Abstract
Serotonin (5HT) is a critical modulator of neural circuits that support diverse behaviors and physiological processes, and multiple lines of evidence implicate abnormal serotonergic signaling in psychiatric pathogenesis. The significance of 5HT underscores the importance of elucidating the molecular pathways involved in serotonergic system development, function, and plasticity. However, these mechanisms remain poorly defined, owing largely to the difficulty of accessing 5HT neurons for experimental manipulation. To address this methodological deficiency, we present a transgenic route to selectively alter 5HT neuron gene expression. This approach is based on the ability of a Pet-1 enhancer region to direct reliable 5HT neuron-specific transgene expression in the CNS. Its versatility is illustrated with several transgenic mouse lines, each of which provides a tool for 5HT neuron studies. Two lines allow Cre-mediated recombination at different stages of 5HT neuron development. A third line in which 5HT neurons are marked with yellow fluorescent protein will have numerous applications, including their electrophysiological characterization. To demonstrate this application, we have characterized active and passive membrane properties of midbrain reticular 5HT neurons, which heretofore have not been reported to our knowledge. A fourth line in which Pet-1 loss of function is rescued by expression of a Pet-1 transgene demonstrates biologically relevant levels of transgene expression and offers a route for investigating serotonergic protein structure and function in a behaving animal. These findings establish a straightforward and reliable approach for developing an array of tools for in vivo and in vitro studies of 5HT neurons.
Keywords: Cre recombinase, pet-1, transgenic, yellow fluorescent protein, monoamine
Serotonin (5HT) is a transmitter of broad relevance to nervous system development and function (1-4). Serotonergic pathways innervate most cytoarchitectonic structures of the CNS, and accordingly they have been implicated in the modulation of circuitry involved in nearly all behaviors and physiological processes (3, 5-7). Additionally, 5HT neurotransmission is modulated through abundant afferent information arising from, for example, other monoaminergic systems (8, 9) and from orexinergic, glutamatergic, and GABAergic pathways (10-12). The remarkably expansive neuromodulatory influence of 5HT is the product of a complex transcriptional cascade that generates 5HT-synthesizing neurons in the ventral hindbrain (13-18). 5HT also figures prominently in mental health disorders as a number of lines of evidence provide strong support for the hypothesis that altered serotonergic signaling contributes to neurological and psychiatric pathogenesis (19-22). Despite considerable progress in understanding the importance of 5HT neurotransmission, however, the mechanisms governing 5HT neuron development and the precise physiological roles of 5HT in the modulation of CNS circuitry are not yet clear.
Studies of 5HT neurons are hindered by their small numbers and scattered distributions in the brain. Moreover, experimental perturbation of 5HT neuron function has relied almost exclusively on the use of dietary or pharmacological means to either deplete 5HT or alter 5HT neurotransmission (4). Although these approaches have contributed significantly to our understanding of the 5HT transmitter system they are often complicated by peripheral effects, variable levels of depletion, lack of target specificity, effects unrelated to depletion of 5HT, and in some instances unclear mechanisms of action (23-26). An additional critical shortcoming is the inability to selectively alter 5HT neuron gene expression. Recent gene-targeting studies of 5HT neurons are also limited in that most of the targeted genes reported to date are ubiquitously expressed and play essential roles in early development. Consequently, propagation of their homozygous null alleles in the mouse results in embryonic or early postnatal death (13, 17). Thus, an approach to conveniently alter gene expression in a 5HT neuron-specific manner would greatly facilitate studies of the 5HT transmitter system.
Here, we present a general strategy for manipulating 5HT neuron-specific gene expression by using a transgenic-based approach. This approach relies on the 5HT neuron-specific expression pattern of the Pet-1 ETS gene. Pet-1 is expressed in all mid-hindbrain 5HT-synthesizing neurons, and its embryonic expression precedes the appearance of serotonergic-specific traits. Moreover, in contrast to other serotonergic-related genes, such as the 5HT transporter (27), Pet-1 transcription in the brain is restricted to 5HT neurons and their postmitotic precursors (28). We recently identified an enhancer region upstream of mouse Pet-1-transcribed sequences that is able to accurately recapitulate the spatiotemporal pattern of Pet-1 expression in the brain. Importantly, the Pet-1 enhancer (ePet) region can direct highly reproducible 5HT neuron-specific transgene expression with little or no ectopic expression among independent founder lines (29). We describe several transgenic lines that demonstrate the reliability and general utility of the ePet region for development of serotonergic tools. The tools described here and additional ones that can now be generated will greatly increase the accessibility of 5HT neurons for investigations of 5HT neuron development, function, and plasticity in culture and behaving animals.
