C. elegans intestines were colonized by fluorescence-labeled (15 min at 4°C) (0.1 mg/ml; Alexa; Molecular Probes) E. faecalis (left panel) but not S. pyogenes (strain A306) (right panel) after 0, 2, 4, 8, or 24 h of incubation in the liquid assay. Results are shown for 4 h.
SPE-B and SLO are the major exotoxins of S. pyogenes. SPE-B is a cysteine proteinase, and SLO is a pore-forming agent binding to immunoglobulin G (IgG) and cholesterol (9). Their contribution to the killing of C. elegans was tested with S. pyogenes exotoxin inhibitors immunoglobulin G (100 μM), Evan's blue (50 μM), and cholesterol (200 μg/ml) and the cysteine proteinase inhibitor cystatin (200 μg/ml) (all inhibitors were from Sigma-Aldrich) (9) These inhibitors did not affect killing of C. elegans, which excludes the involvement of relevant exotoxins. (e.g., strain M1 killed 100% ± 0% [mean ± SD; n = 2] of the worms with and without inhibitors at 24 h in the liquid assay). These results were confirmed by unaltered killing abilities of an SLO-negative mutant and a SPE-B negative mutant compared to the wild-type strains (Fig. 4).
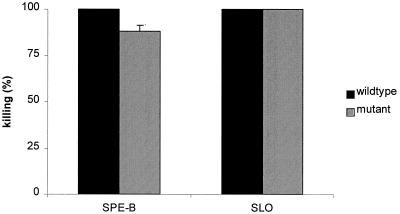
FIG. 4.
S. pyogenes exotoxins SPE-B and SLO are not involved in the killing of C. elegans. Knockout mutants for SPE-B or SLO killed C. elegans like the wild types in the liquid assay at 24 h. Results are presented as means + SDs (error bars) (n = 2).
Killing of C. elegans by S. pyogenes is caused by hydrogen peroxide production.
The assays (liquid and solid) were run under aerobic conditions required for the growth of the nematode. Under these conditions streptococci can produce reactive oxygen species such as hydrogen peroxide. Therefore, we subsequently examined the role of reactive oxygen species in the killing process. Rescue of worms was analyzed with superoxide dismutase and catalase, two enzymes that detoxify the reactive compounds superoxide and hydrogen peroxide, respectively. Catalase, but not superoxide dismutase, fully prevented killing of worms (P = 0.0079; Table 2). C. elegans was found to be very sensitive to hydrogen peroxide exposure: 50% of the nematodes were killed after 4 h of incubation in 1 mM hydrogen peroxide (Fig. 5). Killing of C. elegans by S. pyogenes was paralleled by the accumulation of hydrogen peroxide, measured in control wells without worms (Table 2). There was a highly significant correlation between hydrogen peroxide production and the killing capacity in different bacterial strains (r2=0.77; n = 30; P < 0.0001; Pearson linear regression analysis [Prism]). In addition we compared bacterial hydrogen peroxide production in wells with and without worms, to exclude the worms as an additional source of hydrogen peroxide (e.g., A306 produced 0.0 ± 0.0 mM and 0.0 ± 0.0 mM H2O2 at 0 h, 0.32 ± 0.02 mM and 0.25 ± 0.02 at 4 h, 0.74 ± 0.01 mM and 0.59 ± 0.02 at 6 h, and 0.94 ± 0.01 mM and 1.06 ± 0.37 at 8 h without and with worms, respectively [means ± SD; n = 2).
TABLE 2.
