Abstract
The study of how bacteria respond to and obtain divalent metal ions provides insight into the regulation of virulence factors in the host environment. Regulation of metal permease operons in gram-positive bacteria may involve the binding of metal-responsive repressors to palindromic domains in their control regions. The Streptococcus parasanguis fimA operon, which encodes an ATP-binding cassette (ABC) transporter system with sequence homology to the LraI family of metal transporters, possesses a palindromic regulatory region with high homology to that of the Streptococcus gordonii ScaR binding domain. Mapping of the promoter and regulatory regions of fimA and the divergently transcribed pepO gene, which encodes a zinc metalloendopeptidase, indicated that their promoter and regulatory elements overlap. fimA had one transcriptional start site, whereas pepO had three. Analysis of truncated versions of the pepO promoter suggested that all three transcriptional start sites are functional. Analysis of promoter activity under various environmental conditions indicated that the fimA operon promoter and the pepO promoter are not coordinately regulated. The fimA operon is responsive to changes in Mn2+ concentration, but the pepO promoter is not. A S. parasanguis fimA mutant showed a growth deficiency under conditions of limiting Mn2+. This deficiency was not alleviated by compensation with either Mg2+ or Fe3+. Wild-type S. parasanguis could take up Mn2+ and Fe3+, while the fimA mutant showed a marked reduction in this ability. These data suggested that FimA is a component of a metal transporter system capable of transporting both Mn2+ and Fe3+. FimA expression itself was shown to be responsive to Mn2+ concentration, but not to availability of Fe3+ or Mg2+.
Streptococcus parasanguis, along with other members of the mitis group of oral streptococci, are among some of the most successful colonizers of the human body. These commensal organisms in the oral cavity have the ability to attach, colonize, and thrive in an environment of continual flux of pH, temperature, mechanical stress, and nutrient availability. Introduction of these oral organisms into the bloodstream of individuals with predisposing heart valve damage can result in endocarditis, a life-threatening illness (3). Nutrients, in particular divalent metal ions, are often sequestered by the host in such a way that the colonizing bacteria must actively gain access to these resources in order to survive in the host environment. Iron in the form of ferric and ferrous compounds is essential for the growth and survival of gram-negative bacterial pathogens, and the ability to acquire these nutrients is considered a virulence trait (8, 34). In gram-positive organisms, such as the streptococci, the role of divalent metals in virulence is less well-defined. Evidence indicates that the lipoprotein receptor-associated antigen I (LraI) family of polypeptides found in a variety of streptococci (4, 7, 22, 23, 35) and of which FimA of S. parasanguis FW213 is a member forms part of a new family of solute-binding receptors of ABC metal ion transporters. Previous studies have shown that Streptococcus gordonii (24) and Streptococcus pneumoniae (9) transporters mediate uptake of Mn2+, while recent work indicates that the Streptococcus pyogenes LraI polypeptide binds Zn2+, Cu2+, and Fe3+ (21).
S. parasanguis FimA is a major virulence factor associated with endocarditis, and it has been suggested that FimA functions in the development of the infection by facilitating adherence to fibrin (5). Other members of the LraI family, including PsaA of S. pneumoniae and SloC of Streptococcus mutans, have also been shown to be virulence factors in animal models (4, 23). A potential virulence factor in S. parasanguis-induced endocarditis may be the zinc metallopeptidase PepO, which has been shown to be highly homologous in sequence and activity to the vasoconstriction-associated mammalian endothelin-converting enzyme (14, 28). The divergently transcribed pepO gene is immediately upstream of the fimA operon. It has been suggested that the promoter domains of the fimA operon and pepO gene overlap and that the two may be coordinately regulated (14). Regulation of transcription is often the means by which metal ion acquisition is regulated (15, 20, 33). Classic examples of this are the ferric uptake repressor (Fur) of Escherichia coli and the diphtheria toxin repressor (DtxR) of Corynebacterium diphtheriae. These metal-responsive repressors bind to regulatory regions controlling the transcription of genes involved in metal transport, siderophore production, and expression of virulence factors (11, 19). Recently, it has been shown that S. gordonii possesses a metallorepressor, ScaR, that shares 26% identity with DtxR of C. diphtheriae. This repressor, under conditions of high Mn2+ concentration, binds a regulatory region in the promoter of the S. gordonii fimA operon homolog, sca, thereby repressing transcription (20). The regulatory domain to which ScaR binds appears to have high DNA sequence homology to palindromic domains in the promoter regions of genes encoding other LraI family members, including the ABC-type transporter of S. parasanguis.