Materials and Methods
Transgenic Mice. ePet-Cre and ePet-enhanced yellow fluorescent protein (EYFP). LacZ was removed from BGZA reporter (30) and replaced with 5′-NarI, AgeI, MluI, AflII, HindIII-3′ polylinker. A Cre recombinase cDNA and simian virus 40 polyadenylation sequence (gift from Stephen O'Gorman, Case Western Reserve University) or EYFP coding sequences from pEYFP containing a polyadenylation signal (Clontech) were subcloned downstream of the β-globin minimal promoter by using the AgeI and AflII sites. The β-globin/Cre recombinase/poly(A) and β-globin/EYFP/poly(A) regions were then released from the β-globin lacZ reporter by EagI digestion and subcloned downstream of the 40-kb ePet genomic fragment present in the modified pBACe3.6 vector (29).
ePet-Pet. A 7.7-kb PacI/EcoRI genomic fragment whose sequences include the entire mouse Pet-1 gene including its own poly(A) sequences was subcloned downstream from the β-globin minimal promoter in the PacI/EcoR1 sites of a modified β-globin lacZ reporter (30) in which the lacZ gene was removed. The β-globin/Pet-1 sequences were then subcloned downstream of ePet present in pBACe3.6 after modifying the NotI site with the polylinker 5′-NotI, RsrII, NotI-3′. All transgenes were released from vector with an RsrII digest. Transgenes were purified for pronuclear transgenesis (31) and then injected into hybrid c57B6/SJL F1 or Pet-1 null zygotes. The ePet-Pet transgene was injected into Pet-1-/-- or Pet-1+/--fertilized eggs in present in a mixed C57BL/6 and 129 background.
Histology. Fluorescence immunohostochemistry and X-gal staining were performed as described (29). Horseradish peroxidase reactions to visualize anti-tryptophan hydroxylase (TPH) staining were performed following the Vectastain ABC kit protocol (Vector Laboratories) and visualized with SigmaFast diaminobenzidine tablets (Sigma). Fluorescent images were collected on an Olympus (Melville, NY) BX51 microscope with a Spot R/T color digital camera. Confocal images were obtained with a Zeiss LSM 410 confocal laser microscope with an argon/krypton laser (excitation 488 nm) and a ×25 Plan-Neofluar numerical aperture, 0.81-mm Korr, oil objective. Postprocessing of certain images was performed with photoshop (Adobe Systems, Santa Clara, CA).
Electrophysiology. Coronal slices (250-300 μm thick) through the median raphe nuclei were prepared from P14-18 mice by using a modified Leica VT1000S vibrotome. Slices were incubated at 30°C for 25 min then maintained at room temperature until needed. Whole-cell patch-clamp recordings were made in B9 neurons visualized under IR-differential interference contrast and epifluorescence optics (Zeiss Axioskop 1 FS) with an Axopatch 1D amplifier (Axon Instruments). During recordings, slices were superfused with artificial cerebrospinal fluid that contained 124 mM NaCl, 3 mM KCl, 1.23 mM NaH2PO4, 26 mM NaHCO3, 10 mM dextrose, 2.5 mM CaCl2, and 1.2 mM MgSO4, equilibrated with 95% O2/5% CO2 and warmed to 30°C (flow rate, 1-2 ml/min). Patch electrodes (3- to 5-MΩ resistance) contained 140 mM K-methylsulfate, 4 mM NaCl, 10 mM Hepes, 0.2 mM EGTA, 4 mM MgATP, 0.3 mM Na3GTP, and 10 mM phosphocreatine. Voltage records were low-pass-filtered at 2 kHz and then digitized at 5 kHz by using a 16-bit A/D converter (ITC-18, Instrutech, Mineola, NY) with custom software. Input resistance was calculated from the response to small amplitude hyperpolarizing steps. Membrane potentials indicated are not corrected for the liquid junction potential. Data are presented as mean ± SEM.
Results
Construction of ePet Transgenes. Our previous studies showed that ePet sequences carried on a 40-kb but not a 12-kb genomic fragment were able to direct reproducible 5HT neuron-specific transgene reporter expression among several individual founders (29). Thus, our strategy for construction of 5HT neuron-specific transgenes depends on the larger genomic fragment, which necessitates their propagation as bacterial artificial chromosomes by using the pBACe3.6 vector (32). ePet transgenes are made by using a two-step procedure in which a cDNA or gene sequence is subcloned into a shuttle vector to supply the β-globin minimal promoter followed by subcloning of β-globin/coding region sequences downstream of ePet in pBACe3.6 (see Materials and Methods). To demonstrate the general utility of ePet transgenes we describe several lines, each of which will have unique applications for in vitro and in vivo studies of 5HT neurons.