Killing depends on bacterial hydrogen peroxide production and can be prevented with catalasea
| Strain | H2O2 production− (mM) | Mean killing (%) ± SD with:
|
|
|---|---|---|---|
| No catalase | Catalase | ||
| A40 | 0.28 ± 0.03 | 90.8 ± 4.9 | 4.0 ± 1.4 |
| A153 | ND | 90.4 ± 7.1 | 1.5 ± 1.4 |
| A296 | 0.28 ± 0.03 | 86.3 ± 8.5 | 7.0 ± 1.4 |
| A306 | 1.7 ± 0.02 | 86.4 ± 3.0 | 2.0 ± 1.4 |
| M1 | BDL | 12.8 ± 2.0 | 2.0 ± 1.4 |
| E. faecalis | BDL | 3.3 ± 0.9 | 2.5 ± 0 |
| OP50 | BDL | 6.5 ± 0.7 | 6.5 ± 2.1 |
Killing and hydrogen peroxide production at 8 h in the liquid assay (means ± standard deviations [n = 2]). Catalase (150 U per well; Sigma), but not SOD (e.g., killing of 100% ± 0% with and without SOD at 24 h in liquid assay for strain A296), completely prevented killing in the liquid assay (P = 0.0079). Catalase and SOD were nontoxic for streptococci as measured by OD660 and plating (e.g., OD660 at 16 h of 0.82 ± 0.01 with and 0.79 ± 0.001 without catalase for strain M1). Results were confirmed for other S. pyogenes strains at different time points in three independent experiments.
Abbreviations: BDL, below detection limit (0.05 mM); ND, not done.
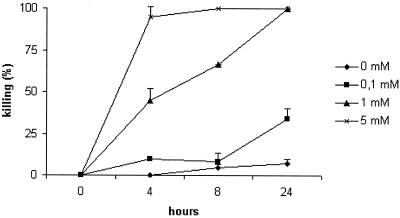
FIG. 5.
C. elegans killing by hydrogen peroxide. Different concentrations of hydrogen peroxide, ranging from 0.1 to 5 mM, in culture medium without bacteria, were applied to the nematodes in the liquid assay. A dose of 1 mM hydrogen peroxide killed all nematodes within 24 h and showed killing kinetics similar to those of S. pyogenes strains (Fig. 2). The experiment was performed in duplicate, and results are presented as means ± SD (error bars).
DISCUSSION
In this study we show that S. pyogenes is able to kill C. elegans and that this killing is only mediated by hydrogen peroxide. The following results support this conclusion. First, the killing mechanism did not depend on infection or the action of major S. pyogenes exotoxins. Second, nematodes were rescued from killing by the addition of catalase. Third, streptococci can produce sufficient amounts of hydrogen peroxide to kill C. elegans, with killing kinetics similar to those of equimolar concentrations of pure hydrogen peroxide. Fourth, there is a highly significant correlation between the killing capacity of a strain and the amount of hydrogen peroxide it produced.
The finding that only live S. pyogenes kill C. elegans suggests that killing depends on the novo synthesis of hydrogen peroxide produced by the bacteria during incubation with the nematodes (25°C under aerobic conditions). In agreement with this, supernatants taken from S. pyogenes grown under normal 37°C conditions did not kill the worms (data not shown).
Since we observed in some strains variations in C. elegans killing in the solid assay, we developed the liquid assay, in which nematodes are exposed to bacteria in culture medium. The liquid assay has three advantages over the more-conventional solid assay: it gives highly reproducible killing patterns; it is more sensitive than the solid assay, as all tested S. pyogenes strains killed C. elegans within 24 h; and it enables better quantitative measurement of toxins. Reproducibility and sensitivity might be increased since exposure of worms to the bacteria is independent of irregularities in the bacterial lawn and mobility of the worms. However, it is important to note that irrespectively of variable killing efficiencies of strains in the solid assay, all controls in the liquid assay (e.g., elimination of killing with catalase or unaltered killing with SPE-B or SLO mutants) could be confirmed with the solid assay.