Studies were undertaken to examine the promoter activity of the fimA operon and pepO gene under conditions of limiting divalent metal ion availability. We show here that the S. parasanguis fimA promoter is repressed under conditions of high Mn2+ but that the divergently transcribed pepO promoter is not. In this paper, we have also characterized the function of FimA further and now have several lines of evidence which indicate that FimA is a metal transporter that is responsive to Mn2+ concentration.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. The wild-type S. parasanguis (formerly called S. sanguis) strain used was FW213 (6). VT930 and VT1393 are isogenic derivatives of FW213. Streptococcal strains were grown statically in the presence of 5% CO2 at 37°C in Todd-Hewitt (TH) (Difco Laboratories, Detroit, Mich.) broth or in a chemically defined medium (CDM) (JRH Biosciences, Denver, Pa.). For analysis of the effects of metals on cell growth, CDM was prepared and incubated with 30 g of Chelex-100 (Sigma) per liter for 4 h at 4°C to remove trace metals (32). The medium was filter sterilized, and metal ions were restored by the addition of MgSO4 52 7H2O, MnCl2·4H2O, or FeCl3·6H2O as described below. Growth in CDM or TH broth was monitored by measuring the optical density at 600 nm (OD600) or 470 nm (OD470) (Spectronic 20D; Milton Roy Company, Rochester, N.Y.), respectively, or by enumerating CFU. VT1548, VT1549, VT1553, and FW213 strains containing the E. coli-streptococcal luciferase shuttle vectors were grown in medium containing 10 μg of erythromycin per ml for plasmid selection.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. parasanguis | ||
| FW213 | Wild-type | 6 |
| VT930 | FW213 fimA::aphA3 | 13 |
| VT1393 | FW213 fapI::aphA3 | 37 |
| VT1548 | FW213 carrying pVT1550 | This study |
| VT1549 | FW213 carrying pVT1551 | This study |
| VT1553 | FW213 carrying pVT1552; control for luciferase assay | This study |
| E. coli HB101 | Host strain for cloning and plasmid propagation | Promega |
| Plasmids | ||
| pT7Blue | 2.9-kb PCR cloning vector; Apr | Novagen |
| pUC18 | 2.7-kb cloning vector; Apr | New England Biolabs |
| pVT1198 | pUC18 containing portions of pepO and fimC; Apr | 14 |
| pVT1327 | pT7Blue containing portions of pepO and fimC; Apr | 14 |
| pSG223 | E. coli-streptococcal luciferase shuttle vector; luciferase gene under the control of the gtf promoter; Err | 16 |
| pVT1550 | pSG223 with the pepO promoter replacing the gtf promoter 5′ of the luciferase gene; Err | This study |
| pVT1551 | pSG223 with the fimA operon promoter replacing the gtf promoter 5′ of the luciferase gene; Err | This study |
| pVT1552 | Promoterless pSG223; control for luciferase assay; Err | This study |
E. coli strain HB101 was grown at 37°C in Luria-Bertani (LB) broth or LB agar plates with the addition of 500 μg of erythromycin per ml to select for the presence of the E. coli-streptococcal luciferase shuttle plasmid encoding an erythromycin resistance marker.
Preparation of whole-cell extracts.
Streptococcal cultures were grown under specific environmental conditions to early logarithmic growth phase (OD470 of 0.3 or OD600 of 0.45) and mid-logarithmic growth phase (OD470 of 0.6 or OD600 of 0.9). Whole-cell extracts were prepared as previously described (28) except that cell pellets were resuspended in 20 mM potassium phosphate buffer for immunoblot analyses or 1× reporter lysis buffer (RLB) (Promega Corporation, Madison, Wis.) for luciferase assays. Protein concentrations were determined by using the bicinchoninic acid protein assay reagent kit (Pierce Chemical Company, Rockford, Ill.) with bovine serum albumin as the standard.
Primer extension analysis.
Total RNA was obtained from FW213 cells grown in TH broth to late log growth phase (OD470 of 0.8). Cells were pelleted by centrifugation and processed using the Bio 101 FastRNA Kit (blue) and the FP120 FastPrep machine (Bio 101, Inc., Vista, Calif.) according to the manufacturer's recommendation. Primer extension analysis was performed as described previously (36). The oligonucleotide 5′-CCCTGGATGGTGAGGGAAAG-3′ was used to map the start of transcription of the fimA operon by primer extension. The oligonucleotide 5′-GTAAAGGAGCAAACTCATG-3′ was used to map the start of transcription of the first pepO transcript, and the oligonucleotide 5′-CATTCCCCGTTGACGTAATC-3′) was used to map the start of transcription of the second and third pepO transcripts and to confirm the transcriptional start site of the first pepO transcript. Oligonucleotide primers were 5′ end labeled using T4 polynucleotide kinase and γ-32P-labeled dATP. The labeled oligonucleotide was hybridized with 10 μg of S. parasanguis total RNA, and extension was performed using SuperScript RNase H− reverse transcriptase (Gibco BRL) for 1 h at 42°C. The extended product was denatured and loaded onto a 6% polyacrylamide gel which contained 7 M urea. DNA sequencing (Sequenase version 2.0, DNA Sequencing Kit; U.S. Biochemicals, Cleveland, Ohio) of pVT1198 or pVT1327 containing the relevant portions of the fimA and pepO loci were performed using the same oligonucleotides as primers, and the products of these reactions were used as molecular size markers for the primer extensions.
Construction of the fimA operon and pepO promoter-luciferase fusion plasmids.