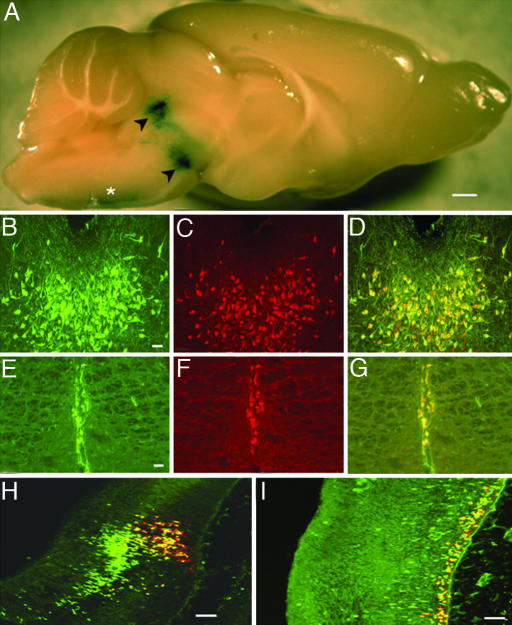
ePet-Cre. A line expressing a site-specific recombinase (33) in a serotonergic-specific manner would provide a powerful tool for manipulation of 5HT neuron chromosomal DNA. Therefore, an ePet-Cre transgene was prepared for pronuclear transgenesis and several ePet-Cre founders were obtained, each of which expressed Cre recombinase in the midbrain raphe (data not shown). To determine whether ePet-Cre is able to excise DNA specifically in 5HT neurons one of these founder lines was crossed to the ROSAR26R indicator strain (34) to generate R26RePet-Cre mice. Brains obtained from R26RePet-Cre mice were examined for LacZ expression by using X-gal histochemical detection. Strong LacZ expression was detected in the midbrain, which corresponded to the location of the dorsal and median raphe nuclei (Fig. 1A, arrowheads). LacZ expression was also present in the ventral medulla where caudal raphe nuclei are located (Fig. 1 A, asterisk). Additional analyses of sections processed for X-gal staining revealed LacZ expression in all serotonergic nuclei but not elsewhere in the brain, spinal cord, or retina (Fig. 1 A and Table 1). To determine whether or not ePet-Cre-induced LacZ expression was restricted to 5HT neurons, sections were processed for double immunostaining with an anti-TPH and anti-β-gal antisera. Importantly, there was an apparent complete concordance between TPH+ cells and β-gal+ ones in each of the midbrain (Fig. 1 B-D) and medullary (Fig. 1 E-G) serotonergic nuclei, suggesting that Cre-mediated excision of the ROSA locus occurred in all 5HT neurons. These findings demonstrate that ePet-Cre-mediated recombination is restricted to 5HT neurons in the CNS, and therefore even transient ePet activity was not evident outside of the 5HT neuron population.
Fig. 1.
ePet-Cre. (A) Whole-mount X-gal stain showing LacZ expression directed by ePet-Cre in adult R26RePet-Cre brain. Black arrowheads, dorsal and median raphe. Asterisk, ventral medullary raphe. (B-I) ePet-Cre activity is restricted to developing and adult 5-HT neurons. Colocalization (D and G)of TPH (B and E) (FITC, anti-TPH 1:200) and β-gal (C and F) (Texas red, anti-β-gal 1:5,000) in B7 dorsal raphe nucleus (B-D) and B2 raphe obscurus nucleus (E-G). (H and I) Colocalization of Cre recombinase (FITC, anti-Cre recombinase 1:100) with 5-HT (Texas red, anti-5-HT 1:10,000) in rostral (H) and caudal (I) E12 hindbrain. (Scale bars: 800 μm, A;50 μm, B-I.)
Table 1. ePet-Cre directed ROSA expression pattern.
| Brain | |
| 5HT neuron | ++++ |
| Other | − |
| Spinal cord | − |
| Retina | − |
| Adrenal gland | − |
| Enterocytes | + |
| Pancreas | + |
| Heart | − |
| Kidney | − |
| Bone marrow | − |
| Liver | − |
| Skin | + |
| Superior cervical ganglion | − |
| Cardiac neural crest | + |
Data were obtained from surveys of individual adult organs and E16 embryonic sagittal sections.
We next sought to determine the onset of ePet-Cre-directed Cre expression by coimmunostaining sections of embryonic hindbrain with anti-Cre recombinase and anti-5HT antisera. This analysis revealed Cre+ cells in the rostral hindbrain at embryonic day (E) 12.5 and then at E13.5 in caudal hindbrain (Fig. 1 H and I). Most of these Cre+ neurons were not yet 5HT+. These findings are consistent with the onset of endogenous Pet-1 expression and ePet activity preceding the appearance of 5HT in the rostral and caudal hindbrain (15, 28, 29). The onset of ePet-Cre expression before the appearance of 5HT indicates that it can be used for gene targeting during 5HT neuron differentiation.
To determine whether ePet-Cre-directed excisions occur outside of the brain we examined LacZ expression in a general survey of R26RePet-Cre whole E16 embryo sagittal sections and sections of adult R26RePet-Cre tissues. This analysis revealed lacZ expression in only small numbers of cells in the skin, gut, pancreas, and cardiac neural crest (Table 1).
