Abstract
Chlamydial IncA localizes to the inclusion membrane and to vesicular fibers extending away from the inclusion. Chlamydial outer membrane components, in the absence of developmental forms, are found within these fibers. This colocalization may explain how chlamydial developmental form antigens are localized outside of the inclusion within infected cells.
Chlamydia are obligate intracellular bacteria that develop inside nonacidified host vacuoles, termed inclusions. Chlamydial inclusion biogenesis remains relatively uncharacterized and, to date, only two host cell macromolecules are known to interact with the inclusion membrane (IM [9, 19]). Chlamydia, however, synthesize and export several proteins to the IM (reviewed in reference 17). IncA, the best characterized of the proteins in the IM, is exposed at the cytoplasmic face of the inclusion by ca. 12 h postinfection, where it remains until lysis of the host cell (10, 15, 16). In addition to its localization on the IM, IncA is also found within fibers that appear as chains of vesicles extending away from the inclusion along distinct routes into the host cytoplasm (2, 17). The significance and composition of the IncA-laden fibers is unknown.
In the present study, we examined the abundance, distribution, and composition of the IncA-laden fibers present in HeLa 229 cells infected with Chlamydia psittaci guinea pig inclusion conjunctivitis, C. trachomatis L2 434/Bu, and C. pneumoniae TWAR. Cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum (MEM-10; standard growth conditions), in MEM-10 containing ampicillin, or in MEM-5 lacking tryptophan (5), as indicated. Methanol-fixed Chlamydia-infected cells cultured under these different environmental conditions were fluorescently labeled (5) and examined by laser scanning confocal microscopy. Antibodies to HSP60 (24), C. trachomatis major outer membrane protein (MOMP) (25), C. psittaci (16), C. trachomatis (2), C. pneumoniae IncA (1), and C. trachomatis Cap1 (8) have been described. Monoclonal antibody (MAb) to chlamydial Mip (MAb 147), chlamydial lipopolysaccharide (LPS; MAb EVI-H1), and C. psittaci MOMP (MAb 62) were kindly provided by John H. Pearce, Harlan Caldwell, and You-Xun Zhang, respectively. Data from three independent experiments were pooled and subjected to statistical analysis (Table 1) using a commercial data analysis program (Minitab, Inc., State College, Pa.).
TABLE 1.
Statistical summary of the antigens localizing within IncA-laden fibers
| Cell type and antigen | % Normal growth conditions (CI) | % Ampicillin exposure (CI) | % Tryptophan depletion (CI) |
|---|---|---|---|
| C. psittaci-infected cells | |||
| IncA-laden fibers | 31 (28-35) | 71 (68-73) | 26 (22-30) |
| LPS in fibers | 27 (19-37) | 94 (92-96) | 0 |
| Mip in fibers | 0 | 27 (24-31) | 0 |
| HSP60 in fibers | 0 | 0 | 0 |
| C. trachomatis-infected cells | |||
| IncA-laden fibers | 47 (41-53) | 70 (64-76) | 6 (4-8) |
| Cap1 in fibers | 12 (9-16) | 30 (25-35) | NEa |
| LPS in fibers | 0 | 8 (6-10) | 0 |
| HSP60 in fibers | 0 | 0 | 0 |
| C. pneumoniae-infected cells | |||
| IncA-laden fibers | 5 (3-7) | 6 (4-8) | NE |
| LPS in fibers | 0 | 27 (20-36) | NE |
| HSP60 in fibers | 0 | 0 | NE |
NE, not examined.