The wild-type pepO and fimA operon promoters from pVT1327 were amplified by PCR using a Techne Genius thermocycler (Techne Ltd., Cambridge, United Kingdom) and the GeneAmp PCR reagent kit (Roche Molecular Systems, Inc., Branchburg, N.J.). As can be seen in Fig. 2B, oligonucleotide primers designated pepO primer 5′ (5′-GGAAGATCTTTTTGATCAAGCTGG-3′) and pepO primer 3′ (5′-GGTCCATGGATCTTCTCGCTTTCATTC-3′) were designed to amplify regions of the pepO promoter, and fim primer 5′ (5′-CCTAGATCTTGCTGGTATAGTCTTC-3′) and fim primer 3′ (5′-GGTCCATGGAGTTTGCTCCTTTAC-3′) were designed to amplify regions of the fimA operon promoter. The primers were also constructed to contain BglII or NcoI restriction sites (shown in bold type) to facilitate cloning of the amplified promoter regions into the respective sites in the E. coli-streptococcal luciferase shuttle plasmid pSG223, thereby replacing the gtf promoter of pSG223 with the fimA operon or pepO promoter and generating pVT1550 or pVT1551, respectively. Plasmids were electroporated into electrocompetent E. coli HB101 cells following previously described protocols (2) using a Gene Pulse apparatus (Bio-Rad Laboratories, Hercules, Calif.). Cloned plasmids were isolated from E. coli using minicolumn plasmid purification kits (Qiagen, Inc., Santa Clarita, Calif.). DNA sequence analyses of the plasmids were performed at the Vermont Cancer Center DNA Analysis Facility at the University of Vermont by the Sanger dideoxynucleotide chain termination method as modified for ABI Prism Dye Terminator cycle sequencing using ampliTaq polymerase on an ABI 373 XL automated DNA sequencer (Perkin-Elmer Cetus, Norwalk, Conn.). Sequencing indicated that the pepO and fimA operon promoters were in frame and in proper orientation for the production of the luciferase protein. The E. coli-streptococcal reporter plasmids were transformed into FW213 cells by electroporation (12). The transformants VT1548 and VT1549 were used in luciferase assays to measure pepO or fimA operon promoter activity, respectively.
FIG. 2.
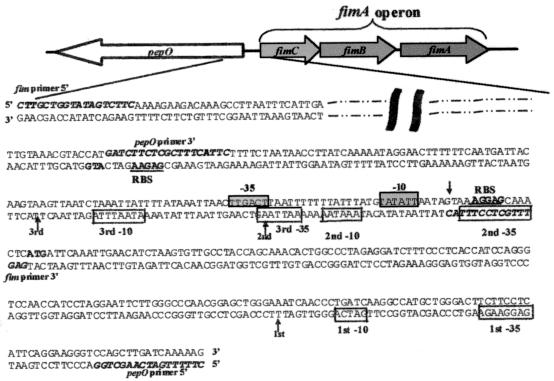
Schematic representation of the orientation of the pepO gene and the fimA operon in S. parasanguis. The expanded region shows the nucleotide sequence encompassing the 5′ coding region of pepO, the intergenic region between pepO and the fimA operon, and the 5′ coding region of the fimA operon. The fimA operon sequence (top sequence) and pepO sequence (bottom sequence) are shown. Sequence reported elsewhere (12) (dashed and dotted line) and sequences to which designated primers were designed for PCR and cloning purposes (bold italic type) are indicated. The arrows indicate the location of the start(s) of transcription for either the fimA operon or pepO gene, as determined by primer extension analysis. For the genes of the fimA operon and pepO gene, the start site of translation (bold type) and putative ribosomal binding sites (RBS) (underlined sequence) are shown. The putative −10 and −35 sites of the fimA operon (boxed sequence on grey background) and the pepO promoter (boxed sequence on white background) are indicated.
Luciferase assay.
Luciferase assays were performed as recommended by the manufacturer of the Luciferase Assay System Kit (Promega Corporation) containing RLB with the following modifications. Bacterial cells were grown under specific environmental conditions, and whole-cell extracts were prepared. Total protein (1 μg/ml) from whole-cell extracts was used for each assay in a total volume of 20 μl of 1× RLB. Assayed samples were equilibrated to room temperature, 100 μl of luciferase assay reagent (Promega Corporation) was added to each sample prior to analysis, and relative light units were determined in a LB9501 Berthold Lumat luminometer (Perkin-Elmer Berthold, Wellesley, Mass.).
Metal uptake and competition.
Cells grown in TH broth to mid-logarithmic phase were diluted in fresh TH broth to approximately 106 cells ml−1 (OD470 of 0.01). For metal uptake experiments, 2.0 μCi of either 54Mn (0.8 μM) (Dupont NEN, Boston, Mass.) or 55Fe (4.0 μM; Dupont NEN) was added to 1.0 ml of culture. Cultures without labeled metals were included to determine the numbers of CFU at the end of the experiment. Bacteria were grown overnight at 37°C and 5% CO2 and washed three times with fresh medium. Cells grown with 54Mn were washed in fresh medium containing 150 μM MnCl2 52> 4H2O; those grown with 55Fe were washed with 300 μM FeCl3·6H2O. After the final washes, cell pellets were resuspended in 100 μl of TH broth, transferred to 5 ml of Cytoscint scintillation fluid (ICN Biomedicals, Inc., Costa Mesa, Calif.), and radioactivity was measured in a Beckman LS6500 scintillation counter using a window calibrated for the appropriate isotope. Competition assays were performed as described above except that excess unlabeled Mn2+ or Fe3+, as indicated, was incubated with the cells along with radiolabeled Mn2+ or Fe3+ in order to evaluate the specificity of the binding interaction.
Immunoblot analysis of FimA and PepO.