In vitro and in vivo studies have shown that hydrogen peroxide produced by S. pyogenes and S. pneumoniae acts as a virulence factor, by exerting direct damage to mammalian cells or other bacteria (7, 9-11, 17). Recently it was shown that hydrogen peroxide, besides pneumolysin, is the key virulence factor in pneumococcal meningitis, by inducing apoptosis in brain cells (3). Since C. elegans is a widely used model organism for studying mammalian apoptosis (reviewed in reference 6) and apoptosis may be involved in the worm defense response to pathogen attack (1), it may be interesting to examine whether apoptosis occurs in C. elegans upon challenge with S. pyogenes and S. pneumoniae. For S. pyogenes, the main mechanism of cell damage during infectious and inflammatory processes may involve a coordinated synergistic cross talk among reactive oxygen species (i.e., hydrogen peroxide), membrane-perforating enzymes (i.e., SLO and SLS), and proteinases (i.e., SPE-B) (9). Synergistic action among these different classes of virulence factors should not play a role in the killing of C. elegans by S. pyogenes, since blocking of membrane perforating enzymes by cholesterol and blocking of proteinases by specific inhibitors did not affect the killing pattern.
The different killing capacities of S. pyogenes in the solid assay could reflect strain dependent differences in hydrogen peroxide production. Although several studies reveal hydrogen peroxide as an S. pyogenes virulence factor, the relationship between the ability to produce hydrogen peroxide of S. pyogenes strains and their severity of infection in humans remains unclear (19). We also could not detect such a relation for the strains used in this study (data not shown). By testing large numbers of clinical S. pyogenes isolates in the C. elegans model, a possible relationship may be detected and more insight might be gained in the role of hydrogen peroxide in S. pyogenes pathogenesis.
Garsin et al. (8) reported killing of C. elegans by S. pneumoniae and E. faecalis but not by S. pyogenes on brain heart infusion plates. At first sight their results seem to contrast with our findings. However, killing of C. elegans by bacteria depends on the culture media and the experimental conditions used. We observed for example that S. pyogenes did not kill C. elegans on blood agar plates (probably because these plates contain catalase) and indeed showed poor killing on brain heart infusion agar plates (data not shown). Therefore, the opposite killing capacities of E. faecalis and S. pyogenes observed in both studies are most probably caused by different experimental conditions and/or strain-specific differences. Nevertheless, the production of (sub)lethal amounts of hydrogen peroxide may be to some extent inevitable, since all experimental set ups have to meet the aerobic conditions needed for the nematodes.
Preliminary experiments in our laboratory also show that killing of C. elegans by S. pneumoniae can be fully prevented by catalase in the solid assay, the liquid assay, and the assay described by Garsin et al. (e.g., S. pneumoniae ATCC serotype 3 killed100% ± 0% without and 6.5% ± 2.1% [mean ± SD; n = 2] with catalase at 24 h in the liquid assay), suggesting that killing of C. elegans by S. pneumoniae is also mediated by hydrogen peroxide. Since hydrogen peroxide production is a common feature of catalase-negative bacteria under aerobic conditions, we speculate that hydrogen peroxide-mediated killing of C. elegans may be a general mechanism by which streptococci and other catalase-negative bacteria can kill C. elegans.