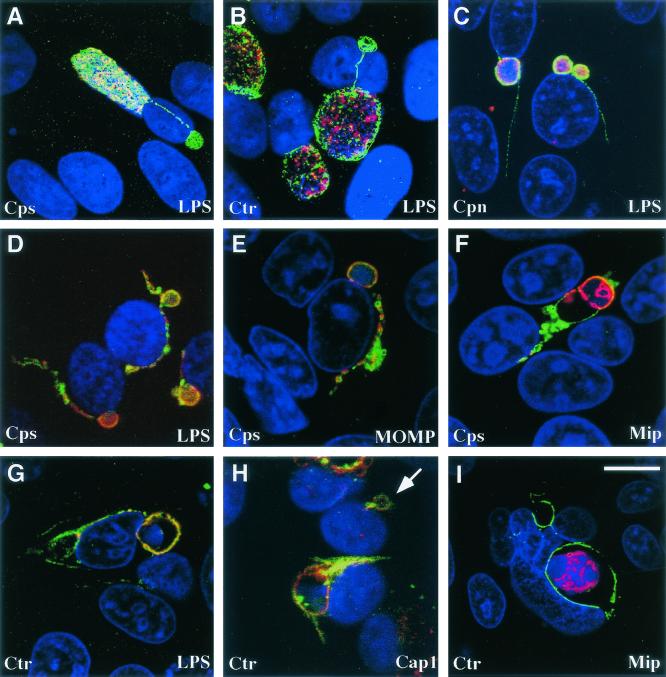
Infection of HeLa cells by each tested chlamydial species led to the formation of IncA-laden fibers that were attached to the intracellular inclusions. Approximately 31% (95% confidence interval [CI] = 28 to 35%) of C. psittaci, 47% (CI = 41 to 53%) of C. trachomatis, and 5% (CI = 3 to 7%) of C. pneumoniae inclusions were associated with IncA-laden fibers (Fig. 1A to C).
FIG. 1.
Antigen distribution within IncA-laden fibers of Chlamydia-infected cells cultured during standard growth conditions and in the presence of ampicillin. C. psittaci (Cps)-, C. trachomatis (Ctr)-, and C. pneumoniae (Cpn)-infected cells were cultured in MEM-10 (A to C) or MEM-10 containing ampicillin (D to I), prior to methanol fixation. All cells were labeled with anti-IncA antibodies (i.e., with fluorescein isothiocyanate). The antigens labeled with TRITC are depicted in the bottom right corner of each panel. Antigen colocalization was examined by using a confocal microscope. DAPI was used to label nucleic acids. The arrow in panel H shows the location of a vacuole embedded with chlamydial antigens that lack developmental forms. Bar, 8 μm (for all images).
There are several reports of chlamydial antigens localizing to areas outside the inclusion during growth (6, 11, 14, 22, 23). It is unclear how these chlamydial macromolecules, generally associated with the developmental forms, become distributed within host cells. The presence of antigens normally found in developmental forms was examined within the IncA-laden fibers of the three chlamydial species under different growth conditions. For C. psittaci-infected cells, these antigens included three membrane-associated antigens (LPS, MOMP, and Mip) and one cytosolic protein (HSP60). Under standard growth conditions, trace amounts of LPS were found within 27% (CI = 19 to 37%) of the IncA-laden fibers. In contrast, MOMP, Mip, and HSP60 remained associated with the developmental forms. The distribution of these same antigens, and Cap1, were examined in C. trachomatis-infected cells. Cap1 is an IM-associated antigen that is a cytotoxic-T-cell target (8). When cultured in MEM-10, C. trachomatis MOMP, LPS, and Mip remained associated with developmental forms, but Cap1 was detected in 12% (CI = 9 to 16%) of the IncA-laden fibers. Within C. pneumoniae-infected cells, LPS and HSP60 were the only antigens examined, both of which were not detected in IncA-laden fibers.
Culturing Chlamydia in the presence of certain antibiotics or the absence of certain amino acids results in the aberrant development of chlamydiae and can lead to a persistent growth state (reviewed in reference 3). These culture conditions also promoted a change in the appearance and frequency of the IncA-laden fibers. C. psittaci-infected cells cultured in the presence of 10 μg of ampicillin/ml showed a significant increase in the size and abundance of the cytosolic fibers (P < 0.0001). Enlarged IncA-laden fibers extended off 71% (CI = 68 to 73%) of the inclusions (Fig. 1D to F, fluorescein isothiocyanate). Exposure to ampicillin also led to an increase in the amount and type of other antigens localizing within the enlarged fibers. Ninety-four percent (CI = 92 to 96%) of the IncA-laden fibers contained increased amounts of LPS (Fig. 1D, TRITC [tetramethyl rhodamine isothiocyanate]). A similar percentage of IncA-laden fibers contained large amounts of MOMP (Fig. 1E, TRITC), whereas only 27% (CI = 24 to 31%) of the fibers contained trace amounts of Mip (Fig. 1F, TRITC). HSP60, however, was never observed outside the chlamydial developmental forms (data not shown), which is consistent with the electron microscopic observations of Raulston et al. (13). The extrainclusionary distribution of LPS, MOMP, and Mip was always associated with the IncA-laden fibers.