S. parasanguis cultures were grown in Chelex-100-treated CDM supplemented with appropriate metal ions to the late logarithmic growth phase (OD600 of 1.2). Cells from 1-ml aliquots were harvested by centrifugation and resuspended in 100 μl of sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol). For immunoblot analysis using whole-cell extracts, 1 μg of total protein was added to sample buffer. Samples were boiled for 10 min, centrifuged, and separated by electrophoresis on 12% polyacrylamide-SDS gels. Proteins were transferred to nitrocellulose (Schleicher and Schuell, Keene, N.H.) and examined by immunoblot analysis (33a) as described previously (37) except that a 1:5,000 dilution of PepO antiserum was used as a probe for PepO and a 1:2,500 dilution of FimA antiserum was used for FimA. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G at a dilution of 1:10,000 was used as the secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Antibody conjugates were detected with a chemiluminescence system as described by the manufacturer (NEN Life Science Products, Boston, Mass.).
RESULTS
The promoter and regulatory regions of S. parasanguis fimA and pepO overlap.
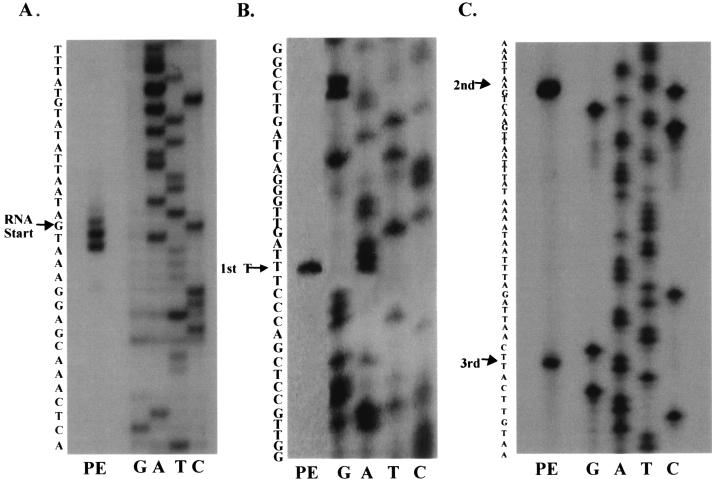
Previous analysis indicated that pepO was transcribed as a monocistronic message with at least two transcriptional start sites, as evidenced by a major and minor transcriptional product in Northern blot analysis (14). Primer extension analysis was performed to further elucidate the exact number of pepO transcriptional start sites and to investigate the potential overlap between the pepO and fimA operon promoters. The analysis indicated that pepO transcription was initiated at three transcriptional start sites producing three transcriptional products (Fig. 1B and C). The first transcriptional start site occurred 267 nucleotides from the start of translation, whereas the second and third sites were 155 and 123 nucleotides from the ATG start site, respectively. The two shorter transcription products were separated by 32 nucleotides and thus were not resolved on the previous Northern hybridization analysis. The first pepO transcription product occurred 118 nucleotides into the fimA coding region and 134 nucleotides from the fimA start of transcription (Fig. 1A), indicating that the promoter regions of pepO and the fimA operon overlap. Using the information provided by primer extension analysis, a schematic diagram indicating the putative −10 and −35 regions of the fimA operon and pepO promoters was constructed (Fig. 2).
FIG. 1.
Primer extension (PE) analysis of the fimA (A) and pepO (B and C) transcripts. In lane PE, oligonucleotide was annealed to 10 μg of S. parasanguis FW213 RNA and extended using SuperScript RNase H− reverse transcriptase. The nucleotide sequences of pVT1198 and pVT1327 (lanes G, A, T, and C) were determined using the same oligonucleotide as a primer.
The fimA operon promoter is responsive to the Mn2+ concentration, but the pepO promoter is not.
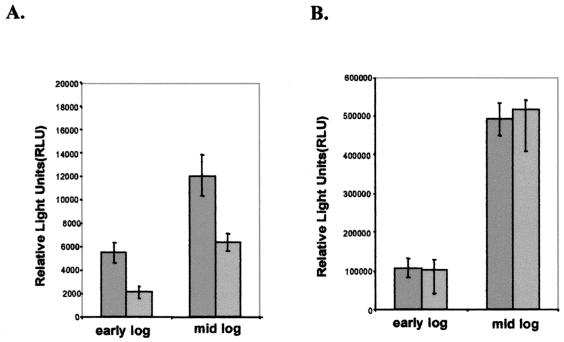
The organization of the fimA operon and the divergently transcribed pepO was described previously (14). A similar gene arrangement is seen with the S. gordonii sca operon and its divergently transcribed pepO (20, 25). On the basis of DNA sequence alignment and homology, it was suggested that the metallorepressor binding region that exists in the promoter of the sca operon of S. gordonii may also be present in the fimA operon of S. parasanguis (20). It was determined (Fig. 3) that the promoter domain of S. parasanguis pepO overlaps the promoter domain of the fimA operon and also includes a potential binding region for a metalloresponsive repressor. This suggests that transcription of both the fimA operon and pepO gene may be responsive to metal availability, since key transcriptional elements for both the fimA operon and pepO promoters would potentially be sequestered by the repressor. This hypothesis was investigated by using E. coli-streptococcal luciferase reporter shuttle plasmids pVT1551 and pVT1550. The gtf promoter of pSG223 was replaced by regions of the fimA operon promoter (Fig. 2), 463 nucleotides 5′ of the fimC start of translation, or the pepO promoter (Fig. 2), 335 nucleotides 5′ of the pepO translational start site, thereby placing the luciferase gene under the control of the fimA operon or pepO promoters, respectively. Luciferase assays of whole-cell extracts of S. parasanguis strains VT1548, VT1549, and VT1553 (control) grown to various growth phases in the presence of 0.1 or 10 μM Mn2+ indicated that the fimA operon promoter activity was repressed as much as threefold if grown in the presence of 10 μM Mn2+ (Fig. 4A). The promoter activity of pepO, however, was unresponsive to changes in Mn2+ concentration (Fig. 4B). The luciferase activity of both promoter constructs increased dramatically in mid-logarithmic growth phase compared to that of early logarithmic growth phase, suggesting that promoter activity of the fimA operon and pepO promoters may be regulated by growth phase. Two other promoter constructs were generated to investigate the possibility that the promoter regions for the two smaller pepO transcripts, the second and third pepO transcripts, may be responsive to Mn2+ concentration. These pepO promoter constructs were also unresponsive to Mn2+, but each did support transcription of the luciferase gene, indicating that each of these domains acts as a functional promoter (data not shown). The pepO promoter constructs also showed luciferase activity at least 10,000 times greater than that of the fimA operon promoter, indicating that the overall strength of the pepO promoter is much greater than that of the fimA operon promoter (Fig. 4).