Acknowledgments
We thank Katja Mummenbrauer and Cathrin Struck for excellent technical assistance; Manfred Rohde for his help with microscopy; Andreas Podbielski and Karl-Hermann Schmidt for their kind gift of the SPE-B and SLO mutant, respectively; and Kadaba Sriprakash for Australian S. pyogenes isolates.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Aballay, A., and F. M. Ausubel. 2001. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc. Natl. Acad. Sci. USA 98:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 3.Braun, J. S., J. E. Sublett, D. Freyer, T. J. Mitchell, J. L. Cleveland, E. I. Tuomanen, and J. R. Weber. 2002. Pneumococcal pneumolysin and H2O2 mediate brain cell apoptosis during meningitis. J. Clin. Investig. 109:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhatwal, G. S., K. T. Preissner, G. Muller-Berghaus, and H. Blobel. 1987. Specific binding of the human S protein (vitronectin) to streptococci, Staphylococcus aureus, and Escherichia coli. Infect. Immun. 55:1878-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driscoll, M. 1996. Cell death in C. elegans: molecular insights into mechanisms conserved between nematodes and mammals. Brain Pathol. 6:411-425. [DOI] [PubMed] [Google Scholar]
- 7.Duane, P. G., J. B. Rubins, H. R. Weisel, and E. N. Janoff. 1993. Identification of hydrogen peroxide as a Streptococcus pneumoniae toxin for rat alveolar epithelial cells. Infect. Immun. 61:4392-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsburg, I., and M. Sadovnic. 1998. Gamma globulin, Evan's blue, aprotinin A PLA2 inhibitor, tetracycline and antioxidants protect epithelial cells against damage induced by synergism among streptococcal hemolysins, oxidants and proteinases: relation to the prevention of post-streptococcal sequelae and septic shock. FEMS Immunol. Med. Microbiol. 22:247-256. [DOI] [PubMed] [Google Scholar]
- 10.Ginsburg, I., and J. Varani. 1993. Interaction of viable group A streptococci and hydrogen peroxide in killing of vascular endothelial cells. Free Radic. Biol. Med. 14:495-500. [DOI] [PubMed] [Google Scholar]
- 11.Hirst, R. A., K. S. Sikand, A. Rutman, T. J. Mitchell, P. W. Andrew, and C. O'Callaghan. 2000. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. Infect. Immun. 68:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin, J., P. E. Kuwabara, and B. Corneliussen. 2000. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10:1615-1618. [DOI] [PubMed] [Google Scholar]
- 13.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 14.Lukomski, S., S. Sreevatsan, C. Amberg, W. Reichardt, M. Woischnik, A. Podbielski, and J. M. Musser. 1997. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Investig. 99:2574-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 16.Medina, E. O. Goldmann M. Rohde A. Lengeling G. S. Chhatwal. 2001. Genetic control of susceptibility to group A streptococcal infection in mice. in press. J. Infect. Dis. 184:846-852. [DOI] [PubMed] [Google Scholar]
- 17.Pericone, C. D., K. Overweg, P. W. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priess, J. R., H. Schnabel, and R. Schnabel. 1987. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell 51:601-611. [DOI] [PubMed] [Google Scholar]
- 19.Saito, M., S. Ohga, M. Endoh, H. Nakayama, Y. Mizunoe, T. Hara, and S. Yoshida. 2001. H2O2-nonproducing Streptococcus pyogenes strains: survival in stationary phase and virulence in chronic granulomatous disease. Microbiology 147:2469-2477. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt, K. H., D. Gerlach, K. Gubbe, A. Geyer, E. Birch-Hirschfeld, E. Straube, and A. Podbielski. 2001. Virulence of group A streptococci in fertile hens eggs is mainly effected by M protein and streptolysin O. Int. J. Med. Microbiol. 291:45-56. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt, K. H., A. Podbielski, W. Reichardt, K. Gubbe, and C. Amberg. 1997. Virulence of Streptococcus pyogenes for chicken embryos after isogenic inactivation of different streptococcal pathogenicity factors. Adv. Exp. Med. Biol. 418:793-795. [DOI] [PubMed] [Google Scholar]
- 22.Stevens, D. L., and E. L. Kaplan. 2000. Streptococcal Infections. Clinical aspects, microbiology, and molecular pathogenesis. Oxford University Press, Oxford, United Kingdom.
- 23.Tan, M. W., and F. M. Ausubel. 2000. Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr. Opin. Microbiol. 3:29-34. [DOI] [PubMed] [Google Scholar]
- 24.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood, W. B. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 27.Yorgey, P., L. G. Rahme, M. W. Tan, and F. M. Ausubel. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol. Microbiol. 41:1063-1076. [DOI] [PubMed] [Google Scholar]