Abstract
Diffusely adhering Escherichia coli strains harboring Afa/Dr adhesins (Afa/Dr DAEC) have been associated with diarrhea and urinary tract infections (UTIs). The present work is the first extensive molecular study of a Afa/Dr DAEC strain using the representational difference analysis technique. We have searched for DNA sequences present in strain C1845, recovered from a diarrheagenic child, but absent from a nonpathogenic K-12 strain. Strain C1845 harbors part of a pathogenicity island (PAICFT073) and several iron transport systems found in other E. coli pathovars. We did not find genes encoding factors known to subvert host cell proteins, such as type III secretion system or effector proteins. Several C1845-specific sequences are homologous to putative virulence genes or show no homology with known sequences, and we have analyzed their distribution among Afa/Dr and non-Afa/Dr clinical isolates and among strains from the E. coli Reference Collection. Three C1845-specific sequences (MO30, S109, and S111) have a high prevalence (77 to 80%) among Afa/Dr strains and a low prevalence (12 to 23%) among non-Afa/Dr strains. In addition, our results indicate that strain IH11128, an Afa/Dr DAEC strain recovered from a patient with a UTI, is genetically closely related to strain C1845.
Diffusely adhering Escherichia coli (DAEC) strains, which are characterized by their diffuse adherence pattern on cultured epithelial HeLa cells (56), have been recognized as the sixth class of diarrheagenic E. coli and appear as a heterogeneous group (12, 41). A subclass of DAEC strains harbors adhesins of the Afa/Dr family (including Afa-I, Afa-III, Dr, Dr-II, and F1845), which recognize the decay-accelerating factor (DAF, or CD55) as a common receptor (43). Afa/Dr DAEC strains are identified in epidemiological studies by hybridization to a specific probe, daaC, which is common to operons encoding Afa/Dr adhesins (5). In addition, a PCR assay to detect Afa/Dr strains has been described recently (35). Afa/Dr DAEC strains have been associated with diarrhea in children (20, 22, 24, 28). However, some studies have reported that Afa/Dr DAEC strains are found equally in children with and without diarrhea (1, 17). Discrepancies between epidemiological studies could be explained in part by age-dependent susceptibility (37). Unlike other diarrheagenic pathovars of E. coli, Afa/Dr DAEC strains are also important in urinary tract infections (UTI) (43).
The alterations induced by wild-type Afa/Dr DAEC strains on polarized host epithelial cells have been studied extensively, but these strains remain poorly characterized at the molecular level. Infection of polarized cultured human intestinal cells by Afa/Dr DAEC strain C1845, isolated from a patient with diarrhea, or IH11128, recovered from a patient with UTI, is followed by elongation of brush border microvilli resulting from rearrangement of cytoskeleton proteins (4, 47), alteration of tight-junction-associated proteins (46), and impairment of several brush border-associated enzymatic activities (45, 47). The Afa/Dr adhesin operon also encodes invasins, AfaD, and DraD (18). Afa/Dr DAEC strains invade epithelial cells at a low rate by CD55- and CD66e-independent mechanisms through interaction with the α5β1 integrin and a pathway involving caveolae and dynamic microtubules (23, 25). A recent study has reported that 50% of DAEC strains hybridize with an irp2 probe, which is part of the yersiniabactin operon, encoding a siderophore-dependent iron transport system (12). In addition, two diverse pathogenicity islands (PAIs) have been described for some DAEC isolates. First, a few DAEC strains contain a homologue of the locus of enterocyte effacement (LEE) pathogenicity island and exhibit pathogenic properties characteristic of enteropathogenic E. coli (EPEC) strains (3). Second, it has recently been shown that the pyelonephritogenic Afa/Dr DAEC strain EC7372 harbors a PAI similar to the one described for the uropathogenic strain CFT073 (PAICFT073) (26), which encodes the classical UTI determinants hemolysin and P pili (27). Other Afa/Dr strains carrying the hly and pap operons and a marker from PAICFT073 have been described recently (32).
To explore Afa/Dr DAEC at the molecular level, we have used a genomic approach, representational difference analysis (RDA) (38, 59), to analyze Afa/Dr strain C1845, recovered from a child with diarrhea (5). We have identified sequences present in strain C1845 but absent from a nonpathogenic E. coli K-12 strain. Our results indicate that E. coli C1845 harbors part of the PAICFT073 and genes from several iron acquisition systems that are common to other enteric bacterial species. We also recovered C1845-specific sequences homologous to putative virulence genes or with no homology to known sequences, and we have analyzed their distribution among Afa/Dr and non-Afa/Dr clinical isolates and among strains from the E. coli Reference Collection (ECOR collection) (44). This analysis allowed us to identify sequences that have a high prevalence in Afa/Dr strains and a low prevalence in non-Afa/Dr strains.
MATERIALS AND METHODS
Bacterial strains.
Strains used for subtractive hybridization were the DAEC strain C1845 (O75:NM), harboring the fimbrial adhesin F1845, isolated from a child with diarrhea (5), and the nonpathogenic laboratory E. coli K-12 strain MG1655, whose genome has been sequenced (6). The uropathogenic DAEC strain IH11128 (O75:H5:K), harboring the Dr adhesin, was isolated from a patient with pyelonephritis (60). EC7372, harboring the Dr-II adhesin, was isolated from a patient with pyelonephritis (50). Prototypes of the different classes of diarrheagenic E. coli are the EPEC strain 2348/69 (36), the enterohemorrhagic E. coli (EHEC) O157:H7 strain EDL931 (Pasteur Institute collection), the enterotoxigenic E. coli (ETEC) strain H10407 (15), and the enteroaggregative E. coli (EAEC) strain 042 (40).
The ECOR collection encompasses 72 strains that are representative of the range of genotypic variation in the E. coli species as a whole (44). We used 42 strains from the ECOR collection that are representative of the four main phylogenetic groups: 10 strains from group A, 8 strains from group B1, 15 strains from group B2, 7 strains from group D, and 2 strains that do not belong to any of the four groups. We have included the three strains (ECOR 64, ECOR 50, and ECOR 37) that hybridize with the daaC probe (29).
Afa/Dr DAEC clinical isolates were from children with diarrhea, asymptomatic children, or patients with pyelonephritis. Twenty E. coli strains that tested positive by colony DNA hybridization to the daaC probe were isolated in Brazil as described previously (55). Ten strains were recovered from infants with acute diarrhea, and 10 strains were recovered from a control group of asymptomatic children. Twenty-five strains that tested positive by the afa PCR assay (35) were recovered from children with diarrhea in New Caledonia (21). In addition, we have included as a control 20 strains isolated from children with diarrhea in the same study (21) that are negative by the afa PCR assay. Twelve strains testing positive by the afa PCR assay were from patients with pyelonephritis (2).