FIG. 3.
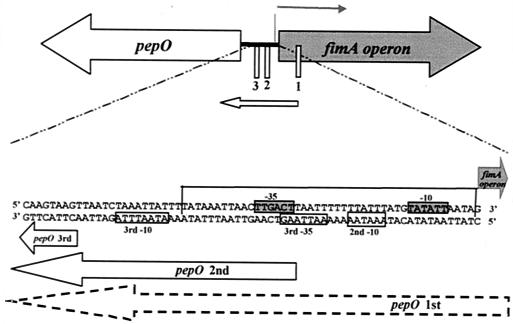
Schematic diagram of the S. parasanguis pepO gene and the fimA operon. The start of transcription of the fimA operon (thick black line) and the three start sites of pepO transcription (thin white boxes) are indicated. The expanded region shows the area of overlap between the fimA operon promoter and the pepO promoter. The fimA operon sequence (top sequence) and pepO sequence (bottom sequence) in the designated 5′ to 3′ orientations are shown. The region that shares high DNA sequence identity with the metallorepressor binding domain in S. gordonii sca operon (20) is boxed on three sides. The putative −10 and −35 sites of the fimA operon (boxed sequence on grey background) and the pepO promoter (boxed sequence on white background) are indicated. The arrows represent transcription products from either the fimA operon promoter or the pepO promoter.
FIG. 4.
Luciferase assay of VT1549 and FW213 strains containing either the fimA operon promoter construct (A) or the pepO promoter construct (B), grown in Chelex-100-treated CDM supplemented with 5.0 μM Mg2+ and 0.1 μM Fe3+ and various concentrations of Mn2+ (0.1 μM [dark grey bars] or 10 μM [light grey bars]). One microgram of total protein of whole-cell extracts in a final volume of 20 μl of RLB was mixed with 100 μl of luciferase assay reagent, and the light produced was measured in a luminometer. These experiments were performed four times; means and standard errors (error bars) are shown and adjusted for controls.
S. parasanguis displays a FimA-dependent growth requirement for Mn2+.
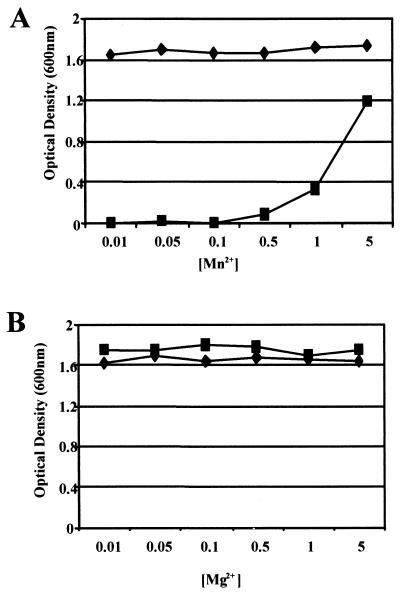
Since the fimA operon promoter of S. parasanguis FW213 was responsive to Mn2+ concentration, further investigation of the effects of divalent metal ions on bacterial growth was investigated. Growth of the fimA mutant strain VT930 in CDM containing low levels of Mn2+ (0.01 to 0.1 μM) was severely inhibited compared with that of wild-type FW213 (Fig. 5A). Growth of the fimA mutant began to be restored when the medium was supplemented with additional Mn2+ (>0.1 μM), and the OD600 of the culture approached that of the wild-type strain when >5.0 μM Mn2+ was added. Variation in the concentration of other ions, such as Mg2+ (Fig. 5B) and Fe3+ (data not shown), did not produce any further growth differences between the wild-type and fimA mutant strains. These data suggest that FimA is a high-affinity transporter of Mn2+ and that there is at least one other low-affinity Mn2+ transporter in S. parasanguis FW213. These data also suggest that FimA is not required for the uptake of Fe3+ or Mg2+, as growth of the fimA mutant is not affected by iron- or magnesium-limited conditions.
FIG. 5.
Effects of Mg2+ and Mn2+ concentrations on growth of wild-type FW213 and fimA mutant. Wild-type FW213 (♦) and fimA mutant (▪) were grown in the presence of 1.0 μM Mg2+ and various concentrations of Mn2+ (A) and 1.0 μM Mn2+ and various concentrations of Mg2+ (B). Cells were grown for 16 h in Chelex-100-treated CDM supplemented with 1.0 μM Fe3+ and metals as indicated, and cell density (OD600) was measured.