Similar to the observations made in C. psittaci-infected cells, the number of C. trachomatis inclusions containing IncA-laden fibers significantly increases to 70% (CI = 64 to 76%) in cells cultured in the presence of 1 μg of ampicillin/ml (P < 0.0001; Fig. 1G to I). LPS (Fig. 1G, TRITC) was found within ca. 8% (CI = 6 to 10%) of the IncA-laden fibers, whereas Cap1 was present in 30% (CI = 25 to 35%) of the fibers (Fig. 1H, TRITC). MOMP was also observed within the fibers, and the abundance of MOMP was similar to that seen with LPS (not shown). During culture in ampicillin, MOMP, LPS, and Cap1 were also found within the membrane of IncA-laden vacuoles that lack developmental forms (assessed by the absence of DAPI [4′,6′-diamidino-2-phenylindole] labeling; Fig. 1G and H). C. trachomatis Mip (Fig. 1I, TRITC) was occasionally localized to the IM but was not found in these empty vacuoles or IncA-laden fibers. Exposure of C. pneumoniae-infected cells to 10 μg of ampicillin/ml had no significant effect on the abundance of the IncA-laden fibers.
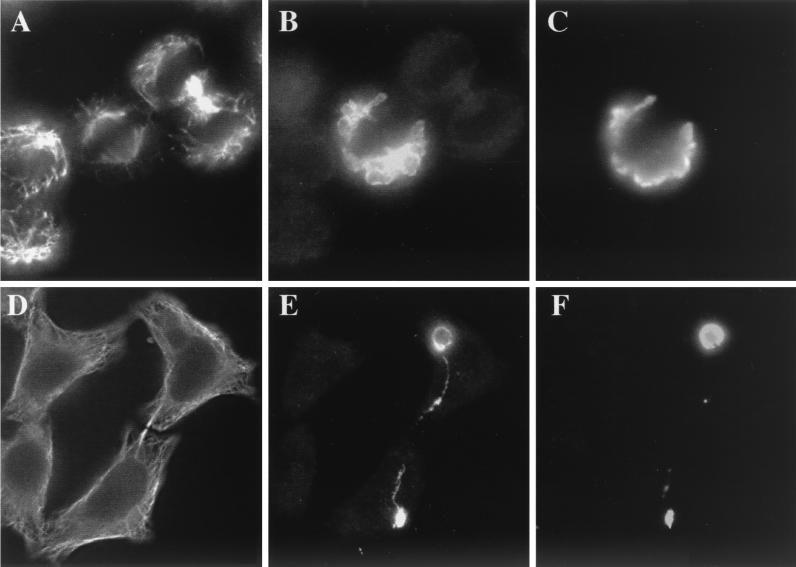
Immunofluorescence of dividing HeLa cells showed that, during host cytokinesis, inclusions containing C. psittaci also divided, distributing the inclusions between the two daughter cells (Fig. 2). IncA-laden fibers commonly formed between the divided inclusions present in the two different cells (Fig. 2E), linking cells that otherwise may appear unassociated and, in some cases, cells that are devoid of chlamydial developmental forms (Fig. 1H). These cells could routinely be found within infected monolayers and, commonly, the vacuole in the apparently uninfected cell was connected via a fiber to an inclusion in a neighboring cell (data not shown). LPS was often found within the fibers extending between the two inclusions (Fig. 2F). These results suggest host cell division may distribute the IncA-laden fibers, and empty IncA-laden vacuoles, between the daughter cells, providing a mechanism for chlamydial antigens to be present in the cytosol of uninfected host cells.