Chromosomal DNA extraction.
Bacterial genomic DNA was extracted as described previously (49).
RDA.
Clones of DNA fragments present in the genome of E. coli C1845 but absent from E. coli MG1655 were prepared as described previously (59). Chromosomal DNA from strain C1845 was digested with the restriction endonuclease MspI, Tsp509I, or Sau3AI. Three subtractive libraries, resulting each time from two rounds of subtraction using first-round adapters (R) and second-round adapters (J) (59), were obtained. For the MspI library, adapters were RMsp10 (5′-CGGTCGGTGA-3′), RMsp24 (5′-CAGCCACTCTCCGACCTCTCACGA-3′), JMsp10 (5′-CGGGTTCATG-3′), and JMsp24 (5′-ACCGACGTCGAC-3′).
Analysis of clones from subtractive libraries.
DNA from the subtractive libraries was cloned into the ClaI (MspI library) or EcoRI (Tsp509I library) site of the pBluescript vector (Stratagene) and then transformed into E. coli DH5α. Clones for the Sau3AI library were obtained by using the pUC18 SmaI/BAP vector of the SureClone ligation kit (Amersham Pharmacia Biotech, Orsay, France) and competent cells of the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). Inserts were amplified by PCRs performed on transformant colonies by using primers P1 (5′-CCCCTCGAGGTCGACGGTAT-3′) and P2 (5′-CCGCTCTAGAACTAGTGGAT-3′) for the MspI and Tsp509I libraries and primers UP (5′-GTAAAACGACGGCCAGT-3′) and RP (5′-CAGGAAACAGCTATGAC-3′) for the Sau3a library.
(i) DNA sequencing.
PCR fragments were sequenced by using the PRISM Ready Reaction Big Dye Terminator kit and an automated ABI PRISM 377 XL DNA sequencer (both from Perkin-Elmer Applied Biosystems, Courtaboeuf, France) according to the manufacturer's instructions. Sequences were analyzed by using the BLASTN and BLASTX computer programs at the National Center for Biotechnology Information (Bethesda, Md.).
(ii) DNA hybridization techniques.
To check for specificity, the amplified difference product from the second subtraction round of each bank was labeled by random-primed incorporation of [α-32P]dCTP and used as a probe against DraI- and EcoRV-digested DNA from C1845 and MG1655 in Southern blot experiments.
To study the distribution of C1845-specific sequences, purified PCR fragments were labeled by using the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Colony blotting was performed by using Hybond-N+ membranes (Amersham Pharmacia Biotech) and the ECL detection system according to the manufacturer's protocol. For Southern hybridization, BamHI-digested chromosomal DNA was applied to an agarose gel and transferred by capillarity onto Hybond-N+ membranes as described elsewhere (54). Hybridizations were performed at 42°C with the ECL kit. Detection by chemiluminescence was performed and revealed by use of X-Omat film (Kodak).
PCR.
Colony PCR was carried out by using PCR Beads Ready To Go (Amersham Pharmacia Biotech) according to the manufacturer's protocol and the Gene Amp PCR system 2400 (Perkin-Elmer Applied Biosystems). Oligonucleotides used in PCR experiments are described in Table 1. PCR was performed as follows: after an initial denaturation (5 min at 94°C), samples were subjected to 30 cycles of amplification, each of which consisted of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The annealing temperature was lowered to 46°C to investigate the presence of C1845-specific sequences among Afa/Dr clinical isolates. PCR fragments were purified from agarose gels by using the Qiaex II gel extraction kit (Qiagen, Courtaboeuf, France).
TABLE 1.
Oligonucleotides used in this study for PCR experiments
| Name | Sequence | Length of PCR fragment (bp) |
|---|---|---|
| L8-F | CAT CAT CCG CTC CAT GC | |
| L6-F | TTC ACG AAG TAA CGC CAG | |
| R15-F | GAT TGC TGG GAA GGC TGG | |
| papF-F | TCG TTG CTT CTG ACA TCG G | 470 |
| papF-R | AAT CAT GCT CAT ACT GGC C | |
| shuA-F | CCG ATC TGC TGC GTC ATG | 460 |
| shuA-R | ATG GAC TCG TCA TTC GGC | |
| daaC-F | AAG GGG GTG GAC CTG AC | 2,380 |
| daaC-R | AGA CGG TAA TCC GCA TG | |
| M008-F | GGT TAT CTT CGG TAT C | 250 |
| M008-R | AGT TAG TGG ACT ACA GC | |
| M014-F | GAT CTA TGG CAA GAC GAG | 240 |
| M014-R | TCG ACA CAA TTC ACA GAC | |
| M020-F | CTT GTC ATC GGT ACC GAG | 172 |
| M020-R | CCA CAC TAT GCA GCC AGC | |
| M030-F | GAT CAG CGC CAC AAT TAC | 120 |
| M030-R | CAG TGG AGT CTG CTG CTC | |
| S014-F | AAG GAG TCA TGG CTG CTG | 150 |
| S014-R | TGC TGT AAC TGG TCG CGG | |
| S044-F | TCT GAC TAC TGC GAC AGG | 220 |
| S044-R | TCT GAG TAG CCG CAG TCG | |
| S070-F | TAT CAC GAA TCA GCT CAC | 130 |
| S070-R | GAG CTA TGT GAG CAA TAC | |
| S081-F | CTG TCT GAG ACT GTA GCG | 120 |
| S081-R | AGC CAT AAA GAG CTT GGC | |
| S094-F | ACC TTC GGG CAG GTT TTC | 190 |
| S094-R | ACC TGT TGT TTA ACG ACC | |
| S109-F | CAC AAC CTG TAG CTG CTG | 65 |
| S109-R | GTG CCG GAA CGT CAC TTC | |
| S111-F | AAT CCT TGA ATA TTG CTG | 220 |
| S111-R | AGC AGA CGA CAA GTT ATG | |
| S164-F | TCA TTA TTC GTG ACA GGC | 60 |
| S164-R | GAC GCG GGT AAT TTA TCC | |
| S177-F | TTG GTA TCT GCA TCG CCG | 220 |
| S177-R | CTT GAA GAT GAA ATT ACC | |
| S184-F | ACG CCG TAT TAT GTG CAG | 133 |
| S184-R | AAT AAC TGA GTG TCG ACC | |
| S199-F | ATC CTG CCG ACA ACG GTC | 150 |
| S199-R | CAG GGT TCT GAG TTC ATG | |
| T007-F | TGC ACC AGA ATA CAC GTC | 192 |
| T007-R | AGC TTC ATG TAG TGA GCG | |
| T018-F | CGC GTA GCG ACC AGT AGC | 110 |
| T018-R | CAA TAA TGG TGA AGG | |
| T011-F | CTG TGC GGC GCA GCG ATC | 150 |
| T011-R | TGG CCG GTA GGA TGA ATG | |
| T024-F | TGA TTC GGA TTG TGA TG | 170 |
| T024-R | ATC ACC TGC CGC TGA C | |
| T027-F | ATG GGC TCA TCT TCA ACG | 140 |
| T027-R | GCG AGA GCT ATG GCT TGG | |
| T033-F | ACG GAT AGG ACT GAT CAG | 217 |
| T033-R | GCG CTA ATG GAT CAG ATG | |
| T034-F | GTA TCA CAT ATC CTG TTG | 260 |
| T034-R | ATT CGT CAC TGA GCG CTG |
Statistical analysis.