A second ePet-Cre founder line, ePet-Cre-2, was also characterized in which recombination was delayed and occurred in a subset of 5HT neurons. We have not determined the reason for these expression characteristics but they might be caused by genomic insertion site or lower transgene copy number. In contrast to R26RePet-Cre mice (Fig. 2 A and B), recombination in R26RePet-Cre-2 was barely detectable at E16 in either the rostral or caudal R26RePet-Cre-2 hindbrain (Fig. 2 C and D), by which time 5HT neuron differentiation has ceased. Maximal levels of recombination generated with ePet-Cre-2 occurred at E18 in both the R26RePet-Cre-2 rostral and caudal domains (Fig. 2 E and F). However, recombination was detectable in only ≈50-70% of 5HT neurons in each of the adult serotonergic nuclei (Fig. 2 G and H). These findings suggest the ePet-Cre-2 line will be useful for investigation of gene function in a subset of 5HT neurons after their differentiation is complete.
Fig. 2.
ePet-Cre-2 directs delayed recombination in a subset of 5HT neurons. (A-F) X-gal staining with neutral red counterstain of ROSA R26RePet-Cre embryos in E15 rostral (A) and E15 caudal (B) or ROSA R26RePet-Cre-2 in E16 rostral (C), E16 caudal (D), E18 rostral (E), and E18 caudal (F) hindbrain. The asterisk in D shows small numbers of LacZ-positive cells at E16 in caudal ROSA R26RePet-Cre-2 hindbrain. (G) Coimmunostaining with anti-TPH (FITC, anti-TPH 1:200) and anti-β-gal (Texas red, anti-β-gal 1:5,000) shows mosaic pattern of recombination directed by ePet-Cre-2 in the P60 adult. (H) High magnification of boxed area in G. (Scale bars: 100 μm, A-F;50 μm, G; and 25 μm I.)
ePet-EYFP. A line in which 5HT neurons are genetically marked with a fluorescent indicator will have a number of in vivo and in vitro applications. Thus, we prepared an ePet-EYFP transgene to develop a line in which 5HT neurons express EYFP (35). Several ePet-EYFP founders were obtained, and in each EYFP fluorescence was observed in mid-hindbrain raphe. One of these lines was selected for detailed analysis of the EYFP expression pattern. As expected from the expression patterns of ePet-lacZ (29) and ePet-Cre (Fig. 1) EYFP fluorescence was restricted to cells in the raphe and cells scattered in the midbrain and medullary reticular formation (Fig. 3 A and E and data not shown). Immunostaining with an anti-TPH antibody showed that EYFP fluorescence was confined to 5HT neurons (Fig. 3 B-D). Confocal imaging of dissociated medullary tissue showed the presence of EYFP fluorescence throughout the entire substructure of the neuron, including cell body, processes, and growth cones (Fig. 3F).
Fig. 3.
ePet-EYFP. (A) EYFP fluorescence in midbrain B7 dorsal raphe. (B-D) Colocalization (D) of TPH (B) and EYFP fluorescence (C) and in ventral medullary B1 raphe pallidus. (E) Colocalization of EYFP fluorescence with TPH (Texas red, anti-TPH 1:200) in lateral B9 5HT neurons. (F) Confocal image of a dissociated EYFP+ 5HT neuron in culture. (Scale bars: 250 μm, A;20 μm, B-D;50 μm, C; and 5 μm, D.)
An obvious application of the ePet-EYFP line will be electrophysiological characterization of 5HT neurons in whole slice and dissociated cultures. To begin to demonstrate this application, brainstem slices of ePet-EYFP mice were prepared for whole-cell patch-clamp recordings with combined epifluorescent and IR/differential interference contrast visualization. We chose to study supralemniscal 5HT neurons within the B9 group (36) as the active and passive membrane properties of these cells have not been reported to our knowledge. 5HT neurons located in the midbrain B9 area generated a characteristic electrophysiological phenotype, observed in five of six EYFP-positive neurons. This phenotype included high input resistance (951 ± 100 MΩ; n = 5), long-duration membrane time constant (75.0 ± 6.7 ms), and a large-amplitude (23.3 ± 1.6 mV) spike afterhyperpolarization (Fig. 4A). EYFP-positive neurons displayed repetitive firing with instantaneous firing rates that varied linearly with current step amplitude (Fig. 4 B and C). Firing rates adapted rapidly during step responses (Fig. 4D). A sixth EYFP-positive neuron in B9 generated intrinsic responses that were distinct from this phenotype, including lower input resistance (380 MΩ), a multiphasic spike afterhyperpolarization, and prominent low-threshold regenerative responses (data not shown), suggesting the presence of electrophysiologically distinct subtypes in area B9. The characterization of these nonraphe 5HT neuron membrane properties demonstrates the utility of the ePet-EYFP line and establishes a convenient approach for the comprehensive electrophysiological analysis of these cells in the genetically tractable mouse brainstem slice preparation.
Fig. 4.