FIG. 2.
Distribution of IncA-laden fibers during host cell division. C. psittaci-infected cells were cultured in MEM-10 (A to C) or MEM-10 containing ampicillin (D to F) for 30 h prior to methanol fixation. Cells were triple labeled with anti-tubulin (A and D), anti-IncA (B and E), and anti-LPS (C and F) antibodies and then examined by using a standard fluorescence microscope. The image in panel E shows an IncA fiber stretching between two inclusions in different cells.
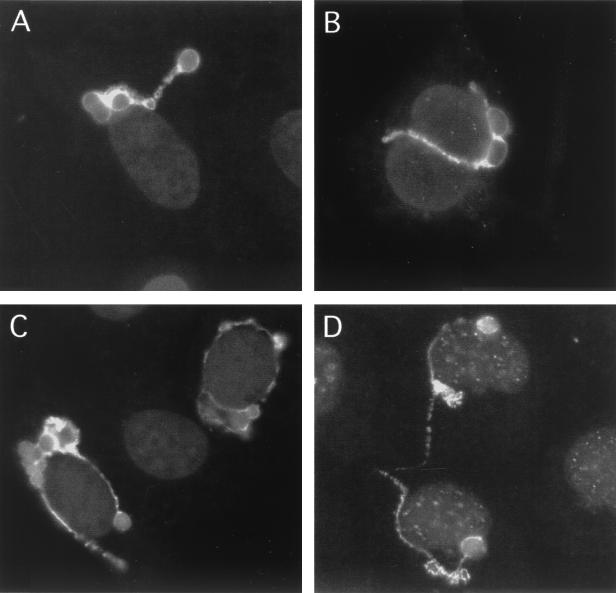
We showed, in immunofluorescence analyses, that chlamydial fibers appeared to follow distinct routes through the host cell, suggesting that fiber distribution may be directed along the host cytoskeletal network. To examine this theory, we examined fiber formation in cells that lacked intact microfilaments (21), microtubules (7, 12), or intermediate filaments (18). Morphologically typical IncA-laden fibers were found extending off 36% (CI = 32 to 40%) of the inclusions in the absence of microfilaments (Fig. 3B) and 55% of infected cells (CI = 51 to 58%) lacking microtubules (Fig. 3C); both percentage values were significantly less than that found in control cells (P < 0.0001). Culture of chlamydiae in a cell line lacking intermediate filaments (SW-13/c1.2 [18]) had no effect on the production of IncA-laden fibers. These results suggest that the production and localization of the IncA-laden fibers is not a function of the host cell cytoskeleton.
FIG. 3.
IncA-laden fibers are present in cells lacking intact cytoskeleton structures. The cytoskeleton of C. psittaci-infected HeLa cells cultured in the presence of MEM-10 and ampicillin was disrupted by using colchicine (A), cytochalasin D (B), or nocodazole (C). (D) Additionally, C. psittaci-infected SW-13/c1.2 intermediate filament-free cells were cultured in the presence of MEM-10 containing ampicillin. Cells were labeled with anti-IncA antibodies and examined by using a standard fluorescence microscope. IncA-laden fibers were still present after the three different drug treatments and in cells lacking intermediate filaments. Dark gray areas represent nucleic acid staining with DAPI.
One possible mechanism of liberating components from the reticulate body surface is through the formation of outer membrane vesicles which have been documented in several bacterial species (reviewed in reference 4) and have been found within the inclusions of Chlamydia-infected cells (20, 23). These vesicles would then fuse with the IM and incorporate, with other material in the IM, into growing IncA-laden fibers extending off the inclusion.
Acknowledgments
We thank M. and R. Hart for their assistance with the statistical analyses. We also thank H. Caldwell, J. Pearce, Y. Zhang, and R. Evans for contributing antibodies and cell lines to this research.