Statistical significance between groups was tested by the χ2 test.
Nucleotide sequence accession numbers.
The sequences of the subtracted DNA fragments have been assigned GenBank accession numbers AZ935556 to AZ935604.
RESULTS
Production of libraries of DNA fragments of Afa/Dr DAEC strain C1845 not found in the genome of E. coli K-12.
By using RDA, we subtracted the genome of the nonpathogenic E. coli K-12 strain MG1655, which has recently been sequenced (6), from that of the pathogenic strain C1845, harboring the F1845 adhesin. Three libraries were produced by using the MspI, Sau3AI, and Tsp509I restriction enzymes. To confirm that the sequences amplified from the second round of subtraction were C1845 specific, the difference product of each bank was labeled and used as a probe against DraI- and EcoRV-digested DNAs from C1845 and the K-12 strain. Strong reactivity was observed with the pathogenic strain C1845, whereas no signal was detected for the K-12 strain (data not shown). Accordingly, the background of nonspecific sequences was very low, since only 5.5% of the sequences recovered were found in the K-12 genome.
Altogether, 172 C1845-specific clones were isolated and sequenced. Of these, 118 clones were unique. Ninety percent of the clones showed significant homology to known sequences, among which, for example, the clone of the F1845 adhesin operon (clone T006) confirms the validity of the RDA method. About 45% of the C1845-specific sequences recovered from RDA had homology to various plasmids (F, R100, R64, pColIb-P9, pO157, and pKYM), including sequences involved in plasmid transfer (tra), plasmid replication, plasmid transposases, insertion sequences, and noncoding plasmid sequences (data not shown). In addition, 8% of the C1845-specific sequences exhibited homology to prophage sequences, including P2 sequences and sequences homologous to O157:H7 EDL933 prophages (data not shown). Plasmid and phage sequences might be associated with virulence sequences, but it is unlikely that these sequences themselves play a role in virulence. Table 2 summarizes the clones that showed significant homology with published sequences other than plasmid and phage sequences. Sequences homologous to PAIs or iron acquisition systems are described in more detail below. A few sequences showed homology to putative virulence genes encoding a putative toxin (M008) or putative proteins involved in fimbrial assembly (S164 and S184). Finally, less than 10% of the C1845-specific clones had no homology with published sequences (Table 2). However, some sequences (S064, S094, S109, T007, and T027) were homologous to sequences from the genome projects of E. coli RS218 and the pyelonephritogenic E. coli strain CFT073 (available at www.genome.wisc.edu).
TABLE 2.
Summary of BLAST search of E. coli C1845-specific clones
| Clonea | Length (bp) | Sequence homologyb | Probability | Accession no. |
|---|---|---|---|---|
| M003 | 308 | r12 (N), PAI of E. coli CFT073 | 5e−14 | AF081285 |
| M008 | 314 | Putative macrophage toxin (P), E. coli O157:H7 | 7e−49 | AAG54519 |
| M014 | 320 | d-Fructokinase (P), E. coli O157:H7 | 6e−47 | AAG57487 |
| M020 | 323 | 2-Deoxy-d-gluconate 3-dehydrogenase (P), E. coli O157:H7 | 6e−08 | BAB37122 |
| M022 | 281 | Malonyl coenzyme A-acyl carrier protein transacylase (P), Salmonella enterica serovar Typhimurium | 2e−08 | O85140 |
| M028 | 398 | ORF78 (P), putative transposase, EPEC | 2e−67 | NP_053140 |
| M029 | 310 | ORF41 (P), putative transposase, EPEC | 8e−57 | NP_053103 |
| M030 | 292 | ORF37 (P), hypothetical protein, 102-kb region of Yersinia pestis | 7e−28 | CAA21360 |
| S013 | 234 | Transposon Tn4311 (N), E. coli | 2e−71 | M22041 |
| S010 | 165 | r6 (N), PAI of E. coli CFT073 | 4e−47 | AF081285 |
| S014 | 217 | Putative protease (P), E. coli O157:H7 | 1e−26 | AAG54523 |
| S044 | 140 | fliC gene for flagellin (N), E. coli U4-41 | 1e−52 | AB028473 |
| S057 | 175 | iucB (N), aerobactin, Shigella flexneri | 3e−91 | AF141323 |
| S058 | 371 | Noncoding downstream pssA (N), E. coli 413189-1 | 2e−92 | Y13614 |
| S064 | 136 | None | ||
| S070 | 269 | None | ||
| S071 | 186 | r3 and malX (N), PAI of E. coli CFT073 | 2e−51 | AF081286 |
| S076 | 286 | Tn3 transposase (P), E. coli | 1e−34 | P03008 |
| S077 | 184 | Probable transposase (P), Y. pestis | 9e−23 | T14971 |
| S080 | 264 | Tn3 transposase (P), E. coli | 1e−12 | P03008 |
| S081 | 235 | None | ||
| S083 | 225 | None | ||
| S094 | 198 | Z0251 (P), hypothetical protein, E. coli O157:H7 | 6e−14 | AAG54520 |
| S097 | 232 | None | ||
| S099 | 294 | Tn3 transposase (P), E. coli | 1e−41 | P03008 |
| S109 | 85 | None | ||
| S111 | 248 | Clone SauE4.C10 (N), E. coli K1 C5 | e−130 | AF222134 |
| S137 | 199 | r3 (N), PAI of E. coli CFT073 | 1e−90 | AF081286 |
| S141 | 286 | Tn3 transposase (P), E. coli | 2e−40 | P03008 |
| S164 | 73 | Putative fimbrial chaperone protein (P), E. coli O157:H7 | 2e−28 | AE005354 |
| S165 | 283 | IS200 transposase (P), serovar Typhimurium | 3e−44 | Q57334 |
| S166 | 219 | None | ||
| S170 | 161 | IS100 transposase (P), Y. pestis | 5e−20 | T17450 |
| S177 | 239 | None | ||