Electrophysiological recordings of EYFP+ 5HT neurons in B9. (A) Responses of an EYFP+ B9 neuron to 40-pA depolarizing and hyperpolarizing current steps at -68 mV. The depolarizing current step evoked an overshooting action potential with a pronounced afterhyperpolarization. (B) Responses from the same B9 neuron to 60-, 80-, and 110-pA step depolarizations from -72 mV. (C) Plot of instantaneous firing frequency (reciprocal of the first interspike interval) versus current step amplitude. (D) Plot of spike frequency adaptation in a EYFP+ B9 neuron during a response to a 250-pA step depolarization from -60 mV (voltage response shown in Inset).
ePet Activity Is Sufficient for Biologically Relevant Levels of Transgene Expression. To determine whether ePet enhancer activity is able to direct biologically relevant levels of gene expression we prepared a transgene, ePet-Pet, in which a 7.8-kb genomic fragment encoding all of the known mouse Pet-1-transcribed sequences was placed downstream of the ePet/β-globin promoter sequences. This transgene was then injected into Pet-1 heterozygous (+/-) or homozygous (-/-) null pronuclei to attempt rescue of Pet-1 loss of function, which is characterized by a 70% deficiency of 5HT neurons (14). In one round of pronuclear injection two Pet-1-/- founders hemizygous for ePet-Pet (Pet-1-/-,ePet-Pet/+) were obtained, and one of these was selected for rescue analysis with an anti-TPH antisera. As expected, the number of TPH+ cells was dramatically reduced in the B7 dorsal raphe of adult -/- compared with +/+ mice (Fig. 5, compare A with B). However, in sections of Pet-1-/-,ePet-Pet/+ dorsal raphe (Fig. 5C) the number of TPH+ cells was not significantly different from that in WT brain (see also Fig. 6, which is published as supporting information on the PNAS web site). HPLC determination of monoamines showed that 5HT and 5-hyroxyindoleacetic acid levels in Pet-1-/-,ePet-Pet/+ brains were also not significantly different from levels in WT brain (see Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site). These findings show that ePet enhancer activity is able to initiate and maintain transcription at a level that is sufficient for rescue of the neurochemical Pet-1 loss-of-function phenotype.
Fig. 5.

ePet directed rescue of Pet-1 loss of function. 5HT neurons in the B7 dorsal raphe were identified by immunostaining of coronal sections with anti-TPH antisera (diaminobenzidine, anti-TPH, 1:200) in +/+ (A), Pet-1-/- (B), and Pet-1-/-,ePet-Pet/+ (C) transgenic mice. (Scale bar: 100 μm.)
Discussion
Development of the transgenic approach described here depended on the identification of a genomic fragment capable of directing reliable 5HT neuron-specific gene expression. Our previous findings (29) indicated that ePet was able to direct transgene reporter expression to all 5HT neurons in the developing and adult brain. The timing of transgene expression precisely coincided with the onset of endogenous Pet-1 expression in the developing hindbrain (28). Importantly, serotonergic transgene expression directed by the 40-kb ePet fragment was highly reproducible as shown by the similar patterns of transgene expression among 16 different ePet-LacZ founder lines (29). Furthermore, in the majority of lines examined ePet transgene expression was not detected elsewhere in the developing and adult brain. This high level of specificity and reproducibility was extended by the findings presented here where we showed that three additional transgenes, ePet-Cre, ePet-EYFP, and ePet-Pet, were also active in developing and adult 5HT neurons. Together these findings demonstrate that the 40-kb ePet can be used as a reliable tool for 5HT neuron-specific transgene expression with little or no ectopic expression.
An ES cell knock-in approach is an alternative way in which the highly restricted expression pattern of the Pet-1 gene can be exploited for manipulation of 5HT neuron gene expression. However, the rapidity and reproducibility of generating ePet transgenes that we report here and previously (29) suggest it may be used to circumvent many of the time-consuming and technically challenging steps involved in ES cell gene targeting. Furthermore, the common need to investigate loss- or gain-of-function phenotypes in defined genetic backgrounds requires backcrossing of an ES cell-targeted allele for several generations. In contrast, ePet transgenes can be injected directly into the pronuclei of defined genetic backgrounds. Indeed, production of ePet-Pet transgenic mice reported here was performed with Pet-1+/- and Pet-1-/- pronuclei, which significantly reduced the time required to investigate the serotonergic phenotype in rescued mice. Thus, ePet transgenic lines in one or more defined genetic backgrounds can be obtained more quickly, which would permit investigation of modifier effects during the initial analyses of the resultant mouse phenotype. An additional advantage of our method is that ePet transgene expression was not detected in the retinal ganglion cell layer or adrenal medulla where endogenous Pet-1 expression is observed. Consequently, the ePet approach affords additional specificity of gene expression compared with that obtained with the ES cell knock-in approach into the Pet-1 locus. The lack of additional genes in ePet other than β-crystallineA2 also minimizes potential gain-of-function phenotypic effects from genes that would be present in a full-length bacterial artificial chromosome.