This work was supported by PHS grant R29AI42869, PHS grant R01AI48679-01, and Oregon Medical Research Fund grant 9823.
Editor: J. D. Clements
Footnotes
Oregon State University Agricultural Experiment Station technical paper 11778.
REFERENCES
- 1.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 1999. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., W. E. Stamm, R. J. Suchland, and D. D. Rockey. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 66:6017-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, W. J., and D. D. Rockey. 2000. Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infect. Immun. 68:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., S. J. Richmond, P. S. Yates, and C. C. Storey. 1994. Lipopolysacchariade in cells infected by Chlamydia trachomatis. Microbiology 140:1995-2002. [DOI] [PubMed] [Google Scholar]
- 7.De Brabander, M., G. Geunens, R. Nuydens, R. Willebrords, F. Aerets, and J. De Mey. 1986. Microtubule dynamics during cell cycle: the effects of taxol and nocodazole on the microtubule system of Pt K2 cells at different stages of the mitotic cycle. Int. Rev. Cytol. 101:215-274. [DOI] [PubMed] [Google Scholar]
- 8.Fling, S. P., R. A. Sutherland, L. N. Steele, B. Hess, S. E. F. D'Orazio, J.-F. Maisonneuve, M. F. Lampe, P. Probst, and M. N. Starnbach. 2001. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 98:1160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 92:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1:119-130. [DOI] [PubMed] [Google Scholar]
- 11.Karimi, S. T., R. H. Schloemer, and C. E. Wilde III. 1989. Accumulation of chlamydial lipopolysaccharide antigen in the plasma membranes of infected cells. Infect. Immun. 57:1780-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis, R. L., and L. Wilson. 1977. Addition of colchicine-tubulin complex to microtubule ends: the mechanism of substoichiometric colchicine poisoning. Proc. Natl. Acad. Sci. USA 8:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raulston, J. E., T. R. Paul, S. T. Knight, and P. B. Wyrick. 1998. Localization of Chlamydia trachomatis heat shock proteins 60 and 70 during infection of a human endometrial epithelial cell line in vitro. Infect. Immun. 66:2323-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richmond, S. J., and P. Stirling. 1981. Localization of chlamydial group antigen in McCoy cell monolayers infected with Chlamydia trachomatis or Chlamydia psittaci. Infect. Immun. 34:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 16.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617-626. [DOI] [PubMed] [Google Scholar]
- 17.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 18.Sarria, A. J., S. K. Nordeen, and R. M. Evans. 1990. Regulated expression of vimentin cDNA in cells in the presence or absence of a preexisting vimentin filament network. J. Cell Biol. 111:553-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 20.Stirling, P., and R. Richmond. 1980. Production of outer membrane blebs during chlamydial replication. FEMS Microbiol. Lett. 9:103-105. [Google Scholar]
- 21.Wodnicka, M., M. Pierzchalska, J. Bereiter-Hahn, and J. Kajstura. 1992. Comparative study on effects of cytochalasin B and D on F-actin content in different cell lines and different culture conditions. Folia Histochem. Cytobiol. 30:107-111. [PubMed] [Google Scholar]
- 22.Wyrick, P. B., J. Choong, S. T. Knight, D. Goyeau, E. S. Stuart, and A. B. MacDonald. 1994. Chlamydia trachomatis antigens on the surface of infected human endometrial epithelial cells. Immunol. Infect. Dis. 4:131-141. [Google Scholar]
- 23.Wyrick, P. B., S. T. Knight, T. R. Paul, R. G. Rank, and C. S. Barbier. 1999. Persistent chlamydial envelope antigens in antibiotic-exposed infected cells trigger neutrophil chemotaxis. J. Infect. Dis. 179:954-966. [DOI] [PubMed] [Google Scholar]
- 24.Yuan, Y., K. Lyng, Y.-X. Zhang, D. D. Rockey, and R. P. Morrison. 1992. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect. Immun. 60:2288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Y.-X., S. Stewart, T. Joseph, H. R. Taylor, and H. D. Caldwell. 1987. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J. Immunol. 138:575-581. [PubMed] [Google Scholar]