| S184 | 201 | TsaC (P), serovar Typhimurium | 1e−6 | BAA82271 |
| S199 | 214 | None | ||
| T002 | 242 | ORF33 (P), hypothetical protein, 102-kb region of Y. pestis | 3e−29 | CAA21356 |
| T006 | 259 | Upstream daaE (N), F1845 adhesin operon, E. coli C1845 | e−139 | M27725 |
| T007 | 201 | None | ||
| T011 | 210 | ShuU (P), heme utilization, Shigella dysenteriae | 6e−32 | AAC27812 |
| T018 | 208 | Sucrose hydrolase, invertase (P), Erwinia amylovora | 9e−17 | CAC14601 |
| T024 | 262 | Z0259 (P), hypothetical protein, E. coli O157:H7 | 4e−22 | AAG54528 |
| T027 | 206 | None | ||
| T028 | 441 | Clone TspE4.A11 (N), E. coli K1 C5 | e−151 | AF222183 |
| T033 | 235 | Clone SauE4.E6 (N), E. coli K1 C5 | 3e−98 | AF222140 |
| T034 | 495 | R4 (P), exogenous ferric siderophore receptor, PAI of E. coli CFT073; Iha (P), adhesin, E. coli O157:H7 | 5e−34 | AAC61730AAF36432 |
| T035 | 252 | Putative molybdenum transport protein (P), E. coli O157:H7 | 2e−41 | AAG56050 |
| ModD (L6) (P), molybdenum transport protein, PAI of E. coli CFT073 | 1e−24 | AAC61710 | ||
| T037 | 256 | Syringomycin synthetase (P), Pseudomonas syringae | 1e−10 | T14593 |
| T039 | 269 | Hypothetical protein PhuW (P), Pseudomonas aeruginosa | 1e−13 | AAC13284 |
Clones are designated by the initial letter of the restriction enzyme used (M, S, or T) and an alphanumeric designation.
Only homologies with at least a probability of e−5 were retained. Homologies to plasmids (n = 58) and phages (n = 11) are not shown. N, similarity at the nucleotide level; P, similarity at the protein level.
E. coli C1845 harbors part of PAICFT073.
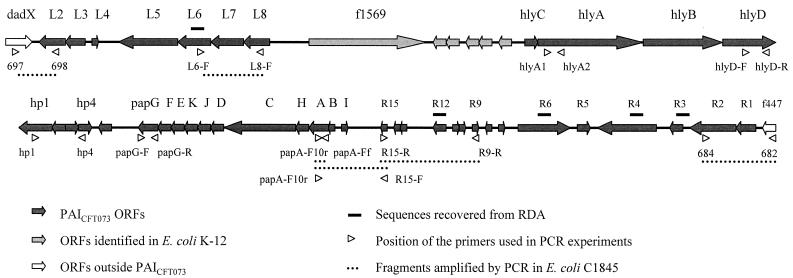
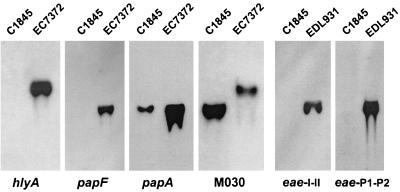
A uropathogenic Afa/Dr DAEC strain, EC7372, which harbors a PAI similar to the one described for the pyelonephritogenic strain CFT073 (PAICFT073), has been described recently (26). PAICFT073 encodes hemolysin (hly), P pili (pap), and several genes of unknown function (27). One gene, R4, is identical to iha, encoding a novel adhesin identified in E. coli O157:H7 (58). RDA revealed that E. coli C1845 also harbors several PAICFT073 genes, since sequences homologous to the L6 (ModD), R3, R4, R6, and R12 open reading frames (ORFs) were recovered (Table 2; Fig. 1). We investigated the extent of similarity between C1845 sequences and PAICFT073 by performing PCR with several pairs of primers covering different areas of PAICFT073 (Fig. 1). Using primers specific for the left and right junctions of PAICFT073 (34), primers specific for L6 and L8 (Table 1), and primers specific for R9 and R15 (26), we amplified fragments of the expected sizes from strain C1845 (Fig. 1). However, strain C1845 does not contain the entire PAICFT073, since we failed to amplify DNA fragments by using oligonucleotides complementary to the hlyA, hlyD, hp1-hp4, papG, and papF sequences (26, 31) (Fig. 1). We have confirmed by Southern blot experiments that C1845 DNA does not hybridize with hlyA or papF probes derived from EC7372 DNA (Fig. 2); on the other hand, a hybridization signal was detected with a papA probe (8) (Fig. 2). The PapA subunit is polymorphic (11 variants), and we have shown that a fragment of the expected size can be amplified from strain C1845 by using primers specific for the F10 papA allele (33), indicating the presence of a remnant of the pap operon. In addition, a fragment of the expected size can be amplified from strain C1845 by using primers specific for the F10 papA allele and R15 (Table 1). Taken together, these results indicate that E. coli C1845 harbors part of PAICFT073, including the R4 gene, but lacks most of the central region encoding the hly and pap operons.
FIG. 1.
E. coli C1845 harbors sequences from the left and right regions of PAICFT073. The schematic representation of PAICFT073 is derived from the work of Guyer et al. (reprinted from reference 27 with permission). Sequences homologous to the L6, R12, R6, R4, and R3 ORFs were recovered by RDA (Table 2). Positions of primers complementary to PAICFT073 sequences used in PCR experiments are indicated. Dotted lines, strain C1845 fragments amplified by PCR.
FIG. 2.
Southern hybridization of BamHI-digested DNA from E. coli C1845, EC7372, or EDL931 with hly, pap, or eae probes. Gene-specific probes were generated by PCR amplification on chromosomal DNA. hlyA, papA, and papF probes are derived from E. coli EC7372. Both eae probes are derived from E. coli 2348/69 (7, 52). Hybridization with a probe derived from E. coli C1845 (M030) is shown as a control.
On the other hand, none of the C1845-specific sequences identified by RDA matched with sequences from the LEE island, which includes a type III secretion system, intimin (Eae), and its receptor (Tir). In addition, genomic DNA from strain C1845 showed no hybridization in a Southern experiment with two different eae probes (7, 52) derived from EPEC DNA (Fig. 2) and showed no DNA amplification in a PCR experiment with degenerated oligonucleotides designed for type III secretion system detection (16). These results indicate that E. coli C1845 does not harbor the LEE island and suggest that C1845 does not encode a type III secretion system.