The ePet transgenic approach we have described here will practically eliminate the accessibility problem for 5HT neurons. Our ePet-Cre transgenic lines can be used to conditionally recombine floxed genes in 5HT neurons either during 5HT neuron differentiation or shortly after differentiation has been completed. For instance, gene-targeting studies have revealed a critical role for the Lmx1b factor in 5HT neuron differentiation (16, 17). However, the neonatal lethality of the Lmx1b null allele (37) has precluded loss-of-function studies in the adult. This phenotype indicates that conditional targeting of Lmx1b with the ePet-Cre lines will be essential for understanding the role of Lmx1b in the development of postnatal 5HT-modulated behaviors. These lines will also enable studies of 5HT neuron circuits in the behaving animal such as the reciprocal orexinergic-serotonergic negative feedback circuit that appears to be involved in stabilizing sleep/wakefulness states (38-40). The ePet-Cre lines could also be crossed to the lacZ wheat germ agglutinin (41) transgenic line for anterograde transneuronal tracing of 5HT circuits. Importantly, ROSA lacZ expression was not detected elsewhere in the developing and adult CNS; therefore, ePet-Cre-mediated excisions appear restricted to this single neuronal cell type. This characteristic identifies a clear advantage of ePet-Cre over the 5HT transporter-Cre knock-in line in which Cre recombinase is much more widely expressed in the brain and periphery (42). It should be kept in mind, however, that ePet does show activity in a small subset of nonneural tissues, which could complicate the interpretation of certain experiments.
Our approach offers a route with which to investigate many other aspects of 5HT neuron function and plasticity. For example, ePet-directed expression of activity monitors and modulators of neuronal excitability could be used to investigate 5HT neuron function and plasticity (43, 44). Temporal control of 5HT neuron gene expression could be developed by introduction of the tetracycline-responsive systems (45) or tamoxifen-inducible Cre recombinase into 5HT neurons (46). The rescue of Pet-1 loss of function with ePet-Pet provides a basis for investigating the structure/function relationships of Pet-1 in its normal cellular environment. Similarly, allelic, mutated, or alternative isoforms of proteins and RNAs involved, for example, in 5HT synaptic signaling and plasticity could be expressed specifically in 5HT neurons to investigate the relationship between primary structure, function, and behavior. The minimal position effects seen with ePet transgenes also increase the likelihood of identifying lines whose level of transgene expression is comparable to the level of expression of the endogenous gene under investigation. Alternatively, variable levels of transgene expression among different founder lines may be useful when exploring gain-of-function dosage effects.
Intracellular recordings of putative 5HT neurons in brain slices have shown that these cells have a high input resistance and a long duration action potential that is followed by a long, slow afterhyperpolarization and they are inhibited from firing by 5HT1a auto-receptor stimulation (47). These studies relied on retrospective formaldehyde-induced fluorescence (48) or more recently on 5HT immunohistochemistry (49) to determine their serotonergic identity. This identification approach has limited the functional analysis of 5HT neurons to those that are positioned in raphe where their higher density increases the chances of being randomly selected. However, estimates in various mammalian species, including mice and humans, indicate that as many as 25% of midbrain/pons 5HT neurons are present in extraraphe locations compared with ≈59% and 16% in the dorsal and median raphe, respectively (36). These lateral 5HT cells are located within and dorsal to the medial lemniscus and in the mesencephalic and pontine reticular formation. Moreover, they are cytoarchitecturally distinct from those in raphe, suggesting possible functional heterogeneity (36). As these scattered reticular neurons are intermingled with large numbers of nonserotonergic neurons they are far less accessible than their raphe counterparts. Consequently, the membrane properties of this 5HT neuron population have not been reported in any species to our knowledge. In the few published reports of mouse raphe 5HT neuron characteristics serotonergic neurochemical identity was not confirmed (50-52). This issue is important as recent studies (49, 53) suggest the functional characteristics of intermingled 5HT and non-5HT neurons in the dorsal raphe are much more similar than previously appreciated. We have shown here that EYFP fluorescence is robust in living brainstem slices and dissociated cells in culture, which permits direct identification of 5HT neuron cell bodies, processes, and growth cones. These findings show that the ePet-EYFP line provides for comparative in-depth studies of mouse raphe and reticular 5HT neuron electrophysiological characteristics and obviates time-consuming retrospective immunohistochemical identification of these cells. The ability to conveniently record from EYFP+ cells combined with genetic approaches available in the mouse should establish the ePet-EYFP line and its derivatives as a powerful system for further studies of 5HT neuron membrane properties.
In conclusion, the ePet transgenic approach will likely change the way 5HT neurons are studied in vivo and in vitro. The ability to reliably alter 5HT neuron gene expression with transgenes tailored for specific experiments should facilitate testing of theories concerning the precise roles of 5HT in various behaviors and physiological processes.
Supplementary Material
Acknowledgments
We thank Dr. Jerry Silver for important discussions of potential applications and Dr. Stephen O'Gorman for the ROSAR26R strain and Cre recombinase cDNA. This research was supported by National Institutes of Health Grants NS047752 (to S.H.), NS33590 (to B.W.S.), and MH62723 (to E.S.D.).