The S. parasanguis fimA mutant showed reduced metal uptake.
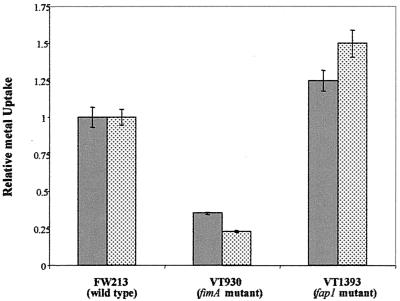
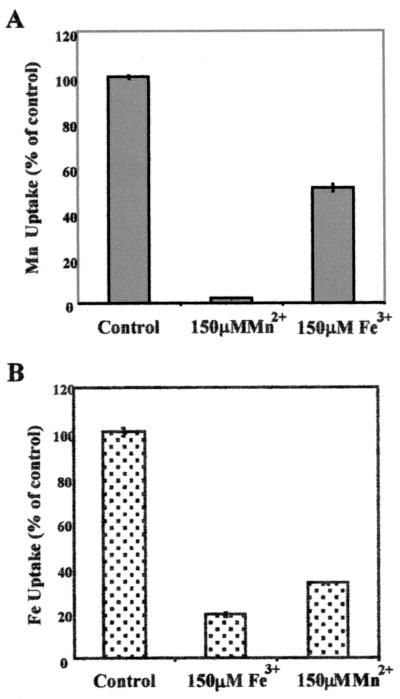
Metal uptake in wild-type FW213 and the fimA mutant was compared (Fig. 6). Bacteria were grown overnight in TH broth containing either 55Fe or 54Mn, washed in fresh media containing an excess of either cold Fe3+ or Mn2+, and radioactivity of the bacterial pellet was measured. Uptake of both isotopes was significantly lower in the fimA mutant; 55Fe uptake was reduced by approximately 80%, while uptake of 54Mn was reduced by approximately 65%. Growth of the fimA mutant was unaffected in TH broth, thus providing additional evidence that other metal transporters are present and functional. The fap1 mutant strain VT1393, an isogenic mutant of FW213 lacking fimbriae, was not affected in its ability to take up 55Fe or 54Mn and serves as a control for this assay. The results of the uptake assay suggest that the ABC-type transporter encoded by the S. parasanguis fimA operon is a metal transporter with multiple specificities. This conclusion was supported by data from competition assays (Fig. 7) in which uptake of radiolabeled manganese or iron in wild-type FW213 was inhibited by the addition of excess unlabeled Fe3+ or Mn2+.
FIG. 6.
Uptake of 54Mn (grey bars) or 55Fe (stippled bars) by wild-type FW213 and fimA and fap1 mutant S. parasanguis strains. Cell-associated counts were corrected for cell number as determined by CFU analysis. All assays were done in duplicate; means and standard errors (error bars) are shown. This experiment was performed twice; a representative data set is shown.
FIG. 7.
Competition of manganese (A) and iron (B) uptake. The indicated amount of MnCl2 or FeCl3 was added to the uptake assay. All assays were done in duplicate; means and standard errors (error bars) are shown. This experiment was performed twice; a representative data set is shown.
Mn2+ concentration affects FimA expression, but not PepO expression.
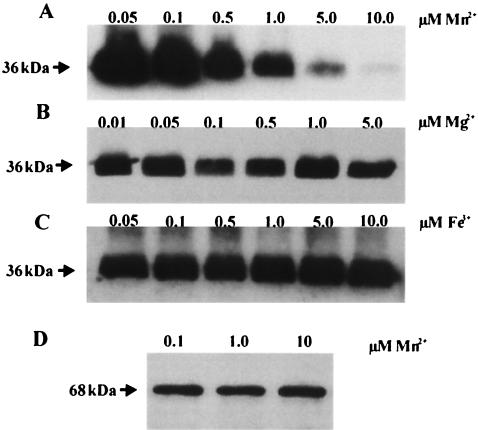
The expression of FimA in wild-type FW213 cells grown in the presence of different concentrations of divalent metals was examined by immunoblot analysis. Immunoblot analysis using FimA polyclonal antiserum indicated that FimA expression, as indicated by the presence of a 36-kDa band, was repressed in cells grown in media containing >0.1 μM Mn2+ (Fig. 8A, B, and C). Variation in the concentration of either Mg2+ or Fe3+ did not result in a change in FimA expression. PepO expression was unaffected by the range of Mn2+ concentrations that produced repression of FimA expression, as shown by the intensity of the 68-kDa band (Fig. 8D). These data were supported by the results of an enzyme-linked immunosorbent assay, which showed an approximate 25% reduction in FimA expression by cells grown in 10 μM Mn2+ compared to those cells grown in 0.1 μM Mn2+, yet no detectable change in PepO expression occurred (data not shown).
FIG. 8.