E. coli C1845 encodes several iron acquisition systems.
Pathogenic bacteria have adapted to the host iron-limiting environment by developing a variety of iron assimilation systems. A recent study has reported that strain C1845 harbors irp2, which is part of the yersiniabactin operon, encoding a siderophore-dependent iron transport system (12). Three DNA sequences recovered from RDA, S057, T011, and T035, exhibited significant homology to other iron acquisition systems found in pathogenic enteric bacteria (Table 2). Clone S057 carries the sequence of the iucB gene, which is part of the aerobactin operon. The siderophore aerobactin is produced by a variety of enteric bacteria, including Shigella spp. and some E. coli strains (13). Clone T011 is homologous to the shuU sequence. The shu operon, which allows the use of hemin as a carbon source, has been identified in Shigella spp. and is found in pathogenic E. coli strains (chu locus) (39, 63). We have shown by PCR using specific primers that another gene of the shu operon, shuA (Table 1), is also present in strain C1845. Clone T035 is homologous to a putative molybdenum transport protein encoded by PAICFT073. In addition, we have shown by using specific oligonucleotides (30) the presence in E. coli C1845 of a putative virulence gene, iroN, encoding a siderophore catechole receptor that is prevalent among E. coli isolates from patients with UTI or bacteremia (30, 53). Hence, strain C1845 contains multiple genes involved in iron acquisition systems which probably play roles during host infection.
Distribution of C1845-specific sequences among Afa/Dr and non-Afa/Dr clinical strains and strains from the ECOR collection.
We have investigated the distribution of several C1845-specific sequences, including putative virulence genes and sequences with no homology to sequences in the databases, among E. coli Afa/Dr and non-Afa/Dr isolates and strains from the ECOR reference collection. We have used 20 Afa/Dr E. coli isolates recovered from children with or without diarrhea in Brazil, 25 Afa/Dr E. coli isolates recovered from children with diarrhea in New Caledonia, and 14 Afa/Dr E. coli isolates recovered from patients with pyelonephritis (see Materials and Methods). As a control, we have used strains from the ECOR collection and non-Afa/Dr clinical isolates recovered from children with diarrhea in New Caledonia. We have chosen 42 strains from the ECOR collection, including the 3 strains which hybridize with the daaC probe (29). ECOR strains belong to four main phylogenetic groups (A, B1, B2, and D), and most of the pathogenic E. coli isolates are concentrated in groups B2 and D (29). Using specific oligonucleotides (11), we have shown by PCR that E. coli C1845 most likely belongs to the B2 group.
In a preliminary experiment, we investigated by PCR the presence of 22 C1845-specific sequences in the 20 Afa/Dr isolates from Brazil (data not shown). These sequences (oligonucleotides are described in Table 1) include putative virulence genes (M008, S164, S184, T011 [shuU], and T034 [iha]), putative metabolic genes (M014 and T018), and sequences with no homology to known proteins. Only four sequences had a high prevalence (>70%) among the Afa/Dr isolates (M030, S109, S111, and S164). The frequencies of the putative virulence sequences M008, S184, T011 (shuU), and T034 (iha) were 30, 25, 55, and 45%, respectively. Interestingly, some sequences (M008, S014, S094, and T024; M030 and S111; S081 and S184) had the same distribution, suggesting a genetic linkage. We then analyzed by colony hybridization on Afa/Dr and non-Afa/Dr strains the distribution of several sequences including M030, S109, S111, S164, and C1845-specific sequences with low or no homology with the published sequences that are not found in the databases of the E. coli RS218 and CFT073 genome projects (M020, S070, S081, S177, S184, S199, and T018). The results shown in Table 3 indicate that three clones (M030, S109, and S111) have a high prevalence among Afa/Dr strains and a low prevalence among non-Afa/Dr strains. These three clones did not hybridize with EPEC, EHEC, ETEC, or EAEC prototype strains (data not shown). In addition, these results confirmed that M030 and S111 have an identical distribution among E. coli strains.
TABLE 3.
Distribution of C1845-specific sequences among Afa/Dr and non-Afa/Dr strains
| Strain origina | No. of strains | No. (%) of isolates positive by colony hybridizationb for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M030 | S109 | S111 | S164 | M020 | S070 | S081 | S177 | S184 | S199 | T018 | ||
| Afa/Dr | ||||||||||||
| ECOR | 3 | 1 (33.5) | 3 (100) | 1 (33.5) | 2 (66.5) | 1 (33.5) | 0 (0) | 1 (33.5) | 0 (0) | 1 (33.5) | 0 (0) | 1 (33.5) |
| Diarrhea | 35 | 30 (85.5) | 28 (80) | 30 (85.5) | 24 (68.5) | 10 (28.5) | 0 (0) | 12 (34.5) | 0 (0) | 12 (34.5) | 0 (0) | 5 (14.5) |
| Asymptomatic | 10 | 8 (80) | 6 (60) | 8 (80) | 5 (50) | 2 (20) | 0 (0) | 1 (10) | 0 (0) | 1 (10) | 1 (10) | 2 (20) |
| Pyelonephritis | 14 | 11 (78.5) | 11 (78.5) | 11 (78.5) | 12 (85.5) | 7 (50) | 0 (0) | 6 (43) | 0 (0) | 7 (50) | 0 (0) | 5 (35.5) |
| Total | 62 | 50 (80.5c) | 48 (77.5c) | 50 (80.5c) | 43 (69.5) | 20 (32c) | 0 (0) | 20 (32) | 0 (0) | 21 (34) | 1 (1.5) | 13 (21) |
| Non-Afa/Dr | ||||||||||||
| ECOR | 39 | 4 (10) | 5 (13) | 4 (10) | 25 (64) | 4 (10) | 0 (0) | 12 (31) | 1 (2.5) | 12 (31) | 2 (5) | 2 (5) |
| Diarrhea | 20 | 3 (15) | 9 (45) | 3 (15) | 8 (40) | 1 (5) | 0 (0) | 4 (20) | 0 (0) | 4 (20) | 1 (5) | 1 (5) |
| Total | 59 | 7 (12) | 14 (23.5) | 7 (12) | 33 (56) | 5 (8.5) | 0 (0) | 16 (27) | 1 (1.5) | 16 (27) | 3 (5) | 3 (5) |
The Afa/Dr ECOR strains are ECOR 64, ECOR 50, and ECOR 37. Afa/Dr clinical isolates from patients with diarrhea include 10 strains from Brazil and 25 strains from New Caledonia (35, 55). The 10 Afa/Dr strains from asymptomatic children were from Brazil (55). Afa/Dr clinical isolates from pyelonephritis patients include 12 strains positive by the afa PCR assay (35), IH11128, and EC7372. The non-Afa/Dr clinical isolates from children with diarrhea (New Caledonia) were negative by the afa PCR assay (35).