Author contributions: M.M.S., C.J.W., J.K.L., S.H., B.W.S., and E.S.D. designed research; M.M.S., C.J.W., J.K.L., R.M., K.L., W.J., R.A.C., and B.W.S. performed research; M.M.S., C.J.W., J.K.L., B.W.S., and E.S.D. analyzed data; and M.M.S. and E.S.D. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: 5HT, serotonin; ePet, Pet-1 enhancer; EYFP, enhanced yellow fluorescent protein; TPH, tryptophan hydroxylase; E(n), embryonic day (n).
References
- 1.Gaspar, P., Cases, O. & Maroteaux, L. (2003) Nat. Rev. Neurosci. 4, 1002-1012. [DOI] [PubMed] [Google Scholar]
- 2.Mason, P. (2001) Annu. Rev. Neurosci. 24, 737-777. [DOI] [PubMed] [Google Scholar]
- 3.Richerson, G. B. (2004) Nat. Rev. Neurosci. 5, 449-461. [DOI] [PubMed] [Google Scholar]
- 4.Sodhi, M. S. & Sanders-Bush, E. (2004) Int. Rev. Neurobiol. 59, 111-174. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, N. M. & Sharp, T. (1999) Neuropharmacology 38, 1083-1152. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs, B. L. & Azmitia, E. C. (1992) Physiol. Rev. 72, 165-229. [DOI] [PubMed] [Google Scholar]
- 7.Vinay, L., Brocard, F., Clarac, F., Norreel, J. C., Pearlstein, E. & Pflieger, J. F. (2002) Brain Res. Brain Res. Rev. 40, 118-129. [DOI] [PubMed] [Google Scholar]
- 8.Baraban, J. M. & Aghajanian, G. K. (1980) Neuropharmacology 19, 355-363. [DOI] [PubMed] [Google Scholar]
- 9.Baraban, J. M. & Aghajanian, G. K. (1981) Brain Res. 204, 1-11. [DOI] [PubMed] [Google Scholar]
- 10.Brown, R. E., Sergeeva, O. A., Eriksson, K. S. & Haas, H. L. (2002) J. Neurosci. 22, 8850-8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, R., Ding, Y. & Aghajanian, G. K. (2002) Neuropsychopharmacology 27, 329-340. [DOI] [PubMed] [Google Scholar]
- 12.Levine, E. S. & Jacobs, B. L. (1992) J. Neurosci. 12, 4037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattyn, A., Simplicio, N., Van Doorninck, J. H., Goridis, C., Guillemot, F. & Brunet, J. F. (2004) Nat. Neurosci. 7, 589-595. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks, T. J., Fyodorov, D. V., Wegman, L. J., Lelutiu, N. B., Pehek, E. A., Yamamoto, B., Silver, J., Weeber, E. J., Sweatt, J. D. & Deneris, E. S. (2003) Neuron 37, 233-247. [DOI] [PubMed] [Google Scholar]
- 15.Pattyn, A., Vallstedt, A., Dias, J. M., Samad, O. A., Krumlauf, R., Rijli, F. M., Brunet, J. F. & Ericson, J. (2003) Genes Dev. 17, 729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, L., Chen, C. L., Luo, P., Tan, M., Qiu, M., Johnson, R. & Ma, Q. (2003) J. Neurosci. 23, 9961-9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, Y. Q., Marklund, U., Yuan, W., Yin, J., Wegman, L., Ericson, J., Deneris, E., Johnson, R. L. & Chen, Z. F. (2003) Nat. Neurosci. 6, 933-938. [DOI] [PubMed] [Google Scholar]
- 18.Craven, S. E., Lim, K. C., Ye, W., Engel, J. D., De Sauvage, F. & Rosenthal, A. (2004) Development (Cambridge, U.K.) 131, 1165-1173. [DOI] [PubMed] [Google Scholar]
- 19.Kinney, H. C., Filiano, J. J. & White, W. F. (2001) J. Neuropathol. Exp. Neurol. 60, 228-247. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, J. A. & Hen, R. (2004) Annu. Rev. Neurosci. 27, 193-222. [DOI] [PubMed] [Google Scholar]
- 21.Mann, J. J. (2003) Nat. Rev. Neurosci. 4, 819-828. [DOI] [PubMed] [Google Scholar]
- 22.Scott, M. M. & Deneris, E. S. (2005) Int. J. Dev. Neurosci. 23, 277-285. [DOI] [PubMed] [Google Scholar]
- 23.Choi, S., Jonak, E. & Fernstrom, J. D. (2004) Brain Res. 1007, 19-28. [DOI] [PubMed] [Google Scholar]
- 24.Chang, N., Kaufman, S. & Milstien, S. (1979) J. Biol. Chem. 254, 2665-2668. [PubMed] [Google Scholar]
- 25.Knuth, E. D. & Etgen, A. M. (2004) Brain Res. Dev. Brain Res. 151, 203-208. [DOI] [PubMed] [Google Scholar]
- 26.Stokes, A. H., Xu, Y., Daunais, J. A., Tamir, H., Gershon, M. D., Butkerait, P., Kayser, B., Altman, J., Beck, W. & Vrana, K. E. (2000) J. Neurochem. 74, 2067-2073. [DOI] [PubMed] [Google Scholar]