Immunoblot detection of FimA expression in wild-type FW213 cells grown in Chelex-100-treated CDM supplemented with metal ions. The cells were grown with the following concentrations of Mn2+, Mg2+, and Fe3+: Mn2+ (0.05 to 10 μM), 5.0 μM Mg2+, and 1.0 μM Fe3+ (A); Mg2+ (0.01 to 5.0 μM), 1.0 μM Fe3+, and 1.0 μM Mn2+ (B); and Fe3+ (0.05 to 10 μM), 5.0 μM Mg2+, and 1.0 μM Mn2+ (C). Proteins were extracted from an equal number of cells as measured by OD490, subjected to SDS-polyacrylamide gel electrophoresis, blotted onto nitrocellulose, and probed with polyclonal antiserum directed toward FimA. (D) Immunoblot detection of PepO expression in wild-type FW213 cells grown in Chelex-100-treated CDM supplemented with Mn2+ (0.1, 1.0, or 10 μM), 5.0 μM Mg2+, and 0.1 μM Fe3. One microgram of total protein per lane was subjected to SDS-polyacrylamide gel electrophoresis, blotted onto nitrocellulose, and probed with polyclonal antiserum directed toward PepO.
DISCUSSION
Colonization of a host niche by bacteria requires delicate, highly regulated expression of proteins. Cues that trigger the expression of virulence factors tend to be simple environmental changes, such as nutrient availability, pH, and the presence of reactive oxygen species (17). The ability of the bacterium to sense its environment while in the host is paramount to its survival. It has been established that low concentrations of divalent metal ions can induce the production of various virulence traits, such as toxin production in C. diphtheriae in the case of low Fe2+ availability. The C. diphtheriae toxin regulator DtxR is a metalloregulator that is responsive to Fe2+ and Fe3+ and regulates the transcription of various genes involved in virulence by binding to a palindromic consensus sequence in the control regions of these genes (27). Palindromic operator elements which bind metallo-responsive regulators also exist in other bacterial species, such as the binding domains of Staphylococcus epidermidis SirR, Mycobacterium tuberculosis IdeR, Treponema pallidum TroR, Bacillus subtilis MntR, and S. gordonii ScaR (10, 18, 20, 30, 31). The cis-acting palindromic elements that bind the iron-responsive DtxR, SirR, and IdeR share homology. This homology, however, is not shared with the palindromic elements which bind the Mn2+-responsive regulators TroR and MntR. ScaR is also a Mn2+-responsive repressor, but its binding domain appears to belong to a family different from that of even TroR and MntR. The domain is characterized by a 46-bp region which includes two palindromic domains that are also present in the promoter regions of other members of the LraI family of permeases (20).
The S. parasanguis fimA operon encodes an ABC-type transport system belonging to the LraI family as indicated by DNA sequence homology (13). The fimA operon also possesses two palindromic domains that have 94 and 81% sequence identity to palindrome I and II, respectively, of the ScaR binding region of S. gordonii (20). The S. parasanguis pepO gene, which encodes a zinc metalloendopeptidase, is immediately upstream of and divergently transcribed from the fimA operon. Mapping of the S. parasanguis fimA operon by primer extension analysis revealed that the transcriptional start site of fimA, like that of the S. gordonii sca operon transcriptional start site, occurs within 16 nucleotides of the translational start site (1). Mapping of the pepO promoter showed that pepO has three transcriptional start sites, with the first starting within the fimA operon coding region. Mammalian ECE-1 also has multiple transcripts that differ in their 5′ untranslated regions as a result of alternative promoter usage (29). These data also indicated that the promoter and regulatory elements of the fimA operon and pepO gene overlap.
Analysis of the promoter activities of the fimA operon and pepO promoters under conditions of limiting Mn2+ availability indicated that the fimA operon is responsive to Mn2+ concentration. The responsiveness of the fimA operon promoter to Mn2+ availability suggests that the transcriptional regulation of the fimA operon is similar to the regulation of the sca operon of S. gordonii (20). In S. gordonii, regulation of transcription of the sca operon is determined by the binding of ScaR, the Mn2+-responsive metallorepressor, to the cis-acting palindromic ScaR binding domain. In S. parasanguis, this putative repressor domain, also present in the promoter region of the fimA operon, did not affect the activity of the divergently transcribed pepO promoter as measured by luciferase activity. The pepO promoter, irrespective of Mn2+ concentration, showed luciferase activity levels 10,000 times greater than that associated with the fimA operon promoter. The high level of pepO promoter activity may be due to the cumulative effects of multiple pepO transcriptional start sites. In S. parasanguis, RNA stability, as well as promoter activity, may also play a role in the robust luciferase activity of the pepO-luciferase constructs. Computational analysis of the RNA folding of the pepO transcripts revealed the formation of a strong stem-loop structure in the 5′ untranslated region of the first and second transcripts, which could possibly protect the RNA from degradation by 5′ exonucleases (data not shown).
Further elucidation of the role of the fimA operon indicated that FimA is involved in the uptake of transition metal ions. Although FimA has specificity for at least two metal ions, Mn2+ and Fe3+, evidence presented here suggests that FimA is involved primarily in high-affinity uptake of Mn2+. Interestingly, the crystal structure of the PsaA metal binding site of the S. pneumoniae ABC-type transporter reveals that the active site was associated with Zn2+, although in vivo studies suggest that it is predominantly a Mn2+ transporter (9, 26). The ability of FimA to transport both Mn2+ and Fe3+ may be explained by the active site having a greater affinity for Mn2+, but conformational shifting of the solute binding domain can accommodate the binding of other divalent metal ions.
In summary, the fimA operon of S. parasanguis encodes a metal transporter with activity similar to those of other members of the LraI family. Transcription and expression of these genes are responsive to Mn2+ availability, but not to Fe3+ or Mg2+ availability. The divergently transcribed pepO does not show the same response to metal availability as the fimA operon, which suggests that the two are not coordinately regulated under these conditions.