PCR products amplified with specific oligonucleotide couples (Table 1) were used as probes. Numbers in bold show the percentages of positive isolates for all 62 Afa/Dr strains and all 59 non-Afa/Dr strains.
The difference between the distribution of the sequence in Afa/Dr strains and that in non-Afa/Dr strains was significant (P < 0.01 by the χ2 test).
Presence of C1845-specific sequences in the uropathogenic Afa/Dr DAEC strain IH11128.
In in vitro experiments on cultured polarized epithelial cells, the structural and functional alterations induced by the pyelonephritogenic Afa/Dr DAEC strain IH11128 are similar to those induced by E. coli C1845 (45-47). It has been shown previously that strain IH11128, which harbors the Dr adhesin, does not carry classical uropathogenic E. coli virulence sequences such as hly, cnf, or cdt (26). To evaluate the similarity of strains C1845 and IH11128 at the molecular level, we investigated the presence of C1845-specific sequences recovered by RDA in strain IH11128 by PCR experiments or colony blotting (Table 3; also data not shown). E. coli IH11128 was found to harbor the diverse iron acquisition systems (irp2, iucB, shuU-shuA, and iroN) and the portion of the PAICFT073 that are found in strain C1845. In addition, like E. coli C1845, strain IH11128 harbors a remnant of the pap operon with the F10 papA allele. All 22 clones whose presence in Afa/Dr clinical isolates was investigated are present in strain IH11128, with the exception of three clones (S070, S177, and S199) that seem restricted to strain C1845, since they were absent or very rare among Afa/Dr clinical isolates (Table 3). Taken together, these results indicate that E. coli C1845 and IH11128 are closely related at the molecular level.
DISCUSSION
In the present study, we provide the first extensive molecular analysis of an Afa/Dr DAEC strain. Using the RDA approach, we have identified sequences from the genome of the Afa/Dr DAEC strain C1845, harboring the F1845 adhesin and recovered from a child with diarrhea, that are absent from the nonpathogenic E. coli K-12 strain MG1655. E. coli C1845 harbors several iron acquisition genes, genes from PAICFT073, and other putative virulence genes. However, our results indicate that C1845 does not harbor genes encoding factors known to subvert host cell proteins to induce brush border lesions, such as type III secretion system and effector proteins. An epidemiological approach allowed us to identify sequences that have a high prevalence in Afa/Dr strains and a low prevalence in non-Afa/Dr strains. Our results indicate that diarrheagenic E. coli C1845 is very close to Afa/Dr DAEC uropathogenic isolates.
Because iron is essential for bacterial growth and free iron is limiting within the host, bacterial pathogens have acquired diverse systems to acquire iron. In addition to the yersiniabactin siderophore (irp2), we have shown that E. coli C1845 harbors sequences encoding several iron transport systems found in other pathotypes of E. coli, including the aerobactin siderophore (iuc), a catechole siderophore receptor (iroN), a heme transport system (shu), and a molybdenum transport system (modD). However, the reason for the redundancy of iron acquisition systems is unclear, and it has been postulated that the yersiniabactin system might have an alternative function, other than iron acquisition, in pathogenic E. coli strains (57). In addition, the diverse systems might play roles at different stages of the infection.
Several PAIs, which contribute to the rapid evolution of bacterial pathogens, have been described for pathogenic E. coli strains (14). We have shown that strains C1845 and IH11128 harbor the left and right ends of a PAI identified in a pyelonephritogenic E. coli strain, PAICFT073 (27), but not the middle part of the island, encoding hly and pap operons. However, a remnant of the pap operon, carrying the F10 papA allele, was detected. Interestingly, the presence of a partial copy of the pap operon, with the F10 papA allele, has been found in several urosepsis strains belonging to the same serogroup as C1845 and IH11128, O75 (33), suggesting that this genetic organization is commonly found among O75 strains. One can hypothesize that the entire PAICFT073 has been inserted into these strains and that a deletion has subsequently taken place. Whether the internal region of the PAI has been simply deleted or replaced by different genetic material remains unknown. Among the PAICFT073 ORFs present in E. coli C1845 and IH11128, R4 is of particular interest because its translated sequence is identical to a novel adhesin of E. coli O157:H7, Iha, that mediates diffuse adherence to epithelial cells (58).
Besides iron acquisition systems and PAICFT073 genes, we have identified a few C1845-specific sequences that show homology to putative virulence genes (M008, S164, and S184). Except for S164, these clones have only a narrow distribution among the Afa/Dr clinical isolates tested. Clone S164, encoding a putative fimbrial chaperone protein, is frequently found in Afa/Dr clinical isolates but is not specific for Afa/Dr strains. It is of interest that M008 is part of a group of four clones (M008, S014, S094, and T024) that are likely to be linked on the C1845 chromosome, because they exhibit the same distribution among the diverse E. coli strains and show homology to O157:H7 sequences that are part of an island not found in E. coli K-12 MG1655 (48). Sequences highly homologous (>98%) to these clones are also found in the database of the RS218 genome project. Because this group of clones encodes a putative virulence factor, it may be part of a novel PAI that would be present in diverse E. coli pathovars. In addition, we have identified three sequences (M030, S109, and S111) that appear with a high frequency among Afa/Dr E. coli strains and a low frequency among non-Afa/Dr E. coli strains. These three Afa/Dr-specific sequences are found with similar frequencies in Afa/Dr isolates recovered from patients with diarrhea or pyelonephritis. Our results indicate that M030 and S111 are genetically linked, because they exhibit identical distribution among 121 E. coli strains. M030 exhibits low homology to an ORF of unknown function (ORF37) encoded by the 102-kb region of Yersinia pestis (9). Sequences highly homologous to M030, S109, and S111 are found in the database of the genome project for RS218, a K1 derivative responsible for meningitis, but these clones do not hybridize with EPEC, EHEC, ETEC, or EAEC prototype strains. Because of the high homology with RS218 sequences, these clones could encode factors that are important for both Afa/Dr and K1 pathogenic strains.