- 27.Hansson, S. S., Mezey, E. & Hoffman, B. J. (1998) Neuroscience 83, 1185-1201. [DOI] [PubMed] [Google Scholar]
- 28.Hendricks, T., Francis, N., Fyodorov, D. & Deneris, E. (1999) J. Neurosci. 19, 10348-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, M. M., Krueger, K. C. & Deneris, E. S. (2005) J. Neurosci. 25, 2628-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helms, A. W., Abney, A. L., Ben-Arie, N., Zoghbi, H. Y. & Johnson, J. E. (2000) Development (Cambridge, U.K.) 127, 1185-1196. [DOI] [PubMed] [Google Scholar]
- 31.Monani, U. R., Sendtner, M., Coovert, D. D., Parsons, D. W., Andreassi, C., Le, T. T., Jablonka, S., Schrank, B., Rossol, W., Prior, T. W., et al. (2000) Hum. Mol. Genet. 9, 333-339. [DOI] [PubMed] [Google Scholar]
- 32.Frengen, E., Weichenhan, D., Zhao, B., Osoegawa, K., van Geel, M. & de Jong, P. J. (1999) Genomics 58, 250-253. [DOI] [PubMed] [Google Scholar]
- 33.Branda, C. S. & Dymecki, S. M. (2004) Dev. Cell 6, 7-28. [DOI] [PubMed] [Google Scholar]
- 34.Soriano, P. (1999) Nat. Genet. 21, 70-71. [DOI] [PubMed] [Google Scholar]
- 35.Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K. & Miyawaki, A. (2002) Nat. Biotechnol. 20, 87-90.11753368 [Google Scholar]
- 36.Vertes, R. P. & Crane, A. M. (1997) J. Comp. Neurol. 378, 411-424. [DOI] [PubMed] [Google Scholar]
- 37.Chen, H., Lun, Y., Ovchinnikov, D., Kokubo, H., Oberg, K. C., Pepicelli, C. V., Gan, L., Lee, B. & Johnson, R. L. (1998) Nat. Genet. 19, 51-55. [DOI] [PubMed] [Google Scholar]
- 38.Li, Y., Gao, X. B., Sakurai, T. & van den Pol, A. N. (2002) Neuron 36, 1169-1181. [DOI] [PubMed] [Google Scholar]
- 39.Liu, R. J., van den Pol, A. N. & Aghajanian, G. K. (2002) J. Neurosci. 22, 9453-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakurai, T., Nagata, R., Yamanaka, A., Kawamura, H., Tsujino, N., Muraki, Y., Kageyama, H., Kunita, S., Takahashi, S., Goto, K., et al. (2005) Neuron 46, 297-308. [DOI] [PubMed] [Google Scholar]
- 41.Braz, J. M., Rico, B. & Basbaum, A. I. (2002) Proc. Natl. Acad. Sci. USA 99, 15148-15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang, X., Masson, J., Gingrich, J. A., Rayport, S. & Hen, R. (2005) J. Neurosci. Methods 143, 27-32. [DOI] [PubMed] [Google Scholar]
- 43.Marek, K. W. & Davis, G. W. (2003) Curr. Opin. Neurobiol. 13, 607-611. [DOI] [PubMed] [Google Scholar]
- 44.Miyawaki, A. (2003) Curr. Opin. Neurobiol. 13, 591-596. [DOI] [PubMed] [Google Scholar]
- 45.Gossen, M. & Bujard, H. (2002) Annu. Rev. Genet. 36, 153-173. [DOI] [PubMed] [Google Scholar]
- 46.Vallier, L., Mancip, J., Markossian, S., Lukaszewicz, A., Dehay, C., Metzger, D., Chambon, P., Samarut, J. & Savatier, P. (2001) Proc. Natl. Acad. Sci. USA 98, 2467-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandermaelen, C. P. & Aghajanian, G. K. (1983) Brain Res. 289, 109-119. [DOI] [PubMed] [Google Scholar]
- 48.Aghajanian, G. K. & Vandermaelen, C. P. (1982) J. Neurosci. 2, 1786-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck, S. G., Pan, Y. Z., Akanwa, A. C. & Kirby, L. G. (2004) J. Neurophysiol. 91, 994-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Yacoubi, M., Bouali, S., Popa, D., Naudon, L., Leroux-Nicollet, I., Hamon, M., Costentin, J., Adrien, J. & Vaugeois, J. M. (2003) Proc. Natl. Acad. Sci. USA 100, 6227-6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trulson, M. E. & Frederickson, C. J. (1987) Brain Res. Bull. 18, 179-190. [DOI] [PubMed] [Google Scholar]
- 52.Froger, N., Gardier, A. M., Moratalla, R., Alberti, I., Lena, I., Boni, C., De Felipe, C., Rupniak, N. M., Hunt, S. P., Jacquot, C., et al. (2001) J. Neurosci. 21, 8188-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirby, L. G., Pernar, L., Valentino, R. J. & Beck, S. G. (2003) Neuroscience 116, 669-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.