Acknowledgments
We thank Diane Hutchins Meyer for critical review of the manuscript.
This work was supported in part by Public Health Service grant R37-DE11000 from the National Institutes of Health.
Editor: E. I. Tuomanen
REFERENCES
- 1.Andersen, R. N., R. D. Lunsford, and P. E. Kolenbrander. 1997. Determination of the transcript size and start site of the putative sca operon of Streptococcus gordonii ATCC 51656 (formerly strain PK488). Adv. Exp. Med. Biol. 418:657-660. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. 1988. Current protocols in molecular biology. Greene Publishers and Associates and Wiley-Interscience, New York, N.Y.
- 3.Baddour, L. M. 1994. Virulence factors among gram-positive bacteria in experimental endocarditis. Infect. Immun. 62:2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64:5255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnette-Curley, D., V. Wells, H. Viscount, C. L. Munro, J. C. Fenno, P. Fives-Taylor, and F. L. Macrina. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, R. M., G. B. Calandra, E. Huff, and K. M. Nugent. 1976. Attributes of potential utility in differentiating among “group H” streptococci or Streptococcus sanguis. J. Dent. Res. 55:A142-A153. [DOI] [PubMed] [Google Scholar]
- 7.Correia, F. F., J. M. DiRienzo, T. L. McKay, and B. Rosan. 1996. scbA from Streptococcus crista CC5A: an atypical member of the lraI gene family. Infect. Immun. 64:2114-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 10.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. W. Yu, S. Z. Sabol, R. K. Holmes, W. R. J. Jacobs, and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escolar, L., J. Pérez-Martín, and V. Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenno, J. C., A. Shaikh, and P. Fives-Taylor. 1993. Characterization of allelic replacement in Streptococcus parasanguis: transformation and homologous recombination in a “nontransformable” streptococcus. Gene 130:81-90. [DOI] [PubMed] [Google Scholar]
- 13.Fenno, J. C., A. Shaikh, G. Spatafora, and P. Fives-Taylor. 1995. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol. Microbiol. 15:849-863. [DOI] [PubMed] [Google Scholar]
- 14.Froeliger, E. H., J. Oetjen, J. P. Bond, and P. Fives-Taylor. 1999. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect. Immun. 67:5206-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman, S. D., and Q. Gao. 1999. Firefly luciferase as a reporter to study gene expression in Streptococcus mutans. Plasmid 42:154-157. [DOI] [PubMed] [Google Scholar]
- 17.Guiney, D. G. 1997. Regulation of bacterial virulence gene expression by the host environment. J. Clin. Invest. 99:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, P. J., A. Cockayne, P. Landers, J. A. Morrissey, C. M. Sims, and P. Williams. 1998. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66:4123-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181:S156-S167. [DOI] [PubMed] [Google Scholar]
- 20.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 21.Janulczyk, R., J. Pallon, and L. Björck. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34:596-606. [DOI] [PubMed] [Google Scholar]
- 22.Jenkinson, H. F. 1994. Cell surface protein receptors in oral streptococci. FEMS Microbiol. Lett. 121:133-140. [DOI] [PubMed] [Google Scholar]
- 23.Kitten, T., C. L. Munro, S. M. Michalek, and F. L. Macrina. 2000. Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence. Infect. Immun. 68:4441-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolenbrander, P. E., R. N. Andersen, R. A. Baker, and H. F. Jenkinson. 1998. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J. Bacteriol. 180:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolenbrander, P. E., R. N. Andersen, and N. Ganeshkumar. 1994. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect. Immun. 62:4469-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, M. C., P. A. Pilling, V. C. Epa, A. M. Berry, A. D. Ogunniyi, and J. C. Paton. 1998. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure 6:1553-1561. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. H., T. Wang, K. Ault, J. Liu, M. P. Schmitt, and R. K. Holmes. 1997. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oetjen, J., P. Fives-Taylor, and E. Froeliger. 2001. Characterization of a streptococcal endopeptidase with homology to human endothelin-converting enzyme. Infect. Immun. 69:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orzechowski, H. D., A. Gunther, S. Menzel, H. Funke-Kaiser, M. Richter, H. Bohnemeier, and M. Paul. 1998. Endothelial expression of endothelin-converting enzyme-1 beta mRNA is regulated by the transcription factor Ets-1. J. Cardiovasc. Pharmacol. 31:S55-S57. [DOI] [PubMed] [Google Scholar]
- 30.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 32.Spatafora, G., and M. Moore. 1998. Growth of Streptococcus mutans in iron-limiting medium. Methods Cell Sci. 20:217-221. [Google Scholar]
- 33.Spatafora, G., M. Moore, S. Landgren, E. Stonehouse, and S. Michalek. 2001. Expression of Streptococcus mutans fimA is iron-responsive and regulated by a DtxR homologue. Microbiology 147:1599-1610. [DOI] [PubMed] [Google Scholar]
- 33a.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 35.Viscount, H. B., C. L. Munro, D. Burnette-Curley, D. L. Peterson, and F. L. Macrina. 1997. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect. Immun. 65:994-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, H., and P. M. Fives-Taylor. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070-1081. [DOI] [PubMed] [Google Scholar]
- 37.Wu, H., K. P. Mintz, M. Ladha, and P. M. Fives-Taylor. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol. Microbiol. 28:487-500. [DOI] [PubMed] [Google Scholar]