Afa/Dr DAEC strains have been associated with both diarrheagenic and uropathogenic infections. Recent data indicate that AfaE1, AfaE2, AfaE3, and F1845 adhesins are found in both diarrheagenic and uropathogenic human isolates (35). While strain C1845 was recovered from a child with diarrhea (5), our results indicate that this strain has several characteristics that have been associated with extraintestinal E. coli strains. These include the B2 phylogenetic group (51), the O75 serotype (42), the production of aerobactin (10), the presence of iroN (53), and the presence of sequences from PAICFT073 (27). On the other hand, multilocus enzyme electrophoresis indicated that Afa/Dr DAEC strain C1845, as well as other DAEC strains, is phylogenetically close to EAEC strains (12); however, the phylogenetic relationship between DAEC strains isolated from patients with diarrhea and DAEC strains isolated from patients with UTIs has not been studied. To further investigate the relation of E. coli C1845 to uropathogenic isolates, we investigated the presence of C1845-specific sequences in the Afa/Dr strain IH11128, recovered from a patient with pyelonephritis. Despite their different origins, both strains appear very similar at the molecular level. The observation that E. coli C1845, isolated from a child with diarrhea, is very close to uropathogenic isolates is in agreement with the hypothesis developed by Germani et al. (19) that a single Afa/Dr strain can be responsible for diarrhea or UTI and/or with the hypothesis that isolation of Afa/Dr DAEC strains from patients with diarrhea may be related to the presence of uropathogenic E. coli in the colon without a causative link to the disease state (17).
It has recently been extensively documented that many enterovirulent bacteria subvert functional membrane-bound proteins as receptors to colonize epithelia and exploit the cell-signaling pathways to cross talk with the host cells and cause disease. The prototype of such subversive enterovirulent pathogens is EPEC, which colonizes the intestinal epithelium by translocating bacterial proteins through a type III secretion system to activate a signaling pathway within the underlying cell and cause the reorganization of the host actin cytoskeleton (61). Like EPEC, wild-type Afa/Dr DAEC strains promote dramatic lesions in the brush border of cultured human differentiated Caco-2 intestinal cells (4, 47). Some DAEC strains contain a homologue of the LEE PAI, which encodes a type III secretion system, and exhibit pathogenic properties characteristic of EPEC strains (3, 62). Our results indicate that strain C1845 does not encode the LEE PAI and lacks type III secretion system and effector proteins, implying that the brush border cytoskeleton is altered by a mechanism different from the one developed by EPEC strains. No known virulence factors involved in cytoskeleton injuries have been found in the RDA conducted in this study, suggesting that interaction between the Afa/Dr adhesins and membrane-bound receptors is the major mechanism by which Afa/Dr DAEC pathogens subvert host cell-signaling pathways to develop pathogenicity. This interpretation is consistent with previous results (47) and with recent data showing that interaction of Afa/Dr DAEC adhesins with CD55 induces the transepithelial migration of polymorphonuclear leukocytes in human intestinal T84 cell monolayers and a proinflammatory response. This phenomenon follows the adhesin-dependent tyrosine phosphorylation of several T84 proteins and activation of the mitogen-activated protein kinases (F. Bétis, P. Brest, V. Hofman, J. Guignot, M.-H. Bernet-Camard, B. Rossi, A. L. Servin, and P. Hofman, unpublished data).
Acknowledgments
We thank E. Bingen (Service de Microbiologie, Hopital Robert Debré, Paris, France), A. Darfeuille-Michaud (Laboratoire de Bactériologie, Clermont-Ferrand, France), B. B. Finlay (University of British Columbia, Vancouver, Canada), S. L. Moseley (University of Washington, Seattle, Wash.), and B. J. Nowicki (The University of Texas Medical Branch, Galveston, Tex.) for providing some strains used in this study.
A.-B.B.-P. is supported by a postdoctoral grant from the Fondation pour la Recherche Médicale (FRM). The laboratory of X.N. is supported by INSERM and the Université René Descartes Paris 5.
Editor: V. J. DiRita
REFERENCES
- 1.Albert, M. J., A. S. Faruque, S. M. Faruque, R. B. Sack, and D. Mahalanabis. 1999. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J. Clin. Microbiol. 37:3458-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambaud, M., P. Courcoux, and A. Labigne-Roussel. 1988. Detection by molecular hybridization of pap, afa, and sfa adherence systems in Escherichia coli strains associated with urinary and enteral infections. Ann. Inst. Pasteur Microbiol. 139:575-588. [DOI] [PubMed] [Google Scholar]
- 3.Beinke, C., S. Laarmann, C. Wachter, H. Karch, L. Greune, and M. A. Schmidt. 1998. Diffusely adhering Escherichia coli strains induce attaching and effacing phenotypes and secrete homologs of Esp proteins. Infect. Immun. 66:528-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernet-Camard, M. F., M. H. Coconnier, S. Hudault, and A. L. Servin. 1996. Pathogenicity of the diffusely adhering strain Escherichia coli C1845: F1845 adhesin-decay-accelerating factor interaction, brush border microvillus injury, and actin disassembly in cultured human intestinal epithelial cells. Infect. Immun. 64:1918-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 180:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbonetti, N. H., S. Boonchai, S. H. Parry, V. Vaisanen-Rhen, T. K. Korhonen, and P. H. Williams. 1986. Aerobactin-mediated iron uptake by Escherichia coli isolates from human extraintestinal infections. Infect. Immun. 51:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., and J. L. Martinez. 1988. Aerobactin production as a virulence factor: a reevaluation. Eur. J. Clin. Microbiol. Infect. Dis. 7:621-629. [DOI] [PubMed] [Google Scholar]
- 14.Dozois, C. M., and R. Curtiss III. 1999. Pathogenic diversity of Escherichia coli and the emergence of ′exotic' islands in the gene stream. Vet. Res. 30:157-179. [PubMed] [Google Scholar]
- 15.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauconnier, A., A. Veithen, P. Gueirard, R. Antoine, L. Wacheul, C. Locht, A. Bollen, and E. Godfroid. 2001. Characterization of type III secretion locus of Bordetella pertussis. Int. J. Med. Microbiol. 290:693-705. [DOI] [PubMed] [Google Scholar]
- 17.Forestier, C., M. Meyer, S. Favre-Bonte, C. Rich, G. Malpuech, C. Le Bouguenec, J. Sirot, B. Joly, and C. De Champs. 1996. Enteroadherent Escherichia coli and diarrhea in children: a prospective case-control study. J. Clin. Microbiol. 34:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, M. I., M. Jouve, J. P. Nataro, P. Gounon, and C. Le Bouguenec. 2000. Characterization of the AfaD-like family of invasins encoded by pathogenic Escherichia coli associated with intestinal and extra-intestinal infections. FEBS Lett. 479:111-117. [DOI] [PubMed] [Google Scholar]
- 19.Germani, Y., E. Begaud, P. Duval, and C. Le Bouguenec. 1997. An Escherichia coli clone carrying the adhesin-encoding afa operon is involved in both diarrhoea and cystitis in twins. Trans. R. Soc. Trop. Med. Hyg. 91:573.. [DOI] [PubMed] [Google Scholar]
- 20.Germani, Y., E. Begaud, P. Duval, and C. Le Bouguenec. 1996. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in New Caledonia. J. Infect. Dis. 174:1124-1126. [DOI] [PubMed] [Google Scholar]
- 21.Germani, Y., M. Morillon, E. Begaud, H. Dubourdieu, R. Costa, and J. Thevenon. 1994. Two-year study of endemic enteric pathogens associated with acute diarrhea in New Caledonia. J. Clin. Microbiol. 32:1532-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giron, J. A., T. Jones, F. Millan-Velasco, E. Castro-Munoz, L. Zarate, J. Fry, G. Frankel, S. L. Moseley, B. Baudry, J. B. Kaper, G. K. Schoolnick, and L. W. Riley. 1991. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J. Infect. Dis. 163:507-513. [DOI] [PubMed] [Google Scholar]
- 23.Goluszko, P., V. Popov, R. Selvarangan, S. Nowicki, T. Pham, and B. J. Nowicki. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J. Infect. Dis. 176:158-167. [DOI] [PubMed] [Google Scholar]
- 24.Gomes, T. A. T., M. A. M. Vieira, C. M. Abe, D. Rodrigues, P. M. Griffin, and S. R. T. S. Ramos. 1998. Adherence patterns and adherence-related DNA sequences in Escherichia coli isolates from children with and without diarrhea in Sao Paulo City, Brazil. J. Clin. Microbiol. 36:3609-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guignot, J., M. F. Bernet-Camard, C. Pous, L. Plancon, C. Le Bouguenec, and A. L. Servin. 2001. Polarized entry of uropathogenic Afa/Dr diffusely adhering Escherichia coli strain IH11128 into human epithelial cells: evidence for α5β1 integrin recognition and subsequent internalization through a pathway involving caveolae and dynamic unstable microtubules. Infect. Immun. 69:1856-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guignot, J., J. Breard, M. F. Bernet-Camard, I. Peiffer, B. J. Nowicki, A. L. Servin, and A. B. Blanc-Potard. 2000. Pyelonephritogenic diffusely adhering Escherichia coli EC7372 harboring Dr-II adhesin carries classical uropathogenic virulence genes and promotes cell lysis and apoptosis in polarized epithelial Caco-2/TC7 cells. Infect. Immun. 68:7018-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jallat, C., V. Livrelli, A. Darfeuille-Michaud, C. Rich, and B. Joly. 1993. Escherichia coli strains involved in diarrhea in France: high prevalence and heterogeneity of diffusely adhering strains. J. Clin. Microbiol. 31:2031-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, J. R., A. E. Stapleton, T. A. Russo, F. Scheutz, J. J. Brown, and J. N. Maslow. 1997. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the class I and class III alleles of papG. Infect. Immun. 65:2153-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, J. R., A. L. Stell, F. Scheutz, T. T. O'Bryan, T. A. Russo, U. B. Carlino, C. Fasching, J. Kavle, L. Van Dijk, and W. Gaastra. 2000. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect. Immun. 68:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao, J. S., D. M. Stucker, J. W. Warren, and H. L. Mobley. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Bouguenec, C., L. Lalioui, L. du Merle, M. Jouve, P. Courcoux, S. Bouzari, R. Selvarangan, B. J. Nowicki, Y. Germani, A. Andremont, P. Gounon, and M. I. Garcia. 2001. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J. Clin. Microbiol. 39:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed]
- 37.Levine, M. M., C. Ferreccio, V. Prado, M. Cayazzo, P. Abrego, J. Martinez, L. Maggi, M. M. Baldini, W. Martin, D. Maneval, B. Kay, L. Guers, H. Lior, S. S. Watermann, and J. P. Nataro. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 38.Lisitsyn, N., and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 39.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560-565. [DOI] [PubMed] [Google Scholar]
- 41.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimmich, W., W. Voigt, and G. Seltmann. 1997. Characterization of urinary Escherichia coli O75 strains. J. Clin. Microbiol. 35:1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nowicki, B., R. Selvarangan, and S. Nowicki. 2001. Family of Escherichia coli Dr adhesins: decay-accelerating factor receptor recognition and invasiveness. J. Infect. Dis. 183(Suppl. 1):S24-S27. [DOI] [PubMed] [Google Scholar]
- 44.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peiffer, I., M.-F. Bernet-Camard, M. Rousset, and A. L. Servin. 2001. Impairments in enzyme activity and biosynthesis of brush border-associated hydrolases in human intestinal Caco-2/TC7 cells infected by members of the Afa/Dr family of diffusely adhering Escherichia coli. Cell. Microbiol. 3:341-357. [DOI] [PubMed] [Google Scholar]
- 46.Peiffer, I., A. B. Blanc-Potard, M. F. Bernet-Camard, J. Guignot, A. Barbat, and A. L. Servin. 2000. Afa/Dr diffusely adhering Escherichia coli C1845 infection promotes selective injuries in the junctional domain of polarized human intestinal Caco-2/TC7 cells. Infect. Immun. 68:3431-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peiffer, I., J. Guignot, A. Barbat, C. Carnoy, S. L. Moseley, B. Nowicki, A. L. Servin, and M. F. Bernet-Camard. 2000. Structural and functional lesions in brush border of human polarized intestinal Caco-2/TC7 cells infected by members of the Afa/Dr diffusely adhering family of Escherichia coli. Infect. Immun. 68:5979-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 49.Perrin, A., X. Nassif, and C. Tinsley. 1999. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect. Immun. 67:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham, T. Q., P. Goluszko, V. Popov, S. Nowicki, and B. J. Nowicki. 1997. Molecular cloning and characterization of Dr-II, a nonfimbrial adhesin-I-like adhesin isolated from gestational pyelonephritis-associated Escherichia coli that binds to decay-accelerating factor. Infect. Immun. 65:4309-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid, S. D., D. J. Betting, and T. S. Whittam. 1999. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J. Clin. Microbiol. 37:2719-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Scaletsky, I. C., S. H. Fabbricotti, R. L. Carvalho, C. R. Nunes, H. S. Maranhao, M. B. Morais, and U. Fagundes-Neto. 2002. Diffusely adherent Escherichia coli as a cause of acute diarrhea in young children in Northeast Brazil: a case-control study. J. Clin. Microbiol. 40:645-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scaletsky, I. C., M. L. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaisanen-Rhen, V. 1984. Fimbria-like hemagglutinin of Escherichia coli O75 strains. Infect. Immun. 46:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wachter, C., C. Beinke, M. Mattes, and M. A. Schmidt. 1999. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 31:1695-1707. [DOI] [PubMed] [Google Scholar]
- 63.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]